Introduction
Colorectal cancer (CRC) is one of the most commonly
diagnosed malignancies with an estimated 1.4 million cases and
693,900 mortalities by GLOBOCAN 2012 and a leading cause of
cancer-associated mortality worldwide (1). The pathogenesis of CRC involving
genetic and environmental factors, and their interactions has been
investigated previously (2–4), and
numerous risk factors of CRC, such as tobacco use, unhealthy diet,
obesity and physical inactivity, have been identified (1). It is widely recognized that loss of
genomic stability and alterations in tumor suppressor genes and
oncogenes have a key role in occurrence and progression of CRC
(2,5,6).
Preventive measures, such as maintaining a healthy body weight,
being physically active, minimizing consumption of red, processed
meat and alcohol, and avoidance of smoking, screening options
(fecal test, colonoscopy, stool DNA test, computed tomography), and
improved treatments at an early stage of the disease are most
likely attributed to reducing the CRC mortality rate observed in a
large number of countries worldwide (1,7).
Additionally, advances in prognostic biomarkers that may allow
personalized treatments could have also contributed to improvements
in overall survival of patients with CRC. In addition, various
biomarkers that may be used to assist in the diagnosis of CRC have
been identified (8,9). However, survival outcomes of CRC,
particularly predictive and prognostic biomarkers, remain to be
elucidated (10,11). Therefore, valid prognostic
biomarkers are urgently required to assist the prediction of the
outcomes of CRC, which would result in earlier performances of
preventative interventions or surgery that would improve survival
rates (12).
The advent of high-throughput transcriptomic
profiling, has allowed biomarker identification to be taken to the
genomic level (13,14). Additionally, it is important to
understand the mRNA expression profiles involved in CRC and
identify reliable biomarkers that may predict the survival of
patients with CRC. The development of gene microarray and RNA
sequencing has allowed for the use of gene expression profiling to
identify genes associated with the carcinogenesis and development
of CRC (13). Therefore, screening
differentially expressed genes (DEGs) based on microarray analysis
is an important novel way to investigate the pathogenesis of CRC,
additionally it is a quicker and more effective method of
identification of the gene transcripts involved in energy
metabolism in CRC and the mRNA isoforms used for diagnosis
(15). Additionally, various
databases, such as The Cancer Genome Atlas (TCGA) and PROGgeneV2
also help identify additional, valuable information using the data
mining method.
In the current study, microarray analysis combined
with a protein-protein interaction network was performed to
identify the DEGs associated with CRC. Additionally, nebulette
(NEBL) and complement C1q like 1 (C1QL1) were identified as hub
genes of interest among DEGs using bioinformatics analysis.
Subsequently, NEBL and C1QL1 were validated using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) in 3
CRC cell lines and 30 CRC tissues with paired adjacent
non-cancerous controls from patients. In order to investigate their
prognostic values, the overall survival of patients with NEBL and
C1QL1 in patients with CRC was determined using PROGgeneV2.
Overall, the present study aimed to identify prognostic biomarkers
among DEGs for CRC and allow for earlier treatment strategy
intervention.
Materials and methods
Data preprocessing and DEGs
screening
The mRNA expression profile dataset GSE41258 was
downloaded from the Gene Expression Omnibus (GEO) database
(www.ncbi.nlm.nih.gov/geo/), which was
performed on the platform of Affymetrix Human Genome U133A Array.
GSE41258 dataset contained 182 samples isolated from patients with
CRC and 54 normal colorectal tissues (16). Expression profiling microarrays
were firstly preprocessed by background correction and
normalization, subsequently, the DEGs were statistically analyzed
in both groups. All analyses were conducted using the limma package
in R (17). The following
thresholds were used to identify DEGs: P<0.01 and
|log2 fold-change (FC)| >1.
Construction of PPI network of DEGs
and identification of hub genes
Protein-protein interactions (PPIs) are indirectly
reflected as reciprocal interactions among DEGs. In the present
study, the online server Search Tool for the Retrieval of
Interacting Genes (STRING v10.5) was used to investigate these
interactions (string-db.org). interacting DEGs were
subsequently visualized using Cytoscape v3.4.0 software (18) to identify the hub genes, which were
identified as highly connected genes in the interaction network if
they had an interaction degree (interD) >20. In the present
study, NEBL and C1QL1 were identified as hub genes, whose
association with CRC remains unclear.
Gene Ontology (GO) analysis of NEBL
and C1QL1
In order to obtain additional insight into the
functional enrichment of NEBL and C1QL1, GO analysis was performed.
The Database for Annotation, Visualization, and Integrated
Discovery (DAVID; david.ncifcrf.gov) version 6.8 was used to investigate
the relevant biological meaning of the hub genes of interest. A
P-value <0.05 indicated a statistically significant functional
annotation.
Cell lines and cell culture
The HCT116, HT29, DLD1 human CRC cell lines and the
NCM460 normal human colorectal epithelial cell line were used in
this study. All the cell lines were purchased from the Type Culture
Collection of the Chinese Academy of Sciences (Shanghai, China).
These cell lines were cultured in Dulbecco's modified Eagle's
medium (GE Healthcare Life Sciences, Logan, UT, USA) containing 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). All cells were cultured at 37°C in a humidified
atmosphere containing 95% O2 and 5% CO2.
Clinical sample collection and ethical
approval
Fresh CRC tissue samples and paired adjacent
non-cancerous tissue samples were obtained after surgical resection
prior to radiation or chemotherapy from 30 patients at the
Department of General Surgery, Zhongnan Hospital of Wuhan
University (Wuhan, China) from March 2017 to June 2017. The samples
were frozen and stored in liquid nitrogen. The morphological
classification of the tumor was performed according to the American
Joint Committee on Cancer (AJCC) staging system (19). Written informed consent was
obtained from all patients. The study was approved by the Ethics
Committee of Zhongnan Hospital of Wuhan University. All procedures
performed in studies involving human participants were in
accordance with the ethical standards of the Ethical Committee of
Zhongnan Hospital of Wuhan University, and with the principle of
the 1964 Helsinki declaration and its later amendments or
comparable ethical standards. The sample characteristics for CRC
patients are presented in Table
I.
 | Table I.Characteristics of patients with
colorectal cancer in microarray and validation experiments. |
Table I.
Characteristics of patients with
colorectal cancer in microarray and validation experiments.
| Characteristics | Microarray (n) | Validation (n) | P-value |
|---|
| Age (years) |
|
| 0.074 |
| Mean ±
SD | 63.30±13.98 | 59.33±13.27 |
|
|
Range | 19–87 | 22–87 |
|
| Gender |
|
| 0.156 |
|
Female | 86 | 10 |
|
| Male | 96 | 20 |
|
| AJCC stage |
|
| 0.002 |
| I | 28 | 3 |
|
| II | 48 | 16 |
|
| III | 49 | 11 |
|
| IV | 57 | 0 |
|
| TNM stage |
|
|
|
|
Tumor |
|
| 0.518 |
|
T1 | 4 | 1 |
|
|
T2 | 34 | 3 |
|
|
T3 | 131 | 25 |
|
|
T4 | 13 | 1 |
|
| Node |
|
| 0.245 |
| N0 | 93 | 18 |
|
| N1 | 46 | 9 |
|
| N2 | 43 | 3 |
|
| Metastasis |
|
| <0.01 |
| M0 | 125 | 30 |
|
| M1 | 57 | 0 |
|
RNA extraction and RT-qPCR
Total RNA was extracted from the cells and tissues
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RNA quantification was
performed using a spectrophotometer (NanoDrop 2000, Thermo Fisher
Scientific, Inc.). Total RNA (1 µg) was used with a first-strand
cDNA using a synthesis kit (Thermo Fisher Scientific, Inc.) to
perform the RT reaction (65°C for 5 min, 42°C for 60 min and 70°C
for 5 min). The mRNA expression levels of the selected genes were
subsequently evaluated by qPCR using a QuantStudio™ 6 Flex
Real-Time PCR instrument (Thermo Fisher Scientific, Inc.) with SYBR
Premix Ex Taq™ II mix (Takara Bio, Inc., Otsu, Japan). The qPCR
reaction was performed with an initial denaturation step of 95°C
for 5 min followed by 40 cycles of 95°C for 3 sec and 61°C for 30
sec. The NEBL primers were forward (F)
5′-GGAATGCAAGCTGGCACTGACA-3′and reverse (R)
5′-GAGTGTCTGTGCTCACCTGCAT-3′; C1QL1 F 5′-AGTATGTGGGCAGACCTCTGCA-3′
and R 5′-CCAGCTTGATGAAGACCTCGTC-3′; and GAPDH F
5′-AGAAGGCTGGGGCTCATTTG-3′ and R 5′-GCAGGAGGCATTGCTGATGAT-3′. The
relative mRNA expression levels were calculated using the
2−ΔΔCq method (20).
GAPDH was used as the internal control.
Statistical analysis
All analyses of experimental validation were
performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA).
Student's t-test was used for comparisons between groups.
Differences between the CRC cell lines and control cell line were
analyzed using one-way analysis of variance with a Bonferroni
correction. P<0.05 was considered to indicate a statistically
significant difference. Data are presented as mean ± standard
deviation. Experiments were repeated independently at least three
times.
Survival analysis of NEBL and
C1QL1
The median value of each hub gene in tumor samples
was calculated. Samples with expression higher than the median
value were placed in the high expression group, and the samples
with expression lower than the median value were placed in the low
expression group. Survival analysis was performed using PROGgeneV2
tool for both groups (21), which
allowed the investigation of prognostic implications of gene
expression associated with CRC in the corresponding microarray
datasets. The GSE41258 dataset was used to predict the overall
survival of the hub genes of interest in the present study. In
addition, survival models were adjusted for multiple covariates
such as age, gender and cancer stages. The survival curves were
visualized using Kaplan-Meier plots for the high and low expression
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
DEGs screening and identification of
hub genes
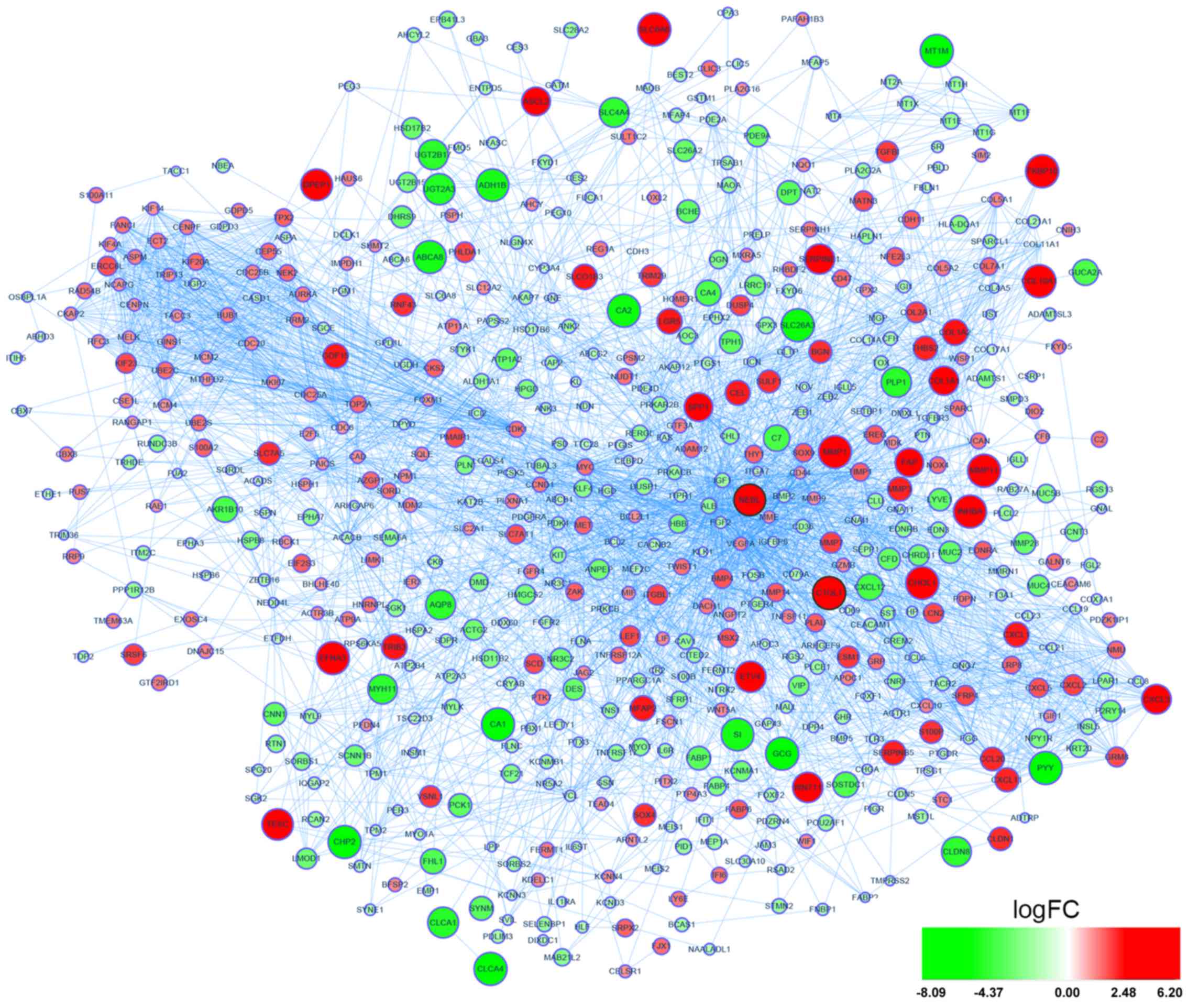
A total of 759 DEGs were identified, including 279
upregulated and 480 downregulated DEGs in primary CRC samples
compared with normal colon samples (Fig. 1). By constructing PPI network of
DEGs, NEBL and C1QL1 were identified as the two hub genes based on
highly connected degree (interD= 34 and 33, respectively) as well
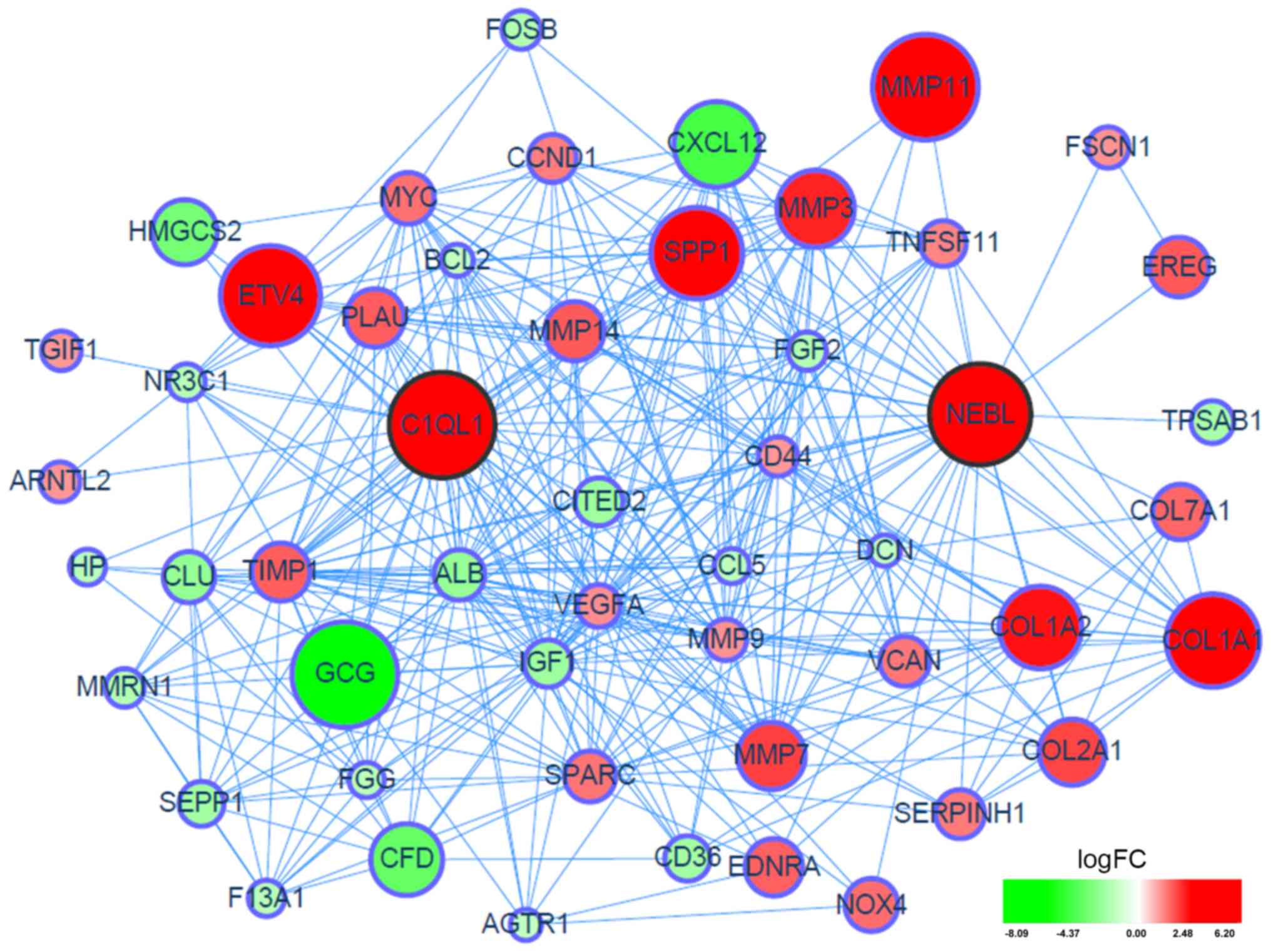
as being overexpressed DEGs in the GSE41258 dataset (Fig. 1). Additionally, a PPI network of
the two hub genes and their linker genes in CRC was presented in
Fig. 2.
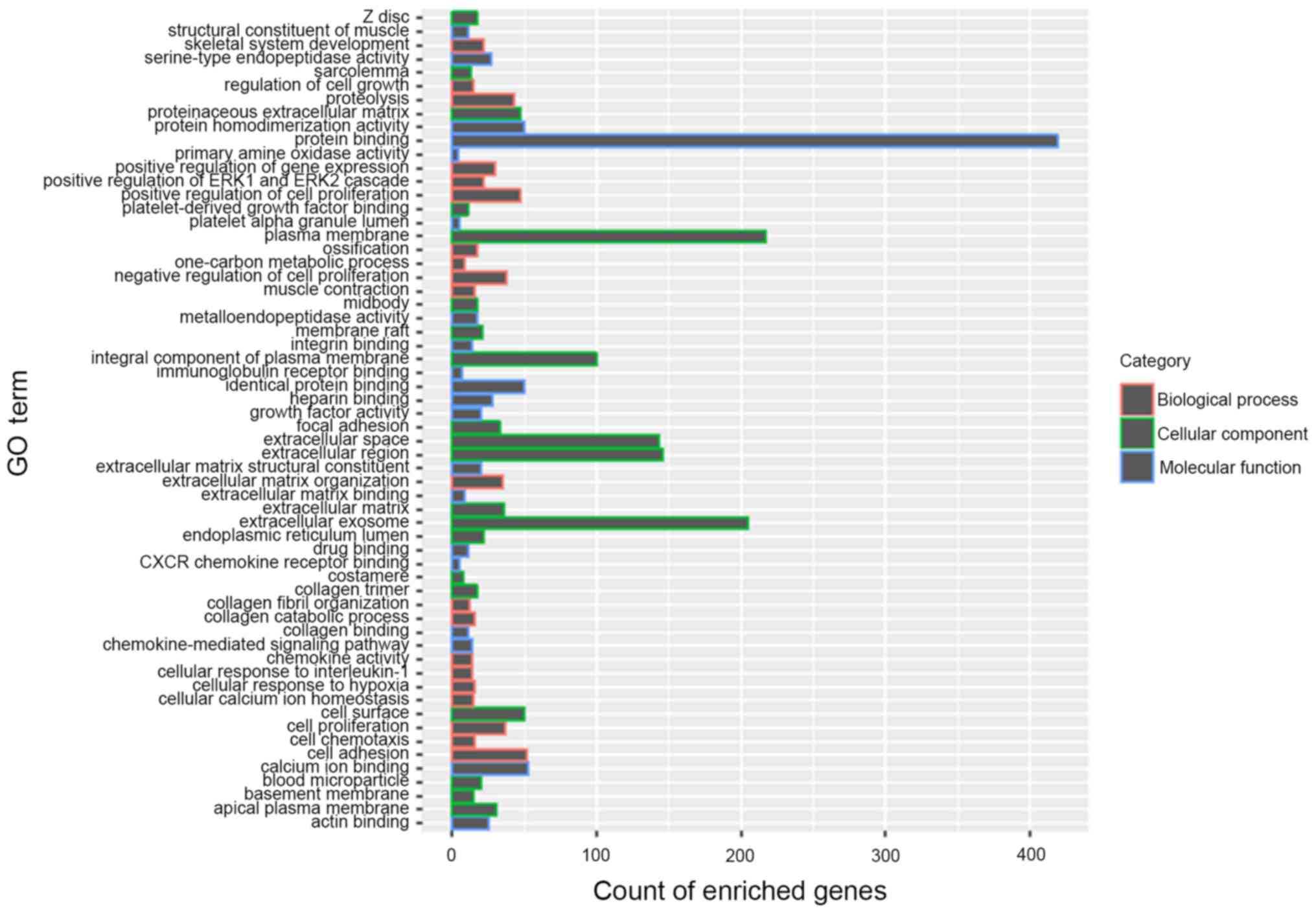
GO analysis of NEBL and C1QL1
Functional enrichment map of DEGs, including NEBL
and C1QL1 was presented in Fig. 3,
which represents a holistic view of the functions of all screened
DEGs. In addition, under the threshold of P<0.05, the biological
processes NEBL was significantly enriched in were ‘regulation of
biological quality’, ‘regulation of cellular component
organization’, ‘biological regulation’, ‘regulation of anatomical
structure size’, ‘regulation of organelle organization’ and
‘regulation of biological process’. The biological processes C1QL1
was enriched in were ‘response to stimulus’, ‘locomotory behavior’
and ‘behavior’ (Table II).
 | Table II.Hub genes with their representative
significantly enriched terms of GO biological processes. |
Table II.
Hub genes with their representative
significantly enriched terms of GO biological processes.
| A, NEBL |
|---|
|
|---|
| GO ID | Description | P-value |
|---|
|
|---|
| 65008 | Regulation of
biological quality |
5.79×10−9 |
| 51128 | Regulation of
cellular component organization |
1.57×10−5 |
| 65007 | Biological
regulation |
1.37×10−4 |
| 90066 | Regulation of
anatomical structure size |
4.28×10−4 |
| 33043 | Regulation of
organelle organization |
4.98×10−4 |
| 50789 | Regulation of
biological process |
3.71×10−3 |
|
| B,
C1QL1 |
|
| GO ID |
Description | P-value |
|
| 50896 | Response to
stimulus |
2.74×10−7 |
| 7626 | Locomotory
behavior |
1.56×10−6 |
| 7610 | Behavior |
1.92×10−6 |
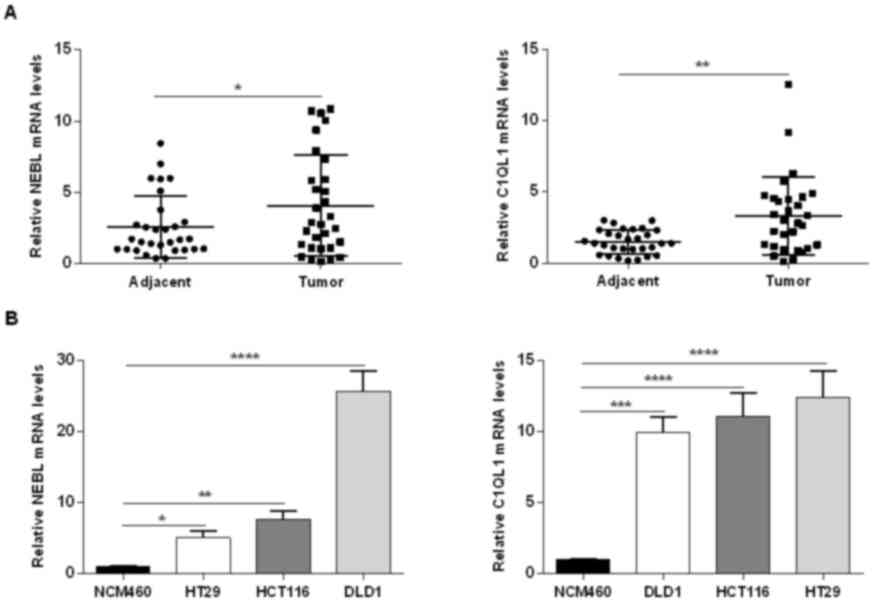
Validation of NEBL and C1QL1 mRNA
expression
The mRNA expression levels of NEBL and C1QL1 in the
present study were confirmed in the tissues of patients with CRC
and CRC cell lines using RT-qPCR. The mRNA expression levels of
NEBL and C1QL1 were higher in CRC tissues when compared with normal
adjacent tissues (NEBL, P=0.049; C1QL1, P=0.001; Fig. 4A). NEBL and C1QL1 expression levels
were upregulated in the 3 CRC cell lines when compared with NCM460
cell line (Fig. 4B).
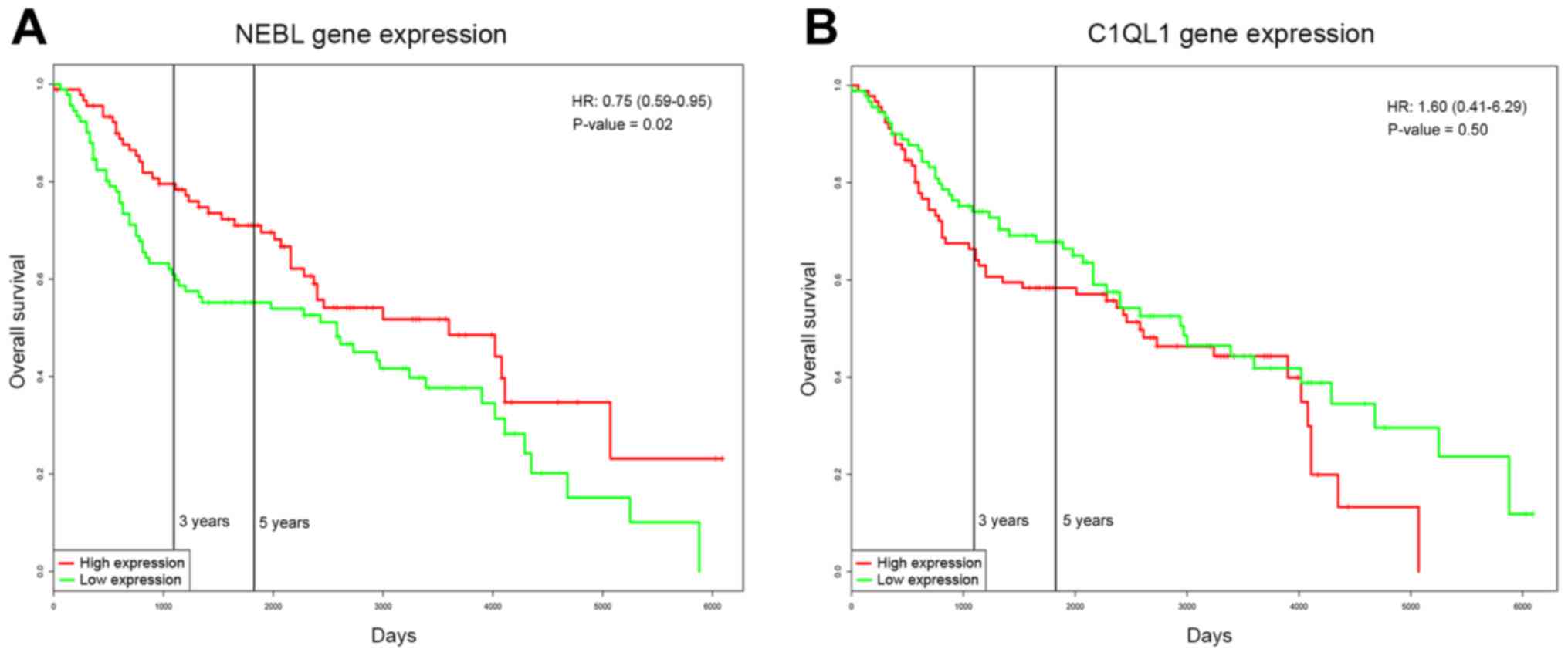
Prognosis of NEBL and C1QL1
Survival analysis was performed for NEBL and C1QL1
using PROGgeneV2 based on the GSE41258 dataset. In the overall
dataset, 91 samples were demonstrated to exhibit a high expression
of NEBL, and a further 91 samples were demonstrated to exhibit a
low expression of NEBL. During >16 years of follow up, the
incidence of death in high and low expression groups was 42 and 60
events, respectively. The median survival of high and low
expression of NEBL were 3,600 and 2,580 days, respectively, which
indicated that the overexpression of NEBL in patients with CRC was
significantly associated with a positive prognosis [P=0.019; hazard
ratio (HR), 0.75; 95% confidence interval (CI), 0.59–0.95; Fig. 5A]. Additionally, the prognosis of
NEBL was associated with age and gender (Table III). However, the expression of
C1QL1 was not associated with the prognosis of CRC (P=0.504; HR,
1.6; 95% CI, 0.41–6.29; Fig.
5B).
 | Table III.Overexpression of NEBL and C1QL1
associated with the prognosis of colorectal cancer using PROGgene
version 2.0 and GSE41258 dataset. |
Table III.
Overexpression of NEBL and C1QL1
associated with the prognosis of colorectal cancer using PROGgene
version 2.0 and GSE41258 dataset.
| Survival
modelsa | NEBL | P-value | C1QL1 | P-value |
|---|
| Overall | 0.75
(0.59–0.95) | 0.02 | 1.60
(0.41–6.29) | 0.50 |
| After adjustment
for age | 0.75
(0.59–0.95) | 0.02 | 1.91
(0.47–7.69) | 0.36 |
| After adjustment
for gender | 0.76
(0.59–0.97) | 0.03 | 1.32
(0.33–5.24) | 0.70 |
| After adjustment
for clinical stage | 1.08
(0.84–1.38) | 0.54 | 0.65
(0.15–2.92) | 0.58 |
| After adjustment
for TMN stage | 0.87
(0.68–1.12) | 0.29 | 0.96
(0.18–4.99) | 0.96 |
Discussion
CRC is one of the most frequent malignant tumors
worldwide (1). The overall
survival of patients with CRC remains poor, primarily due to the
late diagnosis and metastasis (22). There are a few prognostic
biomarkers identified at present that may be applied successfully
in clinical practice; therefore, it is essential to identify novel
potential biomarkers of prognosis in patients with CRC in order to
facilitate early intervention.
The NEBL gene was previously reported to be highly
expressed in cardiac muscle, regulating the attachment and
migration of extracellular matrix (23). It was primarily identified as a
novel mixed lineage leukemia fusion partner gene in an infant that
suffered from acute myeloid leukemia (24). Additionally, NEBL has previously
been demonstrated to have a significant effect on the regulation of
cardiac function (25),
concurrently its mutation may lead to various cardiomyopathies
(26). NEBL was also identified to
be associated with foot processes in podocyte injury and osteogenic
abilities (27,28). The aforementioned validated
biological processes of NEBL were in accordance with the findings
of the GO analysis performed in the present study. However,
information regarding the expression and role of NEBL in tumors,
particularly in CRC is insufficient. The present study revealed
that NEBL was highly expressed in CRC tissues and cell lines. It is
of note that the present study observed that higher NEBL expression
was associated with improved patient prognosis. A prognostic
biomarker refers to a measurable and quantifiable method that is
closely associated with clinical outcomes in the certain context of
patients. Therefore, the findings of the current study suggested
that NEBL overexpression may be a prognostic biomarker for CRC.
However, although NEBL was highly expressed in CRC, its effect on
clinical phenotypes of CRC remains to be elucidated. Instead of
acting as an oncogene, it may have an important role in combating
oncogenes involved in CRC progression, and once a tumor develops it
may be overexpressed in patients with CRC, thus contributing to an
improved prognosis for CRC.
The C1QL1 gene was originally cloned as a
senescence-associated gene that was highly expressed in the brain,
which may also have a role in neuronal differentiation (29). It has been previously demonstrated
that C1QL1 specifically binds to its receptor, the adhesion G
protein-coupled receptor 3, controlling the stereotyped pattern of
connectivity and regulating maturation of synapses between climbing
fibers and Purkinje cells (30,31).
However, its expression and role in CRC have remain to be
investigated. To the best of our knowledge the present study was
the first to demonstrate that C1QL1 was upregulated in CRC;
however, it was unable to identify a significant correlation
between C1QL1 expression and the prognosis of patients with
CRC.
NEBL and C1QL1 have been investigated in various
other diseases in the recent years (25,26,31).
The present study identified NEBL and C1QL1 to be involved in CRC
using microarray analysis, where both DEGs were overexpressed in
CRC tissues. Additionally, the experimental findings revealed were
consistent among CRC cell lines and tissues and confirmed the
outcomes of bioinformatics analysis. However, several limitations
of the present study should be mentioned. Firstly, a larger sample
size of CRC tissues with survival time are required in order to
confirm the current conclusions. In addition, the biological
function of the two hub genes remains unknown, and the relationship
between them and the tumor malignant phenotype should be evaluated
in future studies.
In conclusion, numerous DEGs including NEBL and
C1QL1 were identified in CRC tissues, NEBL was overexpressed in CRC
tissues and may be used as a prognostic biomarker for patients with
CRC.
Acknowledgements
The present study was supported by a research grant
from the National Natural Science Foundation of China (grant no.
81472735).
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
DEG
|
differentially expressed gene
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vogelstein B, Fearon ER, Hamilton SR, Kern
SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM and Bos
JL: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rahmani F, Avan A, Hashemy SI and
Hassanian SM: Role of Wnt/β-catenin signaling regulatory microRNAs
in the pathogenesis of colorectal cancer. J Cell Physiol.
233:811–817. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Louis P, Hold GL and Flint HJ: The gut
microbiota, bacterial metabolites and colorectal cancer. Nat Rev
Microbiol. 12:661–672. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakayama T, Hirakawa H, Shibata K, Nazneen
A, Abe K, Nagayasu T and Taguchi T: Expression of angiopoietin-like
4 (ANGPTL4) in human colorectal cancer: ANGPTL4 promotes venous
invasion and distant metastasis. Oncol Rep. 25:929–935. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gellad ZF and Provenzale D: Colorectal
cancer: National and international perspective on the burden of
disease and public health impact. Gastroenterology. 138:2177–2190.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmed FE: miRNA as markers for the
diagnostic screening of colon cancer. Expert Rev Anticancer Ther.
14:463–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rawson JB and Bapat B: Epigenetic
biomarkers in colorectal cancer diagnostics. Expert Rev Mol Diagn.
12:499–509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Walther A, Johnstone E, Swanton C, Midgley
R, Tomlinson I and Kerr D: Genetic prognostic and predictive
markers in colorectal cancer. Nat Rev Cancer. 9:489–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Verdaguer H, Saurí T and Macarulla T:
Predictive and prognostic biomarkers in personalized
gastrointestinal cancer treatment. J Gastrointest Oncol. 8:405–417.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van der Stok EP, Spaander MCW, Grünhagen
DJ, Verhoef C and Kuipers EJ: Surveillance after curative treatment
for colorectal cancer. Nat Rev Clin Oncol. 14:297–315. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Malapell U, Mayo de-Las-Casas C, Rocco D,
Garzon M, Pisapia P, Jordana-Ariza N, Russo M, Sgariglia R, De Luca
C, Pepe F, et al: Development of a gene panel for next-generation
sequencing of clinically relevant mutations in cell-free DNA from
cancer patients. Br J Cancer. 116:802–810. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Broderick P, Carvajal-Carmona L, Pittman
AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K,
Fielding S, et al: A genome-wide association study shows that
common alleles of SMAD7 influence colorectal cancer risk. Nat
Genet. 39:1315–1317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Snezhkina AV, Krasnov GS, Zaretsky AR,
Zhavoronkov A, Nyushko KM, Moskalev AA, Karpova IY, Afremova AI,
Lipatova AV, Kochetkov DV, et al: Differential expression of
alternatively spliced transcripts related to energy metabolism in
colorectal cancer. BMC Genomics. 17 Suppl 14:10112016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheffer M, Bacolod MD, Zuk O, Giardina SF,
Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA and Domany E:
Association of survival and disease progression with chromosomal
instability: A genomic exploration of colorectal cancer. Proc Natl
Acad Sci USA. 106:pp. 7131–7136. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hari DM, Leung AM, Lee JH, Sim MS, Vuong
B, Chiu CG and Bilchik AJ: AJCC cancer staging manual 7th edition
criteria for colon cancer: Do the complex modifications improve
prognostic assessment? J Am Coll Surg. 217:181–190. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(sT)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goswami CP and Nakshatri H: PROGgeneV2:
Enhancements on the existing database. BMC Cancer. 14:9702014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Keane MG and Johnson GJ: Early diagnosis
improves survival in colorectal cancer. Practitioner. 256:15–18, 2.
2012.PubMed/NCBI
|
|
23
|
Lee EJ, De Winter JM, Buck D, Jasper JR,
Malik FI, Labeit S, Ottenheijm CA and Granzier H: Fast skeletal
muscle troponin activation increases force of mouse fast skeletal
muscle and ameliorates weakness due to nebulin-deficiency. PLoS
One. 8:e558612013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cóser VM, Meyer C, Basegio R, Menezes J,
Marschalek R and Pombo-de-Oliveira MS: Nebulette is the second
member of the nebulin family fused to the MLL gene in infant
leukemia. Cancer Genet Cytogenet. 198:151–154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mastrototaro G, Liang X, Li X, Carullo P,
Piroddi N, Tesi C, Gu Y, Dalton ND, Peterson KL, Poggesi C, et al:
Nebulette knockout mice have normal cardiac function, but show
Z-line widening and up-regulation of cardiac stress markers.
Cardiovasc Res. 107:216–225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Perrot A, Tomasov P, Villard E, Faludi R,
Melacini P, Lossie J, Lohmann N, Richard P, De Bortoli M, Angelini
A, et al: Mutations in NEBL encoding the cardiac Z-disk protein
nebulette are associated with various cardiomyopathies. Arch Med
Sci. 12:263–278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miao J, Fan Q, Cui Q, Zhang H, Chen L,
Wang S, Guan N, Guan Y and Ding J: Newly identified cytoskeletal
components are associated with dynamic changes of podocyte foot
processes. Nephrol Dial Transplant. 24:3297–3305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aino M, Nishida E, Fujieda Y, Orimoto A,
Mitani A, Noguchi T, Makino H, Murakami S, Umezawa A, Yoneda T and
Saito M: Isolation and characterization of the human immature
osteoblast culture system from the alveolar bones of aged donors
for bone regeneration therapy. Expert Opin Biol Ther. 14:1731–1744.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bérubé NG, Swanson XH, Bertram MJ, Kittle
JD, Didenko V, Baskin DS, Smith JR and Pereira-Smith OM: Cloning
and characterization of CRF, a novel C1q-related factor, expressed
in areas of the brain involved in motor function. Brain Res Mol
Brain Res. 63:233–240. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuzaki M: The C1q complement family of
synaptic organizers: Not just complementary. Curr Opin Neurobiol.
45:9–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sigoillot SM, Iyer K, Binda F,
González-Calvo I, Talleur M, Vodjdani G, Isope P and Selimi F: The
secreted protein C1QL1 and its receptor bai3 control the synaptic
connectivity of excitatory inputs converging on cerebellar Purkinje
cells. Cell Rep. pii:S2211–1247, 00059-100065. 2015.
|