Introduction
Enamel is an oral epithelial-derived hard tissue
(1). During enamel development,
ameloblasts synthesize and secrete a tissue-specific extracellular
matrix that facilitates the initiation and orientation of
hydroxyapatite crystallites (2,3).
Amelotin (Amtn) is a recently identified enamel matrix protein that
is expressed and secreted predominantly by ameloblasts during the
transition and maturation stages (4). Amtn knockout mice have
hypomineralized enamel (5) and, in
transgenic mice, overexpression of Amtn disrupts the enamel
microstructure, causing biomineralization defects (6). To understand the role of Amtn in
amelogenesis, it is essential to study the molecular regulatory
mechanisms of Amtn gene expression.
Runt-related transcription factor 2 (Runx2) is an
important molecule in bone and tooth development (7,8). In
mice, targeted disruption of Runx2 results in cessation of
bone and tooth development (9). In
humans, Runx2 mutation causes cleidocranial dysplasia, a
genetic bone disorder (10).
Dental abnormalities, including supernumerary teeth, are also
associated with cleidocranial dysplasia caused by Runx2
mutations (11). It has been
demonstrated that Runx2 binds to the consensus sequences in a
number of gene promoters, including Bone Sialoprotein,
Osteocalcin, Dentin Sialophosphoprotein and Ameloblastin
to regulate the expression of target genes (12,13).
Runx2 is a dimeric transcription complex comprising α unit (Runx2)
DNA binding and a stabilizing core-binding factor β subunit (Cbfβ).
Cbfβ acts as a binding partner for all Runx proteins and
conditional Cbfβ knockout mice exhibit impaired enamel
formation (14).
The results of the present study suggest that Runx2
protein is upregulated in ameloblasts from the transition stage to
the maturation stage. Runx2 specifically regulates Amtn gene
expression in ameloblast lineage cells (ALCs) by interacting with
two functional regions in the mouse Amtn promoter. Results
of electrophoretic mobility shift (EMSA), chromatin
immunoprecipitation (ChIP) and luciferase reporter assays revealed
that Runx2 is associated with the transcription activity of the
Amtn gene in mice via Runx2 binding sites.
Materials and methods
Immunolocalization of Runx2 in mouse
mandibles
A total of 30 Kunming strain male mice (2–5 g; 5, 7
and 10 days old; provided by the Laboratory Animal Center, School
of Medicine, Shandong University, Jinan, China) were housed under
controlled conditions (22±1°C; 50–60% humidity; 12 hour light-dark
cycle). All protocols involving mice were reviewed and approved by
the Ethical Committee of the Institute of Zoology, Chinese Academy
of Sciences (Beijing, China). Mandibles were obtained from mice on
postnatal days (PND) 5, 7 and 10 (10 mice/group). The dissected
mandibles were fixed with 4% paraformaldehyde in PBS for 20 h at
4°C and decalcified at 4°C in a 10% (w/v) Na2-EDTA
solution (pH 7.0) for 10 days. Paraffin samples were prepared as
previously described (15). For
immunohistochemistry, sections were incubated overnight with rabbit
polyclonal antibody against Runx2 (ab23981; 1:300; Abcam,
Cambridge, MA, USA), or Cbfβ (ab72696; 1:300 dilution; Abcam) at
4°C. Antibody binding was detected using the vectastain ABC Elite
kit and a peroxidase substrate kit (both Vector Laboratories, Inc.,
Burlingame, CA, USA). For the control group, the primary anti-Runx2
antibody was replaced with non-immune rabbit IgG (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), and the nuclei were
stained with hematoxylin for 2 min at room temperature.
Plasmid construction
Full-length mouse Runx2 cDNA and Cbfβ cDNA were
amplified by polymerase chain reaction (PCR) using DNA polymerase
(Takara Biotechnology Co., Ltd., Dalian, China) from mouse ALCs
(provided by Dr Toshihiro Sugiyama, Akita University, Akita,
Japan). The following thermocycling conditions were used for the
PCR: Initial denaturation at 94°C for 10 min; 40 cycles of 94°C for
15 sec, 60°C for 30 sec and 72°C for 1 min; and a final extension
at 72°C for 10 min. The following primer sequences were used: Runx2
forward, 5′-AAGGATCCTGATGCGTATTCCTGTAG-3′ and reverse,
5′-AATCTAGAATATGGCCGCCAAACAGA-3′; Cbfβ forward,
5′-GGCGCGGCCTGAGGGCGGGAAGA-3′ and reverse,
5′-CGTTAAGTGGAGCACAGCTTATG-3′. Amplification products were cloned
into a eukaryotic expression vector pcDNA3.1(+) (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The mouse Amtn promoter
region of −1463/+196 was amplified by PCR using DNA polymerase
(Takara Biotechnology Co., Ltd.) with mouse genomic DNA as a
template. The reaction conditions of PCR were the same as above.
The following primer sequences were used for amplification of the
−1463/+196 region, forward, 5′-CCGGTACCCAGTGTAGTATGTCATTCT-3′ and
reverse, 5′-GCCGAGATCTGTTTCGATTTGCTACCT-3′. The amplification
product was cloned into pGL3-basic reporter vector (Promega
Corporation, Madison, WI, USA) to generate pGL3-Amtn (pWT). The
region between bp −1463 and +196 of mouse Amtn promoter was
analyzed using the online software JASPAR (http://jaspar.genereg.net/) to predict potential
binding sites (16). Two putative
Runx2 binding sites, AACCACT (site1: −1342/-1336) and AACCAAA
(site2: −98/-92) were identified. The mutated Amtn promoter-driven
luciferase reporter vectors pGL3-Amtn-mut1 (pmut1), pGL3-Amtn-mut2
(pmut2), and pGL3-Amtn-mut1+2 (pmut1+2) were created using a
site-directed mutagenesis kit (Takara Biotechnology Co., Ltd.). The
sequences of all promoter constructs were verified by DNA
sequencing analyses which were carried out by Takara Biotechnology
Co., Ltd.
DNA transfection
Immortalized mouse ALCs were cultured as previously
described (17,18). All transfections were performed in 6-well
plates. For Runx2 small interfering (si)RNA experiments, ALCs were
transfected with 60 pmol Runx2 siRNA and control siRNA (sc-37146
and sc-37007; Santa Cruz Biotechnology, Inc.), respectively, using
Lipofectamine® RNAi MAX (Invitrogen; Thermo Fisher
Scientific, Inc.). Cultures were maintained for 36 h at 37°C and
cells were collected for reverse transcription-quantitative (RT-q)
PCR and western blot analyses. For Runx2 overexpression
experiments, ALCs were transfected with pcDNA3.1(+), pcDNA3.1-Runx2
(pRunx2), pcDNA3.1-Cbfβ (pCbfβ) or pRunx2 + pCbfβ using
Lipofectamine® Plus (Invitrogen; Thermo Fisher
Scientific, Inc.). At 36 h post-transfection, the cells were
collected for RT-qPCR.
RT-qPCR
Total RNA from mouse ALCs was extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed
using the MMLV-RT system (Promega Corporation) for 15 min at 37°C.
The sequences of the primers used in qPCR are listed in Table I. Amplification was performed for
using the SYBR® PrimeScript® kit (Takara
Biotechnology Co., Ltd.) under the following thermocycling
conditions: Initial denaturation 95°C for 30 sec; 40 cycles of 95°C
for 5 sec, 60°C for 30 sec. GAPDH was used as an internal control.
Relative gene expression levels were determined using the
2−ΔΔCq method (19).
Each experiment was performed in triplicate and repeated three
times.
 | Table I.Primers used in RT-qPCR and ChIP. |
Table I.
Primers used in RT-qPCR and ChIP.
| Name | Direction | Primer sequence (5′
to 3′) | Experiment |
|---|
| Amtn | Forward |
CAAATCAGGTTTTTCCTTCCATAA | RT-qPCR |
|
| Reverse |
CCAGGGTGAACGGGAGAGTA |
|
| Klk4 | Forward |
GTCAGCAGCCGGATCATACAAGG | RT-qPCR |
|
| Reverse |
GCACCAAGACTCCCGAGCAGAAA |
|
| ALP | Forward |
AACCTGACTGACCCTTCGC | RT-qPCR |
|
| Reverse |
CAATCCTGCCTCCTTCCAC |
|
| GAPDH | Forward |
TGTGTCCGTCGTGGATCTGA | RT-qPCR |
|
| Reverse |
TTGCTGTTGAAGTCGCAGGAG |
|
| Runx2 (1) | Forward |
TTCTCCATCAGTGTCCACCTT | ChIP |
|
| Reverse |
TGGGGTCACTGTCACTCATAA |
|
| Runx2 (2) | Forward |
ATACCCTCTAGTCAGATCG | ChIP |
|
| Reverse |
GCTGATTTGTGTTTGTAG |
|
Western blot analyses
At 36 h post-transfection, ALCs were collected and
lysed in radioimmunoprecipitation assay buffer (Sigma Aldrich;
Merck KGaA, Darmstadt, Germany) and lysates were centrifuged at
10,000 × g for 7 min at 4°C. The supernatant was collected and
total proteins were quantified using a bicinchoninic acid protein
assay (Pierce; Thermo Fisher Scientific, Inc.). A total of 30 µg
protein samples were separated for each sample by 12% SDS-PAGE and
transferred onto polyvinylidene fluoride membranes. Following
blocking in 5% bovine serum for 2 h at 20°C, membranes were
incubated with anti-Runx2 antibody (cat. no. ab23981; Abcam,
diluted at 1:1,000) for 1 h at 37°C. Membranes were subsequently
incubated with horseradish peroxidase-conjugated anti-rabbit
antiserum (cat. no. A0545, Sigma Aldrich; Merck KGaA; 1:2,000) for
30 min at 37°C. β-actin antibody (N-21; 1:1,500; Santa Cruz
Biotechnology, Inc.) was used as an internal control. Blots were
developed by using an Amersham enhanced chemiluminescence kit (GE
Healthcare Life Sciences, Little Chalfont, UK). The experiment was
performed three times.
Nuclear protein extraction and
EMSA
293T cells (Wuhan Boster Biological Technology,
Ltd., Wuhan, China) were seeded in 60 mm plates at a density of
0.3×105 cells/cm2. Following 16 h culture,
cells were transfected with 10 µg pcDNA3.1(+), 5 µg pRunx2 plus 5
µg pcDNA3.1(+) or 5 µg pRunx2 plus 5 µg pCbfβ. After 24 h, nuclear
proteins from transfected cells were isolated using a nuclear
protein extraction kit (Pierce; Thermo Fisher Scientific, Inc.).
Protein concentration of the nuclear extracts was determined using
a bicinchoninic acid assay. EMSA was performed using a lightshift
chemiluminescent EMSA kit (Pierce; Thermo Fisher Scientific, Inc.).
Probes were labeled using the Biotin 3′ end DNA labeling kit
(Pierce; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and incubated with nuclear protein extracts
in the binding buffer from the EMSA kit for 20 min at room
temperature. For the competition assay, nuclear proteins were
pre-incubated with a 100-fold molar excess of unlabeled probe or
mutation probe in the binding buffer. For the supershift
experiments, 10 µg of anti-Runx2 antibody or normal rabbit IgG was
incubated with nuclear extracts for 30 min at room temperature
prior to the binding reaction. DNA-protein complexes were resolved
in 5% native polyacrylamide gels, transferred onto Nylon
N+ membranes, and cross-linked for the detection of
biotin-labeled DNA using chemiluminescence. Each experiment was
repeated three times.
ChIP
ChIP analysis was performed using the EZ-ChIP kit
(EMD Millipore, Billerica, MA, USA). The ALCs were seeded at a
density of 0.7×105 cells/cm2 in a 150
mm2 dish and cultured for 24 h at 37°C. Proteins and DNA
were cross-linked with 1% formaldehyde for 10 min at room
temperature. Cross-linking was stopped by the addition of glycine.
Cell pellets were resuspended in 1 ml of 1% SDS lysis buffer
(Beyotime Institute of Biotechnology, Haimen, China) containing
protease inhibitors and fragmented using a sonic dismembrator
(Soniprep 150, Sanyo, London, UK). For immunoprecipitation,
antibodies against Runx2 or normal rabbit IgG (negative control)
were used. The precipitated and input DNAs were subjected to PCR
with two pairs of primers both specific to Runx2 listed in Table I, which generates 165-bp and 179-bp
DNA fragments. The reactions were performed under the following
conditions: 94°C for 1 min; 40 cycles of 94°C for 15 sec, 56°C for
30 sec and 72°C for 15 sec; and a final extension at 72°C for 10
min. PCR products were visualized on a 2% agarose gel following
electrophoresis and analyzed by Matlab 8.6 software (MathWorks,
Inc., Natick, MA, USA). Each experiment was performed three
times.
Luciferase reporter assay
ALCs were cultured in 24-well plates at a density of
1×105 cells/cm2 for 16 h at 37°C, following
which cells were transfected with 400 ng/well of the indicated
reporter plasmids plus 50 ng of pRL-TK. In all transfection
experiments, the amount of plasmid DNAs was normalized as necessary
with the pcDNA3.1(+) expression plasmid so that the total DNA was
constant in each group. Following 30 h transfection, cells were
harvested and luciferase activity was measured using a Luciferase
Assay Reagent (Promega Corporation). To obtain the relative
luciferase activity, luciferase activity values were divided by
Renilla luciferase activity values. Each experiment was performed
in triplicate and repeated three times.
Statistical analysis
Data were analyzed using two-way analysis of
variance and are expressed as the mean ± standard deviation. The
statistical differences between two groups were evaluated using
Student's t test. Dunnett's test for multiple comparisons was
applied when the overall F test result was significant. P<0.05
was considered to indicate a statistically significant
difference.
Results
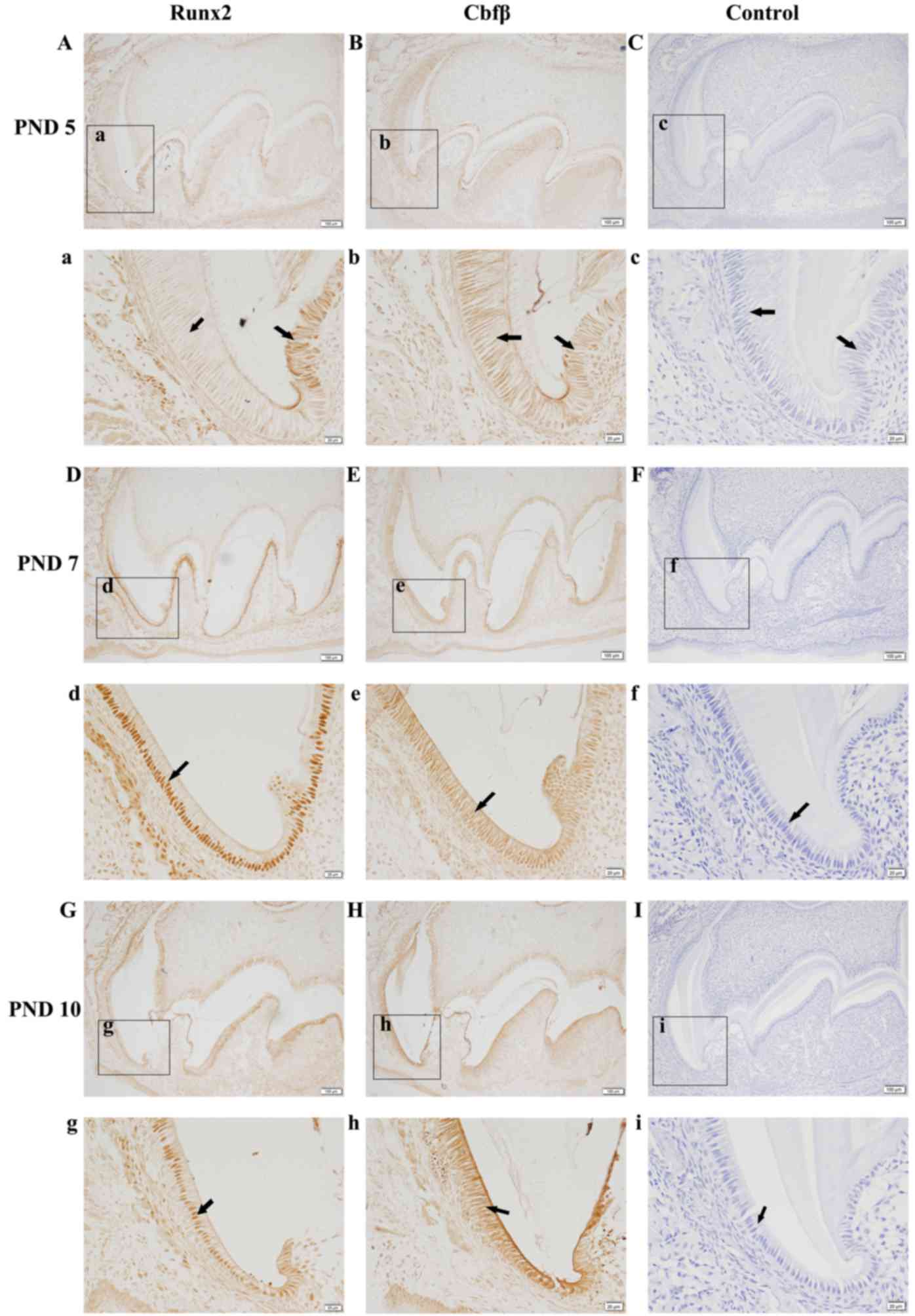
Runx2 and Cbfβ localize in ameloblasts
during amelogenesis
A detailed immunohistochemistry analysis of Runx2
expression in the first mandibular molars revealed that Runx2 is
expressed in ameloblasts in a stage-dependent manner during
amelogenesis (Fig. 1). Strong
staining was also observed in osteoblasts in bone tissue
surrounding the enamel organ. In the late secretory stage, a faint
Runx2 expression was observed in the nucleus of ameloblasts
(Fig. 1Aa). The signal increased
when ameloblasts progressed to the transition stage (Fig. 1Dd) and continued to be observed at
the maturation stage (Fig. 1Gg).
Runx2 is known to function by forming a heterodimer with Cbfβ
(20), so Cbfβ expression was also
investigated using immunohistochemistry. Strong staining for Cbfβ
was observed in ameloblasts at the secretory stage and was evenly
distributed in the nucleus and cytoplasm of first mandibular molars
from PND 5 mice (Fig. 1Bb). With
enamel development, Cbfβ protein accumulated in the nucleus of
ameloblasts at the transition (Fig.
1Ee) and maturation stages (Fig.
1Hh). These results suggest that Runx2 and Cbfβ are
co-localized in the nucleus of ameloblasts from the late secretory
stage to the maturation stage. In the control section of incisors,
no positive signal was observed (Fig.
1C, F and I).
Runx2 regulates Amtn expression in
ALCs
During tooth development, Kallikrein-related
peptidase 4 (Klk4) and alkaline phosphatase (ALP) are predominantly
expressed during the late stages of enamel formation (21,22).
Therefore, the expression of Amtn, Klk4, and ALP in
ALCs by was examined using RT-qPCR. When ALCs were treated with
siRNA targeting Runx2, the expression of Runx2 was
significantly decreased at the protein level as determined by
western blotting (Fig. 2A). A
significant reduction in Amtn mRNA expression was observed, whereas
the expression of Klk4 and Alp was not affected by siRNA targeting
Runx2 (Fig. 2B). These data
suggest that Runx2 may participate in Amtn gene expression.
Next, the effect of overexpressed Runx2 on Amtn gene
expression in ALCs was investigated. As shown in Fig. 2C, Runx2 overexpression alone had no
significant effect on Amtn gene expression, whereas
Amtn gene expression was upregulated by Cbfβ overexpression
in a dose-dependent manner. Combined treatment with overexpressed
Runx2 and Cbfβ further elevated Amtn expression in ALCs
compared with overexpressed Cbfβ alone. These results suggest that
Cbfβ is essential for Runx2-induced Amtn gene expression in
ALCs.
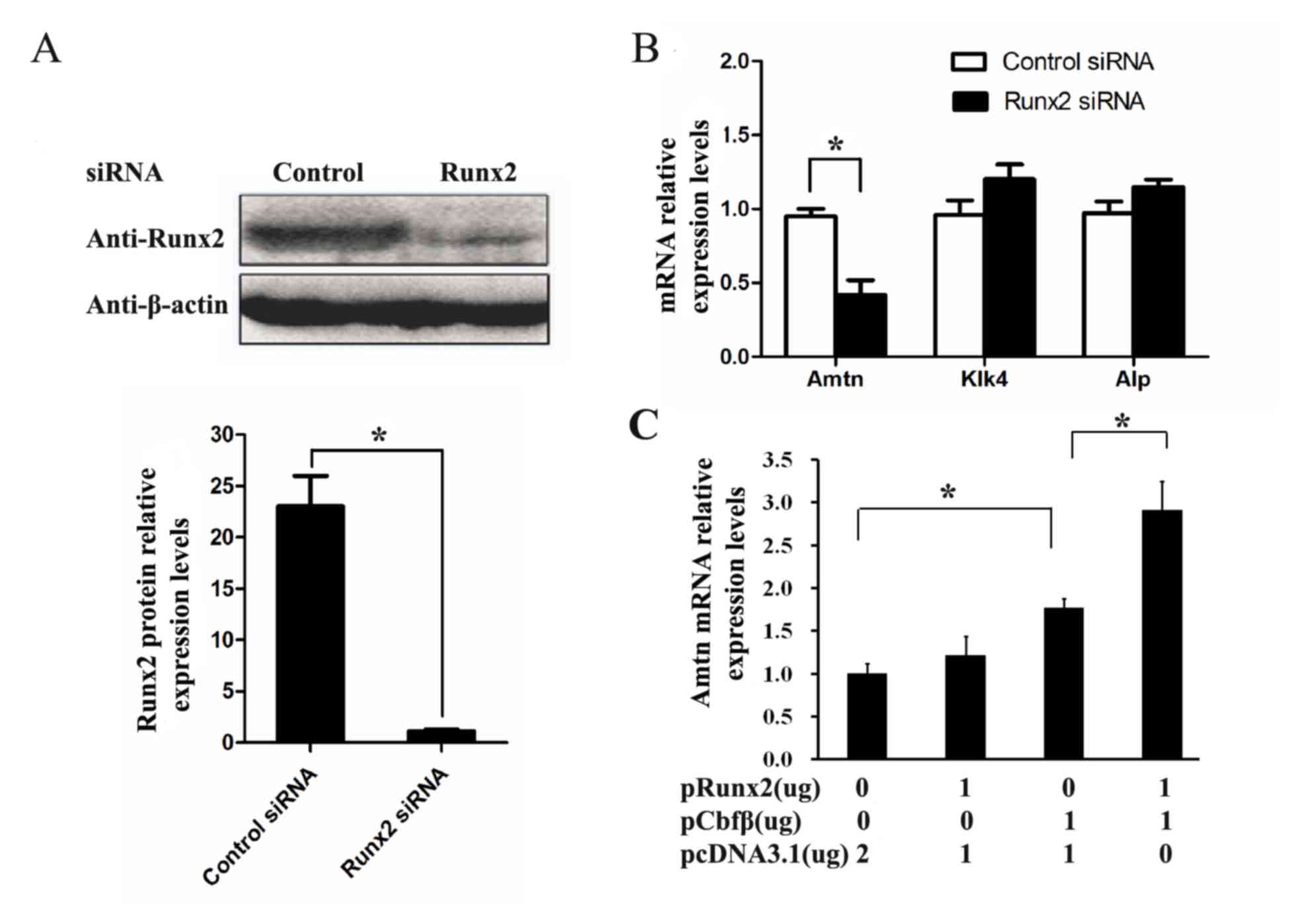 | Figure 2.The regulation of Runx2 on Amtn gene
expression in the ALCs. ALCs were transfected with Runx2 siRNAs or
control siRNAs for 36 h. (A) The expression level of Runx2 was
significantly decreased at the level of protein as determined by
western blotting. β-actin was used as a loading control. (B) The
mRNA levels of Amtn, Klk4, and Alp were analyzed by
RT-qPCR. (C) ALCs were transfected with 1 µg pRunx2, 1 µg pCbfβ, or
1 µg pRunx2 plus 1 µg pCbfβ. After 36 h transfection, the mRNA
level of Amtn gene was analyzed by RT-qPCR. *P<0.05.
Runx2, runt-related transcription factor 2; Amtn, amelotin; ALCs,
ameloblast lineage cells; siRNA, small interfering RNA; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction. |
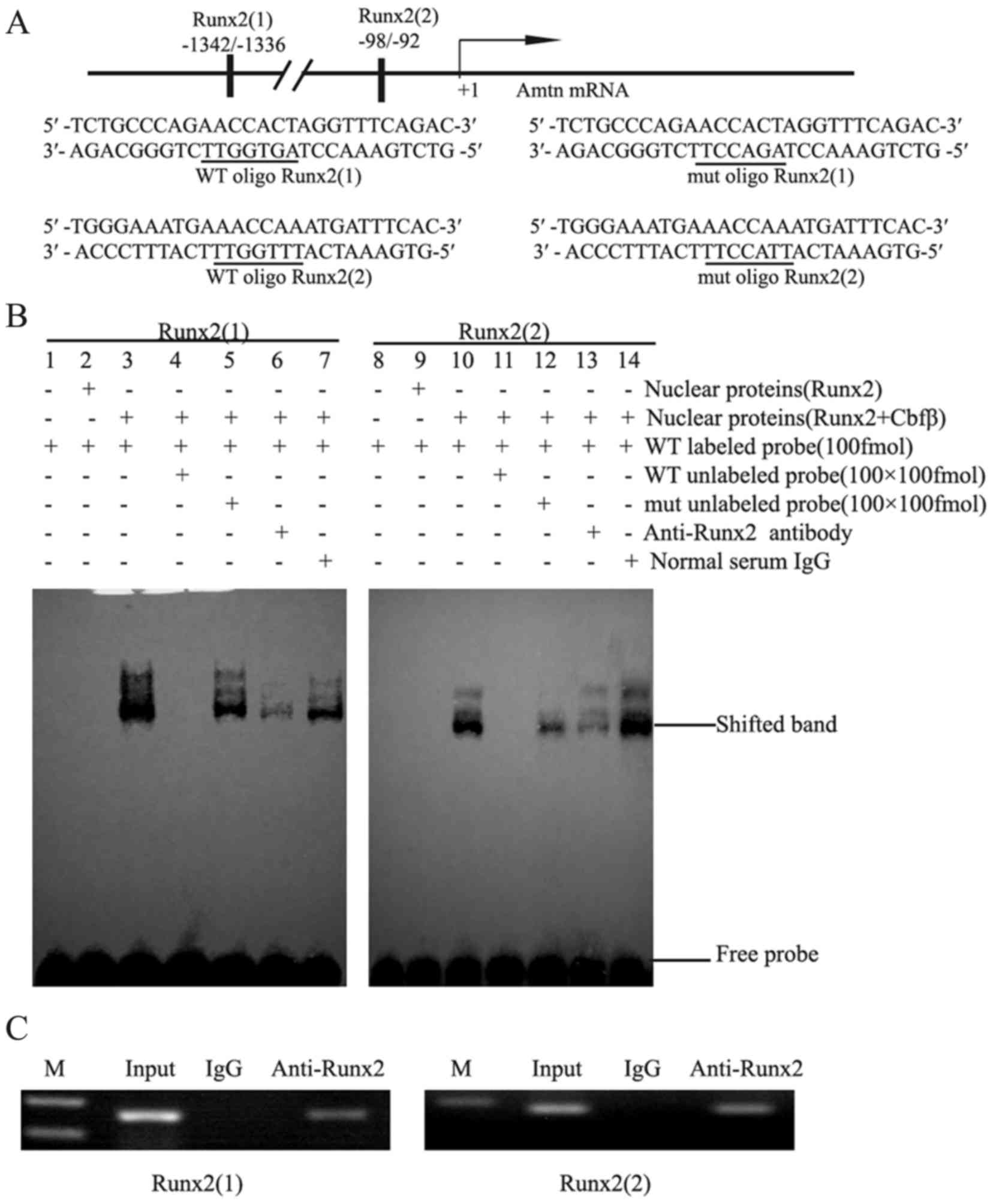
Runx2 binds to the putative Runx2
binding sites in the presence of Cbfβ
To assess whether Runx2 activates Amtn
expression by binding to Runx2 sites. Two putative Runx2 binding
sites, AACCACT (site1: −1342/-1336) and AACCAAA (site2: −98/-92),
were identified and underlined (Fig.
3A). EMSA was performed using nuclear extracts from 293T cells
and probes corresponding to the two putative Runx2 binding
sequences. As negative controls, the nuclear extracts from 293T
cells transfected with pcDNA3.1(+) did not bind to the
oligonucleotide (lanes 1 and 8, Fig.
3B). No oligonucleotide binding was observed for nuclear
extracts from the cells transfected with pcDNA3.1 -Runx2 (lanes 2
and 9, Fig. 3B), whereas the
nuclear extracts from 293T cells cotransfected with Runx2
and Cbfβ plasmids exhibited strong binding to the
oligonucleotide (lanes 3 and 10, Fig.
3B). Binding specificity was confirmed by preincubating nuclear
extracts with a 100-fold excess of unlabeled wild type and mutant
oligonucleotides. As presented in Fig.
3B, the DNA-Runx2/Cbfβ complex was strongly inhibited by the
addition of a 100-fold molar excess of unlabeled wild type probes
(lanes 4 and 11), but only partially inhibited by a 100-fold molar
excess of unlabeled mutated probes (lanes 5 and 12). The
DNA-Runx2/Cbfβ complex was further verified using anti-Runx2
antibody (lanes 6 and 13, Fig.
3B), but not by normal IgG (lanes 7 and 14, Fig. 3B). These results suggest that the
Runx2/Cbfβ complex may specifically bind to the Amtn
promoter to regulate Amtn gene transcription.
Runx2 binds to the endogenous Amtn
promoter in vivo
To confirm that Runx2 binds to the Amtn
promoter, an in vivo ChIP assay was performed (Fig. 3C). When the endogenous Amtn
gene chromatin from ALCs was randomly fragmented by sonication,
antibodies against Runx2 was able to precipitate down the
Amtn promoter sequences, in which a 165-bp fragment
containing Runx2 site 1 and a 179-bp fragment containing Runx2 site
2 were amplified by PCR (Fig. 3C).
As a negative control, normal IgG failed to precipitate the
promoter sequences. As a loading control, the corresponding 165-bp
and 179-bp fragments were also identified in isolated genomic DNA
inputs. These results suggest that two Runx2 binding sites in the
Amtn promoter may serve essential roles in mediating
Amtn transcription in ALCs.
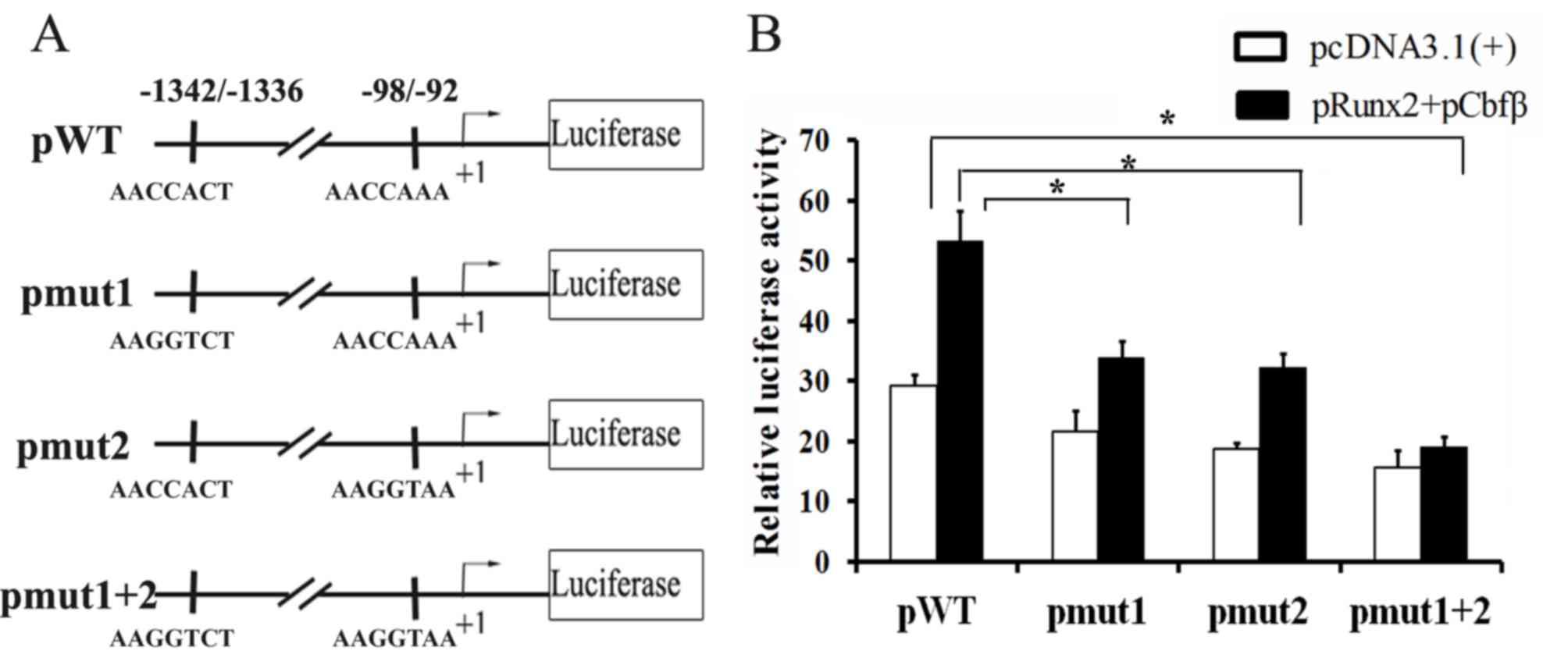
Runx2 activates Amtn promoter activity
via Runx2 binding sites in the presence of Cbfβ
To further confirm that Runx2 regulates Amtn
expression via the Runx2 binding sites, the −1463 bp to +196 bp
region was cloned into the pGL3-basic vector (pGL3-Amtn). Two
putative Runx2 binding sites in pGL3-Amtn (pWT) were mutated to
create pGL3-Amtn-mut1 (pmut1), pGL3-Amtn-mut2 (pmut2), and
pGL3-Amtn-mut1+2 (pmut1+2) (Fig.
4A). These constructs were transfected into ALCs and the
promoter activities were analyzed in the presence or absence of
overexpressed Runx2 and Cbfβ. To investigate Runx2/Cbfβ complex on
Amtn promoter activity, a luciferase reporter assay was
performed. The effects of Runx2/Cbfβ complex on the mutant
Amtn promoter activity were observed. Mutation of either
Runx2 site within the Amtn promoter caused statistically
significant changes in promoter activity (Fig. 4B). Compared with pWT, the basal
promoter activity of pmut1 or pmut2 was lower in the absence of
Runx2/Cbfβ overexpression and the promoter activity was further
decreased following mutation of both sites. Co-transfection with
Runx2 and Cbfβ significantly enhanced the promoter activity of pWT,
but only partially activated the promoter activity of pmut1 or
pmut2. Furthermore, mutation of the two Runx2 binding sites
completely eradicated the promoter activity in the presence of
Runx2 and Cbfβ. These results suggest that Runx2/Cbfβ complex
regulates Amtn gene expression via Runx2 binding sites in
the Amtn promoter.
Discussion
The present study investigated the regulatory
mechanism of Runx2 on Amtn gene expression during
amelogenesis and the following results were obtained: i) Runx2 is
predominantly expressed in the nuclei of ameloblasts at the
transition and maturation stages; ii) Runx2 and Cbfβ are
co-localized in the nucleus of ameloblasts at the transition and
maturation stages; iii) Runx2 regulates Amtn but not
Klk4 and ALP gene expression; iv) Runx2 binds to
specific motifs in the Amtn promoter in the presence of
Cbfβ; and, v) Mutation of the Runx2 binding sites decreased or
eliminated the activation of Amtn promoter activity by the
Runx/Cbfβ complex.
Runx2 is essential in the early stages of bone and
tooth development. In addition to the skeletal defects in
Runx2 knockout mice, developing teeth fail to advance beyond
the bud stage (23). Elevated
Runx2 expression was detected in the ameloblasts at the late
stages of enamel development, which suggests that Runx2 may serve a
role in enamel maturation. The results of the present study are
consistent with a previous report that Runx2 is elevated at the
late stage of enamel development (24). Runx2 is a master transcription
factor of bone and serves a role in all stages of bone formation
(25). It is essential for the
initial commitment of mesenchymal cells to the osteoblastic lineage
and also controls the proliferation, differentiation and
maintenance of these cells (26).
Enamel and bone are mineralized tissues with similar developmental
processes (27,28). Enamel-related gene products (ERPs)
are detected in non-enamel tissues such as bone (29). It has been reported that individual
ERPs affect individual transcription factor (Runx2, Sp7, bone
sialoprotein and Msh homeobox 2) cascades to affect downstream
effects on osteoblast differentiation, mineralization and calvarial
bone development (29). In the
present study, Runx2 was highly expressed in ameloblasts and
osteoblasts, but weakly expressed in pre-odontoblasts, supporting
the critical role of Runx2 in enamel development. It has been
reported that Cbfβ, a partner protein of Runx2 transcription
factor, is expressed in the secretory stage of ameloblasts during
tooth development by in situ hybridization (14), which is consistent with the
findings of the current study that Cbfβ protein is expressed in
cytoplasm and nucleus of secretory ameloblasts as detected by
immunohistochemistry. It is well established that the Runx2/Cbfβ
complex in the nucleus serves an essential role in bone formation
(30). The present study indicated
that Cbfβ protein accumulates in the nucleus of ameloblasts at the
transition and maturation stages, which suggests that Runx2/Cbfβ
complexes may serve an important role in the later stage of enamel
development.
Amtn is a recently identified enamel matrix protein
(4,31). Due to the temporal-spatial
expression pattern of Amtn being similar to that of Runx2 in
late stage enamel development, it was hypothesized that Amtn
gene expression may be regulated by Runx2. This hypothesis was
verified using Runx2 knockdown, in which Runx2 siRNA
abrogated Amtn gene expression. The overexpression
experiment further confirmed that co-expression of Runx2 and Cbfβ
augments Amtn gene expression in the ALCs. These results may
explain the similar spatial-temporal expression pattern of
Amtn as that of Runx2 during amelogenesis. A notable
discovery is that Runx2 downregulates Amtn gene
transcription activity. Runx2 may act as either a positive
regulator or a repressor for expression of the targeted genes, and
Runx2 may downregulate expression of the targeted genes by
interacting with mothers against decapentaplegic homolog (Smad)3
via Smad binding site (32). It is
possible that Runx2 may interact with unknown factors binding to
the promoter to downregulate Amtn gene expression at the
late maturation stage, when Amtn expression evidently
decreases (4). The findings of the
present study suggest that the Amtn gene is specifically
upregulated by the Runx2/Cbfβ complex as a physiological
consequence of the late secretory and maturation stages.
Runx2 regulates gene expression by binding to the
consensus core sequences AACCACA in the promoter of target genes.
To evaluate the regulation of Amtn promoter activity by
Runx2, mouse Amtn promoter (−1463/+96) was analyzed and it
was identified that it contains two putative Runx2 binding sites,
AACCACT (−1342/-1336) and AACCAAA (−98/-92), which are similar to
the AACCACA sequences. The binding of Runx2 to the AACCACT element
was demonstrated using EMSA and ChIP assays, which is consistent
with previous reports that Runx2 binds to the AACCACT sequences in
the promoter of bone- and tooth-associated genes, including dentin
sialophosphoprotein, osteocalcin and collagenase 3 (33,34).
Another putative Runx2 binding site AACCAAA (−98/-92) also
demonstrated strong binding to Runx2. To the best of our knowledge,
this is a new identified sequence binding to Runx2. In the present
study, the Amtn promoter was trans-activated by Runx2 in a
sequence-specific manner, suggesting that these binding sites are
essential for activating the promoter activity of Amtn
gene.
In conclusion, the present study demonstrated that
the expression of mouse Amtn gene in amelogenesis is
mediated by Runx2/Cbfβ complex. Runx2/Cbfβ can bind to the two
Runx2 binding motifs AACCACT (−1342/-1336) and AACCAAA (−98/-92) in
the Amtn promoter and regulate Amtn gene expression.
The present study may contribute to elucidation of the mechanism of
tooth enamel development and prevention of clinical tooth enamel
disease.
Acknowledgements
The authors would like to thank Dr Toshihiro
Sugiyama (Akita University, Akita, Japan) for providing the
ameloblast lineage cells.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant nos. 81670954, 81170927
and 81441107) and the Natural Science Foundation of Shandong
Province (grant no. ZR2017MH103).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
YG and XL designed the experiments. XL, ZX, QC, CX
and YS performed the experiments. YW and LZ analyzed data. XL and
YG wrote the manuscript.
Ethics approval and consent to
participate
All protocols involving mice were reviewed and
approved by the Ethical Committee of the Institute of Zoology,
Chinese Academy of Sciences (Beijing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lumsden AG: Spatial organization of the
epithelium and the role of neural crest cells in the initiation of
the mammalian tooth germ. Development. 103 Suppl:S155–S169
|
|
2
|
Moradian-Oldak J: Protein-mediated enamel
mineralization. Front Biosci (Landmark Ed). 17:1996–2023. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ravindranath RM, Devarajan A and Uchida T:
Patiotemporal expression of ameloblastin isoforms during murine
tooth development. J Biol Chem. 282:36370–36376. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iwasaki K, Bajenova E, Somogyi-Ganss E,
Miller M, Nguyen V, Nourkeyhani H, Gao Y, Wendel M and Ganss B:
Amelotin-a novel secreted, ameloblast-specific protein. J Dent Res.
84:1127–1132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abbarin N, San Miguel S, Holcroft J,
Iwasaki K and Ganss B: The enamel protein amelotin is a promoter of
hydroxyapatite mineralization. J Bone Miner Res. 30:775–785. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lacruz RS, Nakayama Y, Holcroft J, Nguyen
V, Somogyi-Ganss E, Snead ML, White SN, Paine ML and Ganss B:
Targeted overexpression of amelotin disrupts the microstructure of
dental enamel. PLoS One. 7:e352002012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Komori T, Yagi H, Nomura S, Yamaguchi A,
Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH and Inada M:
Targeted disruption of Cbfa1 results in a complete lack of bone
formation owing to maturationalarrest of osteoblasts. Cell.
89:755–764. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Otto F, Thornell AP, Crompton T, Denzel A,
Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen
BR, et al: Cbfa1, a candidate gene for cleidocranial dysplasia
syndrome, is essential for osteoblastdifferentiation and bone
development. Cell. 89:765–771. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
D'Souza RN, Aberg T, Gaikwad J, Cavender
A, Owen M, Karsenty G and Thesleff I: Cbfa1 is required for
epithelial-mesenchymal interactions regulating tooth development in
mice. Development. 126:2911–2920. 1999.PubMed/NCBI
|
|
10
|
Mundlos S, Otto F, Mundlos C, Mulliken JB,
Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et
al: Mutations involving the transcription factor CBFA1 cause
cleidocranial dysplasia. Cell. 89:773–779. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ryoo HM, Kang HY, Lee SK, Lee KE and Kim
JW: RUNX2 mutations in cleidocranial dysplasia patients. Oral Dis.
16:55–60. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen S, Rani S, Wu Y, Unterbrink A, Gu TT,
Gluhak-Heinrich J, Chuang HH and Macdougall M: Differential
regulation of dentin sialophosphoprotein expression by Runx2 during
odontoblast cytodifferentiation. J Biol Chem. 280:29717–29727.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yeung F, Law WK, Yeh CH, Westendorf JJ,
Zhang Y, Wang R, Kao C and Chung LW: Regulation of human
osteocalcin promoter in hormone-independent human prostate cancer
cells. J Biol Chem. 277:2468–2476. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kurosaka H, Islam MN, Kuremoto K, Hayano
S, Nakamura M, Kawanabe N, Yanagita T, Rice DP, Harada H, Taniuchi
I and Yamashiro T: Core binding factor beta functions in the
maintenance of stem cells and orchestrates continuous proliferation
and differentiation in mouse incisors. Stem Cells. 29:1792–1803.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao Y, Wang W, Sun Y, Zhang J, Li D, Wei Y
and Han T: Distribution of amelotin in mouse tooth development.
Anat Rec (Hoboken). 293:135–140. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khan A, Fornes O, Stigliani A, Gheorghe M,
Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni
SR, Tan G, et al: JASPAR 2018: Update of the open-access database
of transcription factor binding profiles and its web framework.
Nucleic Acids Res. 46:D12842018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao Y, Li D, Han T, Sun Y and Zhang J:
TGF-beta1 and TGFBR1 are expressed in ameloblasts and promote MMP20
expression. Anat Rec (Hoboken). 292:885–890. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakata A, Kameda T, Nagai H, Ikegami K,
Duan Y, Terada K and Sugiyama T: Establishment and characterization
of a spontaneously immortalized mouse ameloblast-lineage cell line.
Biochem Biophys Res Commun. 308:834–839. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:1–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Komori T: Requisite roles of Runx2 and
Cbfb in skeletal development. J Bone Miner Metab. 21:193–197.
2003.PubMed/NCBI
|
|
21
|
Simmer JP, Richardson AS, Smith CE, Hu Y
and Hu JC: Expression of kallikrein-related peptidase 4 in dental
and non-dental tissues. Eur J Oral Sci. 119 Suppl 1:S226–S233.
2011. View Article : Google Scholar
|
|
22
|
Yadav MC, de Oliveira RC, Foster BL, Fong
H, Cory E, Narisawa S, Sah RL, Somerman M, Whyte MP and Millán JL:
Enzyme replacement and prevents enamel defects in hypophosphatasia
mice. J Bone Miner Res. 27:1722–1734. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aberg T, Cavender A, Gaikwad JS, Bronckers
AL, Wang X, Waltimo-Sirén J, Thesleff I and D'Souza RN: Phenotypic
changes in dentition of Runx2 homozygote-null mutant mice. J
Histochem Cytochem. 52:131–139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bronckers AL, Engelse MA, Cavender A,
Gaikwad J and D'Souza RN: Cell-specific patterns of Cbfa1 mRNA and
protein expression in postnatal murine dental tissues. Mech Dev.
101:255–258. 2011. View Article : Google Scholar
|
|
25
|
McGee-Lawrence ME, Carpio LR, Bradley EW,
Dudakovic A, Lian JB, van Wijnen AJ, Kakar S, Hsu W and Westendorf
JJ: Runx2 is required for early stages of endochondral bone
formation but delays final stages of bone repair in Axin2-deficient
mice. Bone. 66:277–286. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vimalraj S, Arumugam B, Miranda PJ and
Selvamurugan N: Runx2: Structure, function and phosphorylation in
osteoblast differentiation. Int J Biol Macromol. 78:202–208. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kawasaki K, Buchanan AV and Weiss KM:
Biomineralization in humans: Making the hard choices in life. Annu
Rev Genet. 43:119–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boskey AL: Mineralization of bones and
teeth. Elements. 3:385–391. 2007. View Article : Google Scholar
|
|
29
|
Atsawasuwan P, Lu X, Ito Y, Chen Y,
Gopinathan G, Evans CA, Kulkarni AB, Gibson CW, Luan X and
Diekwisch TG: Expression and function of enamel-related gene
products in calvarial development. J Dent Res. 92:622–628. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lim KE, Park NR, Che X, Han MS, Jeong JH,
Kim SY, Park CY, Akiyama H, Kim JE, Ryoo HM, et al: Core binding
factor β of osteoblasts maintains cortical bone mass via
stabilization of Runx2 in mice. J Bone Miner Res. 30:715–722. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moffatt P, Smith CE, St-Arnaud R, Simmons
D, Wright JT and Nanci A: Cloning of rat amelotin and localization
of the protein to the basal lamina of maturation stage ameloblasts
and junctional epithelium. J Biochem. 399:37–46. 2006. View Article : Google Scholar
|
|
32
|
Ohyama Y, Tanaka T, Shimizu T, Matsui H,
Sato H, Koitabashi N, Doi H, Iso T, Arai M and Kurabayashi M:
Runx2/Smad3 complex negatively regulates TGF-β-induced connective
tissue growth factor gene expression in vascular smooth muscle
cells. J Atheroscler Thromb. 19:23–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiménez MJ, Balbín M, López JM, Alvarez J,
Komori T and López-Otín C: Collagenase 3 is a target of Cbfa1, a
transcription factor of the runt gene family involved in bone
formation. Mol Cell Biol. 19:4431–4442. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Merriman HL, van Wijnen AJ, Hiebert S,
Bidwell JP, Fey E, Lian J, Stein J and Stein GS: The
tissue-specific nuclear matrix protein, NMP-2, is a member of the
AML/CBF/PEBP2/runt domain transcription factor family: Interactions
with the osteocalcin gene promoter. Biochemistry. 34:1–13132. 1995.
View Article : Google Scholar
|