Introduction
Gastric cancer (GC) is the fifth most common type of
cancer worldwide and the third leading cause of global
cancer-associated mortality (1).
GC is a complex solid tumor arising from genetic alterations,
environmental interactions and host-associated factors (2). GC is a major contributor to the
worldwide disability-adjusted life-years among patients with cancer
(3). Achieving greater
understanding of the molecular genomic mutations in GC is pivotal
to improving therapies and outcomes for patients with GC. Recently
great progress has been achieved in GC, including the
identification of novel cellular pathways and molecular components
(4). The Cancer Genome Atlas
(TCGA) project recently classified GC as possessing four genomic
subtypes based on ~300 molecular profiles of patients with GC
(5). Further study of the
molecular mechanisms and cellular pathways of GC may provide novel
insight for the improvement of early diagnostic techniques,
precision therapies and prognostic predictions for patients with
GC.
The gene, zinc finger and BTB domain containing 7A
(ZBTB7A) is also known as lymphoma related factor (6), factor that binds to inducer of short
transcripts of human immunodeficiency virus type 1 (7) and osteoclast-derived zinc finger
(8). ZBTB7A is one member of the
protection of telomeres protein POZ-1/BTB and Kruppel (POK)
transcription factors family (9–11).
The POK transcription factor family has been demonstrated to bind
DNA via a Kruppel-like DNA-binding domain and represses
transcriptional activity by recruiting co-repressor complexes via
the POZ domain (12). ZBTB7A was
reported to promote oncogenesis through its capacity to repress the
transcription of an important tumor suppressor gene alternative
reading frame (ARF) (11).
Previously, aberrant ZBTB7A overexpression has been reported in a
number of different types of human cancer, including breast cancer,
non-small cell lung cancer (NSCL), lymphoma and ovarian cancer
(11,13–17),
suggesting that ZBTB7A acts as novel proto-oncogene in multiple
tissues.
However, the frequent chromosomal deletion of the
ZBTB7A gene locus (19p13.3) in multiple types of human cancer
(18–20) suggests it is not a proto-oncogene.
This evidence implies that the function of ZBTB7A is determined by
its context in solid tumors. A previous study demonstrated that the
loss of ZBTB7A promoted progression of mouse prostate cancer by
activating transcription factor SOX9-dependent signaling pathway in
a phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and
dual-specificity protein phosphatase PTEN (PTEN)-loss background
(21). Another study also reported
that ZBTB7A acts as a transcriptional repressor by binding directly
to the promoter of glycolytic genes and repressing their
transcription (22). Liu et
al (20) also demonstrated
that ZBTB7A can bind directly to the promoter region of the
melanoma cell adhesion molecule to suppress its transcription and
represses melanoma metastasis. These reports suggest that ZBTB7A
can act as a tumor suppressor under certain circumstances. Whether
ZBTB7A acts as oncogene or tumor suppressor is context-dependent in
different types of cancer.
Frequent deletions in the ZBTB7A gene locus have
been reported in a number of different types of cancer (18–20).
In the present study, it was hypothesized that ZBTB7A may function
as a tumor suppressor in GC. Recently, one study demonstrated that
downregulation of ZBTB7A by small interfering (si)RNA suppressed
the migratory ability of GC cells without an impact on cell
proliferation and apoptosis (23).
However, it remains unclear whether overexpression of ZBTB7A in a
GC cell line will affect cell proliferation, apoptosis and
migratory capacity. Therefore, it is necessary to further
investigate the function and potential mechanism of ZBTB7A in GC,
which may be a novel target for treatment and improve clinical
outcome.
Materials and methods
Human cancer genomic analysis
Copy number alterations (CNA) and mRNA data for the
ZBTB7A and PTEN genes in 441 cases of human gastric adenocarcinoma
were downloaded from TCGA database (24,25).
Z-score indicates ZBTB7A mRNA expression levels. For the analysis
of overall patient survival, the ZBTB7A expression data, along with
the survival data, were divided into two groups, ‘ZBTB7A low
expression’ and ‘ZBTB7A high expression’ based on the median
expression level of ZBTB7A.
Cell culture
The gastric adenocarcinoma cell line SGC-7901 was
bought from the Type Culture Collection of the Chinese Academy of
Sciences, (Shanghai, China). SGC-7901 cells were cultured in
RPMI-1640 medium (cat no. 10-013-CVR; Corning Incorporated,
Corning, NY, USA) with 10% fetal bovine serum (FBS, cat no. VS500T;
Ausbian, Vian-Saga Company, Shanghai, China; http://www.viansaga.com/h-pd-1.html#_pp=2_731)
and 1% penicillin/streptomycin. 293 cells (cat no. R79007; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) were cultured in DMEM
(cat no. R10-017-CVR; Corning Incorporated) with 10% FBS. All cells
were cultured at 37°C in a humidified incubator (MCO-15A; Sanyo,
Osaka, Japan) containing 5% CO2.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated with the SuperfecTRI™ reagent
(cat no. 3101-100; Shanghai Pufei Biotech Co., Ltd., Shanghai,
China). A total of 1 µg RNA was reverse transcribed into cDNA using
the Moloney-murine leukemia virus kit (cat no. M1705; Promega
Corporation, Madison, WI, USA). qPCR was performed in a Real-Time
PCR machine system (cat no. MX3000p; Agilent Technologies, Inc.,
Santa Clara, CA, USA), using cDNA and SYBR Master mixture (cat no.
DRR041B; Takara Biotechnology Co., Ltd., Dalian, China) (26–27).
The following cycling conditions were used: One cycle for 30 sec at
95°C, 40 cycles for 5 sec at 95°C and 30 sec at 60°C, then one
cycle of dissociation including 15 sec at 95°C, 30 sec at 60°C and
15 sec at 95°C. GAPDH was used as an endogenous control. The primer
sequences of GAPDH and ZBTB7A genes were as follows: GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse;
5′-CACCCTGTTGCTGTAGCCAAA-3′. ZBTB7A forward,
5′-CATCTGCGAGAAGGTCATCC-3′ and reverse 5′-TGTCCTGCCTGGTGAAGC-3′
(26,28).
Plasmid construction and lentiviral
transfection
The pGV115-ZBTB7A-FLAG-green fluorescent protein
(GFP)-puro plasmid (20 µg; Shanghai GeneChem Co., Ltd., Shanghai,
China) was constructed by inserting a full-length human cDNA of
ZBTB7A-FLAG gene into a pGV115-GFP-puro plasmid vector. The
pGV115-ZBTB7A-FLAG-GFP-puro plasmid along with another two
lentiviral packaging plasmids pHelper1.0 and pHelper2.0 (15 µg
each; both from Shanghai GeneChem Co., Ltd.) was cotransfected into
293 cells using Lipofectamine™ reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The lentiviral supernatant was collected,
concentrated and purified in the 48–72 h following cotransfection.
The SGC-7901 cell line was treated with an equal concentration of
2×108 (PFU/ml) of lentiviral supernatant for
transfection. Cells were observed for GFP expression under a
fluorescence microscope after 72 h viral transfection.
Western blotting
Protein was extracted using 2X
radioimmunoprecipitation assay lysis buffer (cat no. WB-0071;
Dingguo Bio Co., Ltd, Shanghai, China) from whole cells. The
protein concentration was measured using a bicinchoninic acid
protein assay kit (cat no. P0010S). The cell lysate was separated
using 10% SDS-PAGE with loading 30 µg protein and then transferred
onto a polyvinylidene difluoride membrane (cat no. IPVH00010; EMD
Millipore, Billerica, MA, USA) at 4°C and 300 mA for 150 min and
blotted with 5% milk in 1X TBST buffer at room temperature for 1 h.
Membranes were then blotted with diluted primary antibodies at 4°C
overnight for GAPDH (1:5,000; cat no. SC-32233; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), FLAG (1:3,000; cat no.
F1804; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and ZBTB7A
(1:2,000; cat no. Ab175918; Abcam, Cambridge, UK).
Survivin-3FLAG-GFP was used as FLAG positive control. A
goat-anti-rabbit secondary antibody (1:5,000; cat no. sc-2004;
Santa Cruz Biotechnology, Inc.) were then incubated at room
temperature for 1.5 h. Subsequently, Pierce™ ECL western blotting
substrate was added for exposure (Thermo Fisher Scientific, Inc.).
Each western blot analysis was performed at least three times
independently.
MTT assay
A total of 2,000 healthy cells/well were seeded into
a 96-well plate (cat no. 3599; Corning Incorporated) with 100 µl
medium. A total of 20 µl 5 mg/ml MTT reagent (cat no. JT343;
Genview, Beijing, China) was added to each well ~4 h prior to
detection. Next, the culture medium was removed and 100 µl/well
dimethyl sulfoxide was added. Following 5 min incubation, the
optical density of the cells was analyzed at 490/570 nm
emission/absorption wavelength on a Tecan infinite machine (cat no.
M2009PR; Tecan Group, Ltd., Mannedorf, Switzerland).
Cell cycle assay
Cells were seeded into 6-cm dishes with 4 ml medium
following lentiviral transfection. Then cells were collected after
3 days. Transfected cells were trypsinized, washed, fixed for 1 h
at 4°C with 75% ethanol and incubated with propidium iodide (PI)
dye (cat no. P4170; Sigma-Aldrich; Merck KGaA) for 30 min at room
temperature. Stained cells were measured for cell cycle phase
distribution using a flow cytometer (Guava® easyCyte HT;
EMD Millipore). Cell cycle data was analyzed using FlowJo software
(version 7.6.1; FlowJo LLC, Ashland, OR, USA). The cell cycle assay
was repeated three times independently.
Apoptosis assay
Cells were seeded into 6-well plates with 2 ml
medium following transfection and were harvested 2 days later.
Cells were stained using the Annexin V-APC&PI Apoptosis
Detection kit (cat no. 88-8007; eBioscience; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Cells
were stained to measure apoptosis using flow cytometry software
(version 7.6.1; FlowJo LLC). The apoptosis assay was repeated three
times independently.
Scratch assay
An equal number of 3×104 cells/well were
seeded into a 96-well plate following transfection. The cell
monolayer was scratched in a straight line in each well. The line
was marked and images were captured under phase-contrast microscope
(Zeiss; XDS-100). Cells were cultured for 8 and 24 h. Following
incubation, images were retaken in the same region centered on the
line. The width was measured and recorded at 0, 8 and 24 h. The
migratory rate was calculated as [(width at 0 h - width at 8 or 24
h)/width at 0 h]. The rate of migration was analyzed. The scratch
assay was repeated four times independently.
Statistical analysis
All results were analyzed using GraphPad Prism
software (version 5; GraphPad software, Inc., La Jolla, CA, USA)
and data were presented as the mean ± standard error of the mean.
The data were analyzed using a Student's t-test for comparisons
between two groups. Multiple comparison tests were performed using
two-way analysis of variance (ANOVA) and Bonferroni post-hoc tests
to analyze the data from the cell cycle distribution and migration
assays. One-way ANOVA was used to analyze the data from the
apoptosis data, followed by Tukey's test. Log-rank test and
Kaplan-Meier estimators were performed. P<0.05 was considered to
indicate a statistically significant difference.
Results
Frequent loss of ZBTB7A and its
association with patient overall survival in the human gastric
adenocarcinoma database
As frequent chromosomal deletions at the ZBTB7A gene
locus have been reported in a number of different types of human
cancer (18–20), in the present study the ZBTB7A gene
was investigated in human gastric adenocarcinoma. CNA, mRNA
expression and overall survival data from 441 patients were
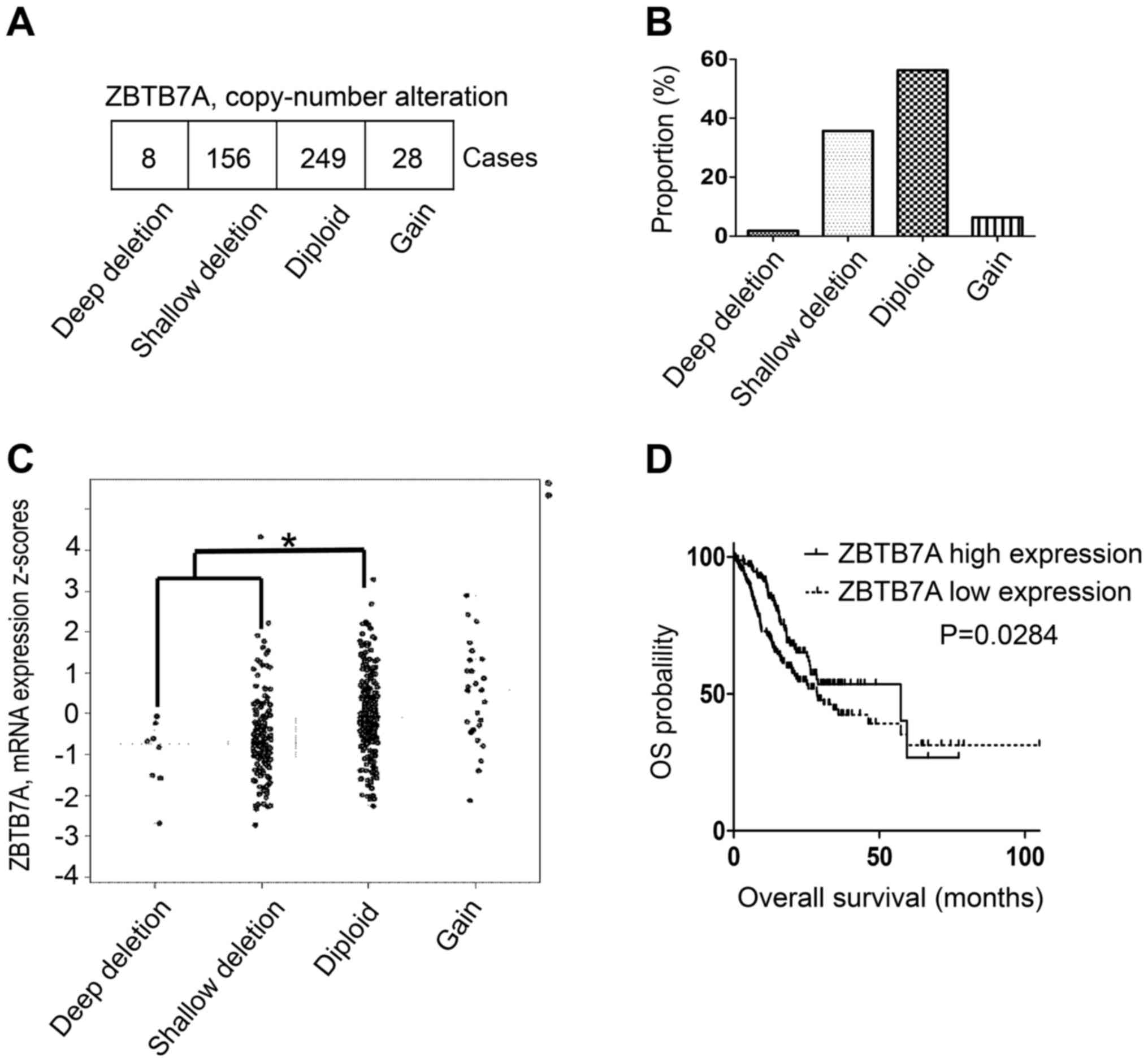
downloaded from the TCGA provisional dataset. A total of 8 patients
(1.8%) presented with a homozygous deletion of ZBTB7A and 156
patients (35.37%) exhibited hemizygous deletions (Fig. 1A and B). A total of 37.17% of
patients with gastric adenocarcinoma exhibited a ZBTB7A gene
deletion, with 56.56% exhibiting no deletion (Fig. 1B). The ZBTB7A mRNA expression level
of the two gene deletion groups was significantly decreased
compared with the normal diploid group (Fig. 1C). The overall survival data were
analyzed and it was demonstrated that the ZBTB7A high expression
group exhibited a median survival of 57.39 months, which was
significantly increased compared with 28.71 months in the ZBTB7A
low expression group (Fig. 1D).
These results implied that low expression of ZBTB7A was associated
with a poor median survival. The data indicated that ZBTB7A may
function as potential tumor suppressor in gastric adenocarcinoma.
To further investigate this, a gain-of-function experiment was
performed for ZBTB7A, and its impact on cell proliferation,
apoptosis and migration in the GC cell line SGC-7901 was
investigated.
Establishment of a ZBTB7A
overexpression system in the SGC-7901 cell line
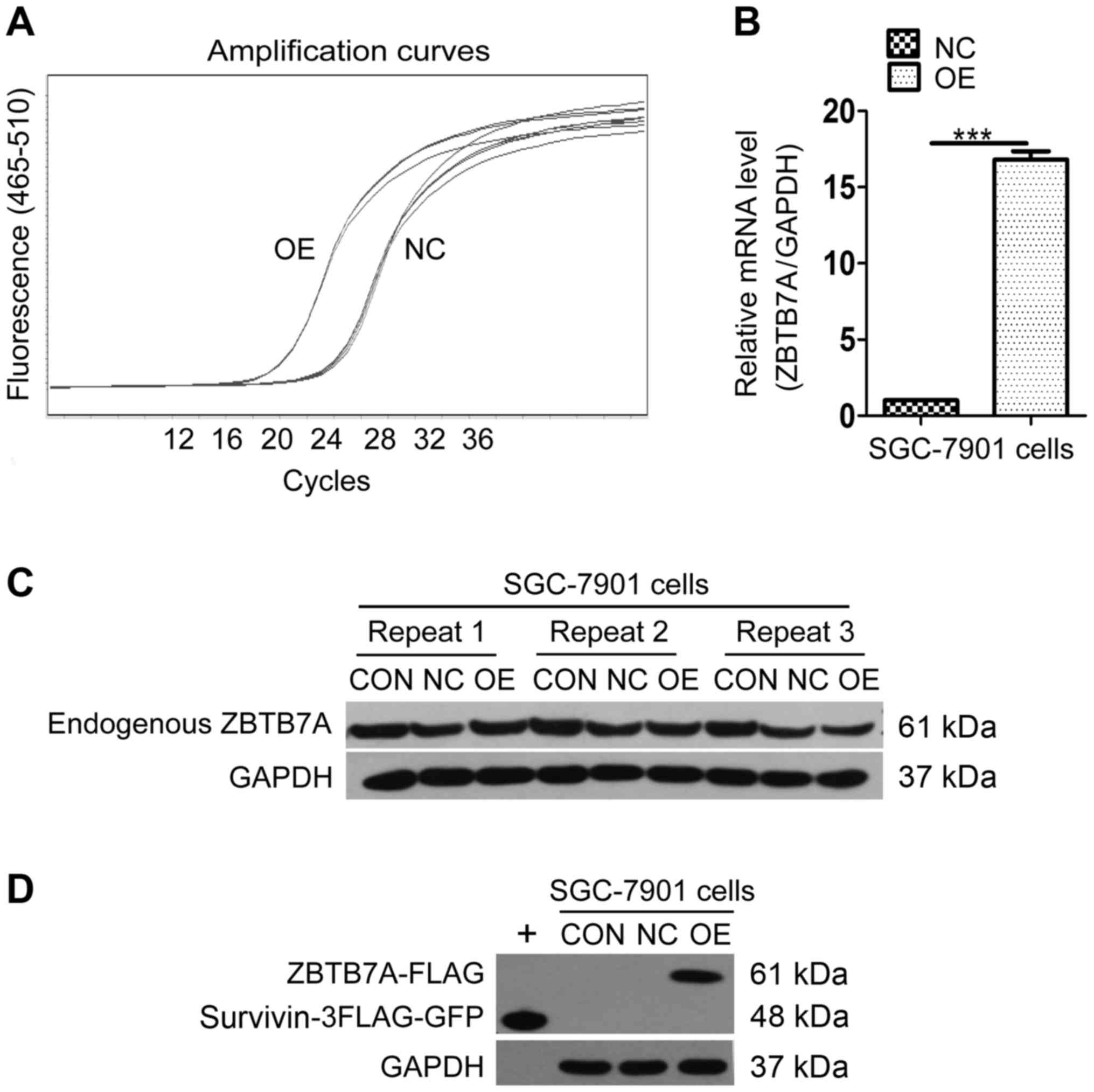
To produce a cell line that overexpressed ZBTB7A,
SGC-7901 cells were transfected using a lentiviral vector. GFP
expression by the negative control (NC) and overexpression (OE)
group confirmed that transfection was successful (data not shown).
The amplification curve of the RT-qPCR results confirmed that the
mRNA level of the OE group reached a peak more rapidly than the NC
group (Fig. 2A). The ZBTB7A/GAPDH
mRNA expression analysis demonstrated that the level of mRNA in the
OE group was ~16 times that of the NC group (Fig. 2B), suggesting that ZBTB7A mRNA was
successfully over-expressed. In the western blot analysis,
endogenous ZBTB7A expression was equal in the control (CON), NC and
OE groups (Fig. 2C). Ectopic
ZBTB7A-FLAG protein was successfully expressed only in OE group
with Survivin-3FLAG-GFP serving as positive control (Fig. 2D). The data indicated that the
ZBTB7A overexpression cell line was successfully constructed. These
cells were used for further assays.
Ectopic ZBTB7A expression results in
cell cycle inhibition at S phase
To investigate whether gain-of-function of ZBTB7A
affects GC cell proliferation or the cell cycle, a cell cycle and
MTT assay were used. In the MTT assay, no significant difference in
proliferation between the CON, NC and OE groups was detected (data
not shown), which suggested that overexpression of ZBTB7A may not
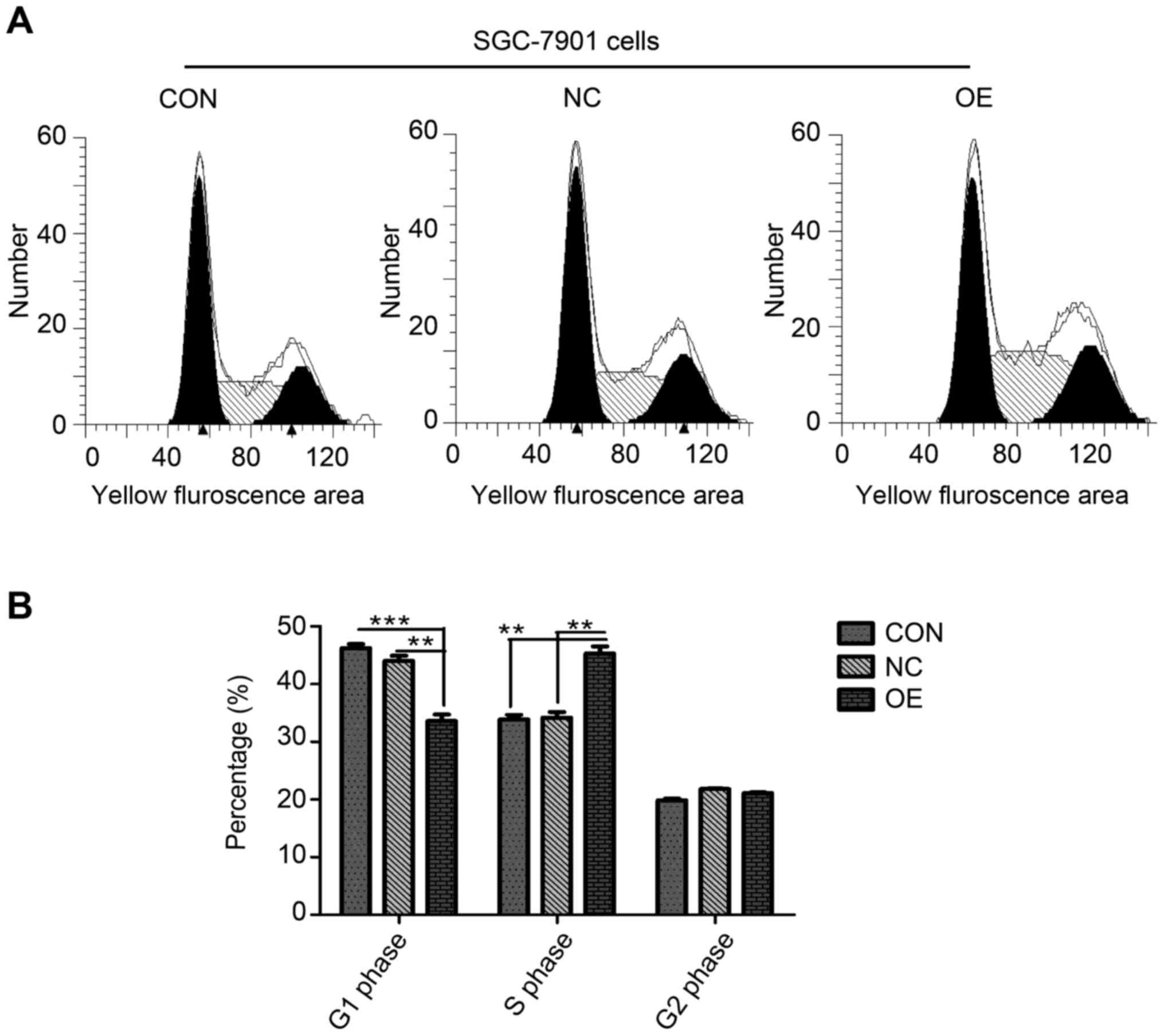
affect cell proliferation. An increased proportion of cells were in
the S phase in the OE group compared with the CON or NC groups
according to the cell cycle assay (Fig. 3A). Statistical analyses were
performed following three repeats. A decreased percentage of cells
in OE group were demonstrated to be in the G1 phase compared with
the CON (P=0.0007) and NC (P=0.0021) groups, with no difference
between the CON and NC groups (Fig.
3B). The P-values for the S phase in the OE group were CON
(P=0.0015) and NC (P=0.0022), with no difference between CON and NC
groups (Fig. 3B). No difference
was detected in the percentage of cells in the G2 phase between the
three groups (Fig. 3B).
Overexpression of ZBTB7A in the SGC-7901 cell line induced an
abnormal number of cells to arrest in the S phase of the cell cycle
but without significant impact on cell proliferation.
Gain-of-function of ZBTB7A in SGC-7901
cell line induces apoptosis
To further investigate the impact on cell death or
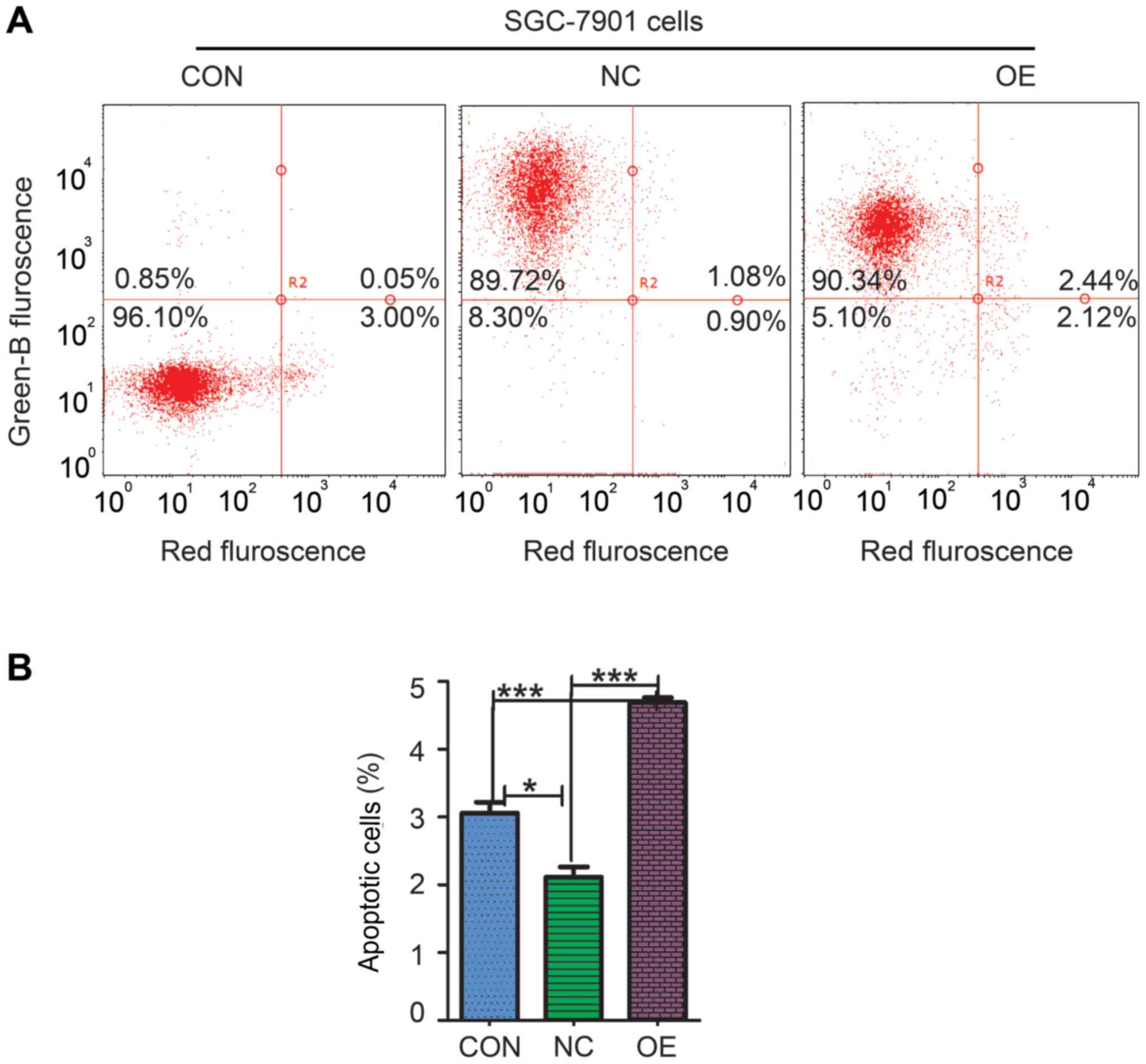
apoptosis of overexpressing ZBTB7A, an apoptosis assay was used. In
the NC group, there were ~90.8% GFP-positive cells and 92.78%
GFP-positive cells in the OE group were detected, with only 0.9%
GFP-positive cells in CON group (Fig.
4A). The apoptosis percentage of CON, NC and OE groups was
3.06±0.27, 2.11±0.26 and 4.69±0.12%, respectively (Fig. 4B). Statistical analysis identified
that percentage of apoptotic cells in the OE group was
significantly increased compared with the CON group (P=0.0007) and
NC group (P=0.0001). These data indicated that gain-of-function of
ZBTB7A in SGC-7901 cell promoted cell apoptosis.
Upregulation of ZBTB7A suppresses cell
migration
The migratory and invasive ability is another key
characteristic of GC cells, which facilitates metastasis to other
organs and results in a poor prognosis in patients (29,30).
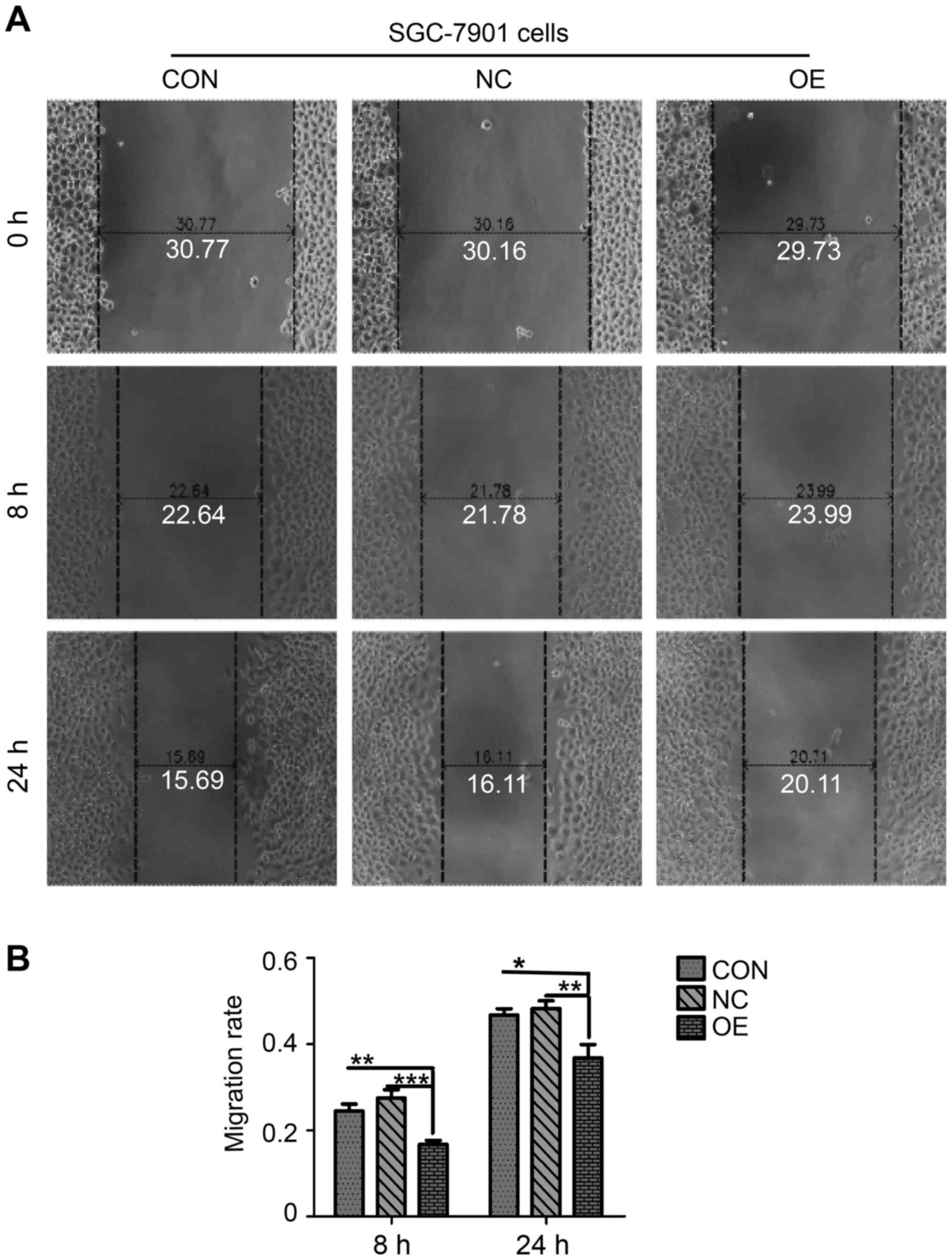
In the present study, the migration assay test was used to
investigate the potential impact of upregulation of ZBTB7A on GC
cell migration (Fig. 5). The
migratory rate of OE group was 0.17±0.02, in contrast with
0.28±0.03 of NC group and 0.24±0.03 of CON group at 8 h (Fig. 5B). Following 24 h, the migratory
rate of the CON, NC and OE groups were 0.46±0.04, 0.49±0.03 and
0.35±0.02, respectively (Fig. 5B).
Another three repeats were conducted independently. Statistical
analysis demonstrated that the migratory rate of cells in the OE
group at 8 and 24 h time points were significantly decreased
compared with the CON group and NC group (Fig. 5B), suggesting that overexpression
of ZBTB7A repressed GC cell migration.
Discussion
TCGA provides a large amount of data, serving an
important resource for the field of cancer research (31). Previously, a number of studies have
demonstrated that the ZBTB7A gene locus exhibited frequent
chromosomal deletions in a number of different types of human
cancer (18–20). In the present study, data mining of
ZBTB7A in TCGA gastric adenocarcinoma dataset was performed. It was
demonstrated that frequent loss of the ZBTB7A gene also occurred in
gastric adenocarcinoma, with 37.17% gene loss compared with 56.56%
normal gene status. Deletion of the ZBTB7A gene resulted in a
decreased in mRNA expression of ZBTB7A. In addition, survival
analysis demonstrated that downregulation of ZBTB7A was associated
with a poor prognosis in patients with gastric adenocarcinoma.
ZBTB7A is a member of the POK transcription factor
family (9–11), which was previously known as a
proto-oncogene that acts by suppressing the transcription of an ARF
of a tumor suppressor (11).
Previously, ZBTB7A has also been reported to be novel
proto-oncogene in different types of cancer, including breast
cancer, NSCL, lymphoma and ovarian cancer (13–17).
However, the frequent chromosomal deletion of the ZBTB7A gene locus
in numerous types of human cancer (18–20)
contradicts its proto-oncogenic role. Previous studies indicate
that ZBTB7A works as a tumor suppressor in melanoma (20), PTEN-loss background prostate cancer
(21) and colonic cancer (22). In the present study, it was
identified that ZBTB7A overexpression induced an abnormal
proportion of cells to be in the S phase; however, this had no
impact on cell proliferation. Furthermore, in the gain-of-function
assay, ZBTB7A promoted cell apoptosis and repressed cell migration
in the SGC-7901 cell line. The present study indicates that ZBTB7A
functions as a tumor suppressor in GC SGC-7901 cell line.
Recently it's been reported that downregulation of
ZBTB7A by siRNA, repressed the migratory ability of GC cells
without an impact on cell proliferation and apoptosis (23), without presenting detailed
mechanisms. In the present study, upregulation of ZBTB7A also
suppressed migratory ability. This phenomenon deserves further
investigation. It was also identified that gain-of-function of
ZBTB7A in the SGC-7901 cell line promotes apoptosis; however, there
was no impact on apoptosis when downregulation of ZBTB7A was
investigated (23). The abnormal
cell cycle S phase accumulation induced by ZBTB7A overexpression
indicated that ZBTB7A may promote the transcription of different
target genes depending on whether it is up- or downregulated. This
implies that the function of ZBTB7A in GC may be background status
dependent.
It was reported that ZBTB7A acted as a
proto-oncogene in certain contexts but also exhibited tumor
suppressive activity in PTEN deficient tumors (29). This suggested that ZBTB7A may
possess onco-suppressive activity in PTEN-deleted gastric
adenocarcinoma as well as in PTEN-deficit prostate cancer (29). This demonstrates that the role of
ZBTB7A in GC may be PTEN deficit associated context-dependent.
In conclusion, the present study identified a novel
genetic event associated with gastric adenocarcinoma, the frequent
loss of ZBTB7A gene. Deletion of ZBTB7A was associated with a poor
prognosis in patients with GC. Gain-of-function of ZBTB7A
demonstrated tumor suppressive-like activity, including inducing
cell cycle arrest at S phase, promoting apoptosis and repressing
cell migration in a GC cell line. The present study indicated that
ZBTB7A functioned as tumor suppressor in GC cells, which may offer
therapeutic or prognostic implications for patients with GC in
future.
Acknowledgements
The authors of the present study would like to thank
Shanghai GeneChem Co., Ltd. (Shanghai, China) for their technical
assistance. In addition, the authors would like to thank the core
lab of the Haikou City Hospital for offering numerous suggestions.
The authors also would like to thank Dr Hongman Wu (Haikou
Municipal People's Hospital) for his advice.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Soerjomataram I, Lortet-Tieulent J, Parkin
DM, Ferlay J, Mathers C, Forman D and Bray F: Global burden of
cancer in 2008: A systematic analysis of disability-adjusted
life-years in 12 world regions. Lancet. 380:1840–1850. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tan P and Yeoh KG: Genetics and molecular
pathogenesis of gastric adenocarcinoma. Gastroenterology.
149:1153–1162.e3. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N
and Di Cesare PE: Leukemia/lymphoma-related factor, a POZ
domain-containing transcriptional repressor, interacts with histone
deacetylase-1 and inhibits cartilage oligomeric matrix protein gene
expression and chondrogenesis. J Biol Chem. 279:47081–47091. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pessler F, Pendergrast PS and Hernandez N:
Purification and characterization of FBI-1, a cellular factor that
binds to the human immunodeficiency virus type 1 inducer of short
transcripts. Mol Cell Biol. 17:3786–3798. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kukita A, Kukita T, Ouchida M, Maeda H,
Yatsuki H and Kohashi O: Osteoclast-derived zinc finger (OCZF)
protein with POZ domain, a possible transcriptional repressor, is
involved in osteoclastogenesis. Blood. 94:1987–1997.
1999.PubMed/NCBI
|
|
9
|
Davies JM, Hawe N, Kabarowski J, Huang QH,
Zhu J, Brand NJ, Leprince D, Dhordain P, Cook M, Morriss-Kay G and
Zelent A: Novel BTB/POZ domain zinc-finger protein, LRF, is a
potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 18:365–375.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Apostolopoulou K, Pateras IS, Evangelou K,
Tsantoulis PK, Liontos M, Kittas C, Tiniakos DG, Kotsinas A,
Cordon-Cardo C and Gorgoulis VG: Gene amplification is a relatively
frequent event leading to ZBTB7A (Pokemon) overexpression in
non-small cell lung cancer. J Pathol. 213:294–302. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maeda T, Hobbs RM, Merghoub T, Guernah I,
Zelent A, Cordon-Cardo C, Teruya-Feldstein J and Pandolfi PP: Role
of the proto-oncogene Pokemon in cellular transformation and ARF
repression. Nature. 433:278–285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Costoya JA: Functional analysis of the
role of POK transcriptional repressors. Brief Funct Genomic
Proteomic. 6:8–18. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang L, Siu MK, Wong OG, Tam KF, Lam EW,
Ngan HY, Le XF, Wong ES, Chan HY and Cheung AN: Overexpression of
proto-oncogene FBI-1 activates membrane type 1-matrix
metalloproteinase in association with adverse outcome in ovarian
cancers. Mol Cancer. 9:3182010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aggarwal A, Hunter WJ III, Aggarwal H,
Silva ED, Davey MS, Murphy RF and Agrawal DK: Expression of
leukemia/lymphoma-related factor (LRF/POKEMON) in human breast
carcinoma and other cancers. Exp Mol Pathol. 89:140–148. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qu H, Qu D, Chen F, Zhang Z, Liu B and Liu
H: ZBTB7 overexpression contributes to malignancy in breast cancer.
Cancer Invest. 28:672–678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vredeveld LC, Rowland BD, Douma S,
Bernards R and Peeper DS: Functional identification of LRF as an
oncogene that bypasses RASV12-induced senescence via upregulation
of CYCLIN E. Carcinogenesis. 31:201–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao ZH, Wang SF, Yu L, Wang J, Chang H,
Yan WL, Zhang J and Fu K: Overexpression of Pokemon in non-small
cell lung cancer and foreshowing tumor biological behavior as well
as clinical results. Lung Cancer. 62:113–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zack TI, Schumacher SE, Carter SL,
Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J,
Mermel CH, et al: Pan-cancer patterns of somatic copy number
alteration. Nat Genet. 45:1134–1140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu XS, Genet MD, Haines JE, Mehanna EK,
Wu S, Chen HI, Chen Y, Qureshi AA, Han J, Chen X, et al: ZBTB7A
Suppresses melanoma metastasis by transcriptionally repressing
MCAM. Mol Cancer Res. 13:1206–1217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang G, Lunardi A, Zhang J, Chen Z, Ala U,
Webster KA, Tay Y, Gonzalez-Billalabeitia E, Egia A, Shaffer DR, et
al: Zbtb7a suppresses prostate cancer through repression of a
Sox9-dependent pathway for cellular senescence bypass and tumor
invasion. Nat Genet. 45:739–746. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu XS, Haines JE, Mehanna EK, Genet MD,
Ben-Sahra I, Asara JM, Manning BD and Yuan ZM: ZBTB7A acts as a
tumor suppressor through the transcriptional repression of
glycolysis. Genes Dev. 28:1917–1928. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi DB, Wang YW, Xing AY, Gao JW, Zhang H,
Guo XY and Gao P: C/EBPα-induced miR-100 expression suppresses
tumor metastasis and growth by targeting ZBTB7A in gastric cancer.
Cancer Lett. 369:376–385. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bengtsson M, Hemberg M, Rorsman P and
Ståhlberg A: Quantification of mRNA in single cells and modelling
of RT-qPCR induced noise. BMC Mol Biol. 9:632008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kubista M, Andrade JM, Bengtsson M,
Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B,
Strömbom L, et al: The real-time polymerase chain reaction. Mol
Aspects Med. 27:95–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang SM, Tie J, Wang WL, Hu SJ, Yin JP, Yi
XF, Tian ZH, Zhang XY, Li MB, Li ZS, et al: POU2F2-oriented network
promotes human gastric cancer metastasis. Gut. 65:1427–1438. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang ZY and Ge HY: Micrometastasis in
gastric cancer. Cancer Lett. 336:34–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garraway LA and Lander ES: Lessons from
the cancer genome. Cell. 153:17–37. 2013. View Article : Google Scholar : PubMed/NCBI
|