|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Corso S and Giordano S: How can gastric
cancer molecular profiling guide future therapies? Trends Mol Med.
22:534–544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bang YJ, Van Cutsem E, Feyereislova A,
Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T,
et al: Trastuzumab in combination with chemotherapy versus
chemotherapy alone for treatment of HER2-positive advanced gastric
or gastro-oesophageal junction cancer (ToGA): A phase 3,
open-label, randomised controlled trial. Lancet. 376:687–697. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aprile G, Giampieri R, Bonotto M, Bittoni
A, Ongaro E, Cardellino GG, Graziano F, Giuliani F, Fasola G,
Cascinu S and Scartozzi M: The challenge of targeted therapies for
gastric cancer patients: The beginning of a long journey. Exp Opin
Investig Drugs. 23:925–942. 2014. View Article : Google Scholar
|
|
5
|
Ritter CA, Perez-Torres M, Rinehart C,
Guix M, Dugger T, Engelman JA and Arteaga CL: Human breast cancer
cells selected for resistance to trastuzumab in vivo overexpress
epidermal growth factor receptor and ErbB ligands and remain
dependent on the ErbB receptor network. Clin Cancer Res.
13:4909–4919. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gottesman MM, Lavi O, Hall MD and Gillet
JP: Toward a better understanding of the complexity of cancer drug
resistance. Annu Rev Pharmacol Toxicol. 56:85–102. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garofalo M and Croce CM: MicroRNAs as
therapeutic targets in chemoresistance. Drug Resist Updat.
16:47–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu Y, Lemon W, Liu PY, Yi Y, Morrison C,
Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al: A gene
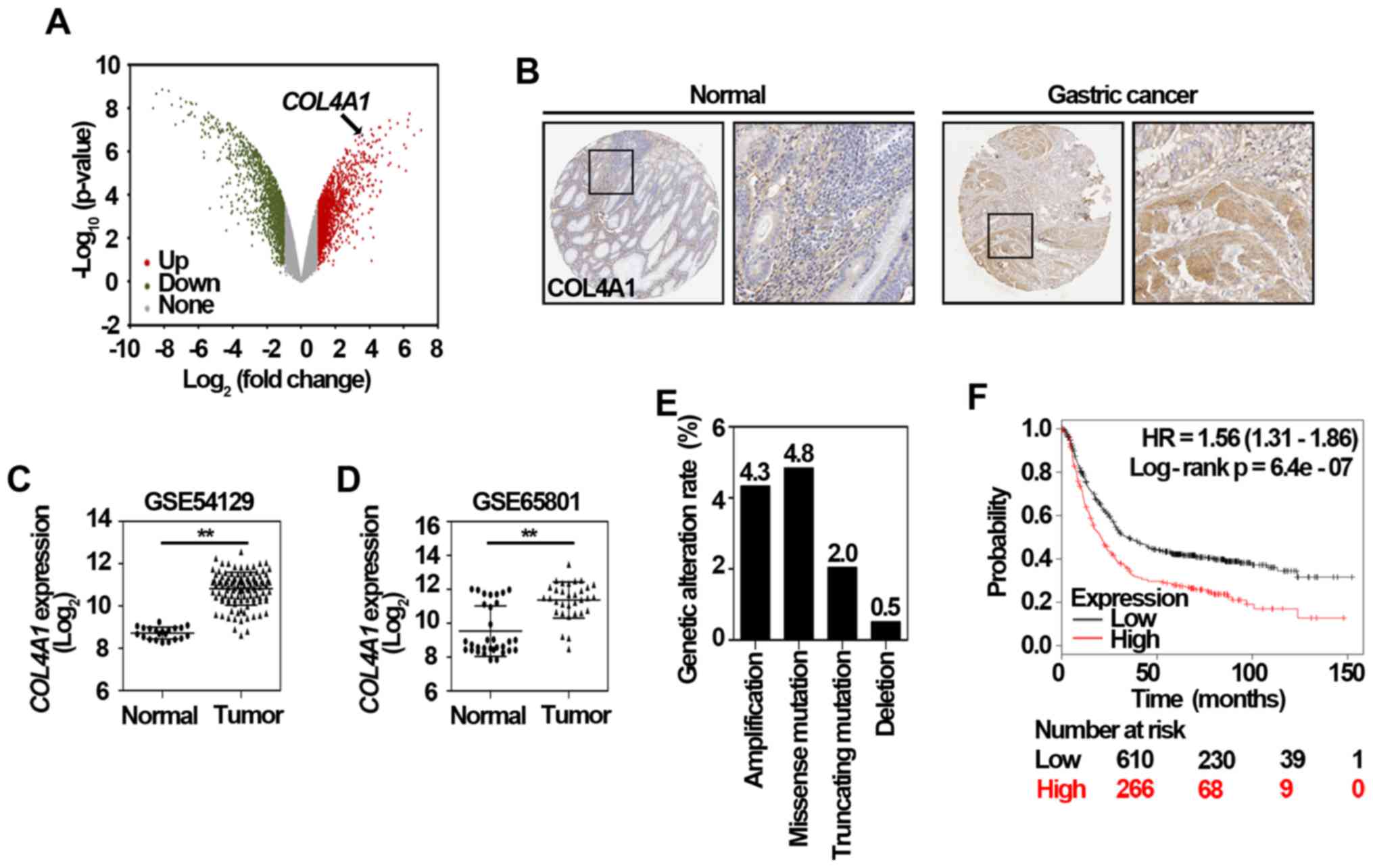
expression signature predicts survival of patients with stage I
non-small cell lung cancer. PLoS Med. 3:e4672006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen F, Xiang CX, Zhou Y, Ao XS, Zhou DQ,
Peng P, Zhang HQ, Liu HD and Huang X: Gene expression profile for
predicting survival of patients with meningioma. Int J Oncol.
46:791–797. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Tumor suppressor genes associated with drug resistance in
ovarian cancer (Review). Oncol Rep. 30:3–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Piro G, Carbone C, Cataldo I, Di
Nicolantonio F, Giacopuzzi S, Aprile G, Simionato F, Boschi F,
Zanotto M, Mina MM, et al: An FGFR3 autocrine loop sustains
acquired resistance to trastuzumab in gastric cancer patients. Clin
Cancer Res. 22:6164–6175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Yu B, Li J, Su L, Yan M, Zhang J, Li
C, Zhu Z and Liu B: Characterization of differentially expressed
genes involved in pathways associated with gastric cancer. PLoS
One. 10:e01250132015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: Two different approaches for Gene Ontology
analysis. BMC Proc. 3 Suppl 4:pp. S102009; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3.0: Deciphering microRNA function
with experimental support. Nucleic Acids Res. 43:W460–W466. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Borst P and Wessels L: Do predictive
signatures really predict response to cancer chemotherapy? Cell
cycle (Georgetown, Tex.). 9:4836–4840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yague E, Arance A, Kubitza L, O'Hare M,
Jat P, Ogilvie CM, Hart IR, Higgins CF and Raguz S: Ability to
acquire drug resistance arises early during the tumorigenesis
process. Cancer Res. 67:1130–1137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xing Z, Chu C, Chen L and Kong X: The use
of Gene Ontology terms and KEGG pathways for analysis and
prediction of oncogenes. Biochim Biophys Acta. 1860:2725–2734.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maier T, Guell M and Serrano L:
Correlation of mRNA and protein in complex biological samples. FEBS
Lett. 583:3966–3973. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Labussiere M, Cheneau C, Prahst C, Gállego
Pérez-Larraya J, Farina P, Lombardi G, Mokhtari K, Rahimian A,
Delattre JY, Eichmann A and Sanson M: Angiopoietin-2 May be
involved in the resistance to bevacizumab in recurrent
glioblastoma. Cancer Invest. 34:39–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Liu Z, Jiang K and Ruan Q:
Angiopoietin2 enhances doxorubin resistance in HepG2 cells by
upregulating survivin and Ref-1 via MSK1 activation. Cancer Lett.
337:276–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sherman-Baust CA, Weeraratna AT, Rangel
LB, Pizer ES, Cho KR, Schwartz DR, Shock T and Morin PJ: Remodeling
of the extracellular matrix through overexpression of collagen VI
contributes to cisplatin resistance in ovarian cancer cells. Cancer
Cell. 3:377–386. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fenouille N, Puissant A, Dufies M, Robert
G, Jacquel A, Ohanna M, Deckert M, Pasquet JM, Mahon FX, Cassuto
JP, et al: Persistent activation of the Fyn/ERK kinase signaling
axis mediates imatinib resistance in chronic myelogenous leukemia
cells through upregulation of intracellular SPARC. Cancer Res.
70:9659–9670. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pannuru P, Dontula R, Khan AA, Herbert E,
Ozer H, Chetty C and Lakka SS: miR-let-7f-1 regulates SPARC
mediated cisplatin resistance in medulloblastoma cells. Cell
Signal. 26:2193–2201. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Januchowski R, Zawierucha P, Rucinski M
and Zabel M: Microarray-based detection and expression analysis of
extracellular matrix proteins in drugresistant ovarian cancer cell
lines. Oncol Rep. 32:1981–1990. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lau CK, Yang ZF, Ho DW, Ng MN, Yeoh GC,
Poon RT and Fan ST: An Akt/hypoxia-inducible
factor-1alpha/platelet-derived growth factor-BB autocrine loop
mediates hypoxia-induced chemoresistance in liver cancer cells and
tumorigenic hepatic progenitor cells. Clin Cancer Res.
15:3462–3471. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Leeuw N, Dijkhuizen T, Hehir-Kwa JY,
Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van
Ravenswaaij-Arts CM, et al: Diagnostic interpretation of array data
using public databases and internet sources. Hum Mutat. 33:930–940.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sayed D and Abdellatif M: MicroRNAs in
development and disease. Physiol Rev. 91:827–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Blenkiron C and Miska EA: miRNAs in
cancer: Approaches, aetiology, diagnostics and therapy. Hum Mol
Genet. 16:R106–R113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu X, Wang H, Lian Y, Chen L, Gu L, Wang J
and Huang Y, Deng M, Gao Z and Huang Y: GTSE1 promotes cell
migration and invasion by regulating EMT in hepatocellular
carcinoma and is associated with poor prognosis. Sci Rep.
7:51292017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC,
Yao L and Qiao PF: miR-34a mediates oxaliplatin resistance of
colorectal cancer cells by inhibiting macroautophagy via
transforming growth factor-beta/Smad4 pathway. World J
Gastroenterol. 23:1816–1827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lanzi C, Zaffaroni N and Cassinelli G:
Targeting heparan sulfate proteoglycans and their modifying enzymes
to enhance anticancer chemotherapy efficacy and overcome drug
resistance. Curr Med Chem. 24:2860–2886. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee B, Sandhu S and McArthur G: Cell cycle
control as a promising target in melanoma. Curr Opin Oncol.
27:141–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wallerand H, Robert G, Pasticier G, Ravaud
A, Ballanger P, Reiter RE and Ferrière JM: The
epithelial-mesenchymal transition-inducing factor TWIST is an
attractive target in advanced and/or metastatic bladder and
prostate cancers. Urol Oncol. 28:473–479. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu FX, Zhao B and Guan KL: Hippo pathway
in organ size control, tissue homeostasis and cancer. Cell.
163:811–828. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lee JE, Park HS, Lee D, Yoo G, Kim T, Jeon
H, Yeo MK, Lee CS, Moon JY, Jung SS, et al: Hippo pathway effector
YAP inhibition restores the sensitivity of EGFR-TKI in lung
adenocarcinoma having primary or acquired EGFR-TKI resistance.
Biochem Biophys Res Commun. 474:154–160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gujral TS and Kirschner MW: Hippo pathway
mediates resistance to cytotoxic drugs. Proc Natl Acad Sci USA.
114:pp. E3729–E3738. 2017; View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Okamoto K, Miyoshi K and Murawaki Y:
miR-29b, miR-205 and miR-221 enhance chemosensitivity to
gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS One.
8:e776232013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu YX, Wang L, Liu WJ, Zhang HT, Xue JH,
Zhang ZW and Gao CJ: MiR-124-3p/B4GALT1 axis plays an important
role in SOCS3-regulated growth and chemo-sensitivity of CML. J
Hematol Oncol. 9:692016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen MJ, Cheng YM, Chen CC, Chen YC and
Shen CJ: MiR-148a and miR-152 reduce tamoxifen resistance in ER+
breast cancer via downregulating ALCAM. Biochem Biophys Res Commun.
483:840–846. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhong S, Li W, Chen Z, Xu J and Zhao J:
MiR-222 and miR-29a contribute to the drug-resistance of breast
cancer cells. Gene. 531:8–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sui C, Meng F, Li Y and Jiang Y: miR-148b
reverses cisplatin-resistance in non-small cell cancer cells via
negatively regulating DNA (cytosine-5)-methyltransferase 1 (DNMT1)
expression. J Transl Med. 13:1322015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM,
Xiao SD and Ran ZH: MicroRNA 506 regulates expression of PPAR alpha
in hydroxycamptothecin-resistant human colon cancer cells. FEBS
Lett. 585:3560–3568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang JX, Qian D, Wang FW, Liao DZ, Wei
JH, Tong ZT, Fu J, Huang XX, Liao YJ, Deng HX, et al: MicroRNA-29c
enhances the sensitivities of human nasopharyngeal carcinoma to
cisplatin-based chemotherapy and radiotherapy. Cancer Lett.
329:91–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Holohan C, Van Schaeybroeck S, Longley DB
and Johnston PG: Cancer drug resistance: An evolving paradigm. Nat
Rev Cancer. 13:714–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Albarello L, Pecciarini L and Doglioni C:
HER2 testing in gastric cancer. Adv Anat Pathol. 18:53–59. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Price-Schiavi SA, Jepson S, Li P, Arango
M, Rudland PS, Yee L and Carraway KL: Rat Muc4 (sialomucin complex)
reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a
potential mechanism for herceptin resistance. Int J Cancer.
99:783–791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Castiglioni F, Tagliabue E, Campiglio M,
Pupa SM, Balsari A and Menard S: Role of exon-16-deleted HER2 in
breast carcinomas. Endocr Relat Cancer. 13:221–232. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Serra V, Markman B, Scaltriti M, Eichhorn
PJ, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Di Cosimo S,
et al: NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K
signaling and inhibits the growth of cancer cells with activating
PI3K mutations. Cancer Res. 68:8022–8030. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nahta R, Yuan LX, Zhang B, Kobayashi R and
Esteva FJ: Insulin-like growth factor-I receptor/human epidermal
growth factor receptor 2 heterodimerization contributes to
trastuzumab resistance of breast cancer cells. Cancer Res.
65:11118–11128. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Boyer J, Allen WL, McLean EG, Wilson PM,
McCulla A, Moore S, Longley DB, Caldas C and Johnston PG:
Pharmacogenomic identification of novel determinants of response to
chemotherapy in colon cancer. Cancer Res. 66:2765–2777. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Denduluri SK, Idowu O, Wang Z, Liao Z, Yan
Z, Mohammed MK, Ye J, Wei Q, Wang J, Zhao L and Luu HH:
Insulin-like growth factor (IGF) signaling in tumorigenesis and the
development of cancer drug resistance. Genes Dis. 2:13–25. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Casa AJ, Dearth RK, Litzenburger BC, Lee
AV and Cui X: The type I insulin-like growth factor receptor
pathway: A key player in cancer therapeutic resistance. Front
Biosci. 13:3273–3287. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sado Y, Kagawa M, Kishiro Y, Sugihara K,
Naito I, Seyer JM, Sugimoto M, Oohashi T and Ninomiya Y:
Establishment by the rat lymph node method of epitope-defined
monoclonal antibodies recognizing the six different alpha chains of
human type IV collagen. Histochem Cell Biol. 104:267–275. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kalluri R: Basement membranes: Structure,
assembly and role in tumour angiogenesis. Nat Rev Cancer.
3:422–433. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Miyake M, Hori S, Morizawa Y, Tatsumi Y,
Toritsuka M, Ohnishi S, Shimada K, Furuya H, Khadka VS, Deng Y, et
al: Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha
1 (COL13A1) produced in cancer cells promote tumor budding at the
invasion front in human urothelial carcinoma of the bladder.
Oncotarget. 8:36099–36114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Jin R, Shen J, Zhang T, Liu Q, Liao C, Ma
H, Li S and Yu Z: The highly expressed COL4A1 genes contributes to
the proliferation and migration of the invasive ductal carcinomas.
Oncotarget. 8:58172–58183. 2017.PubMed/NCBI
|
|
65
|
Sulpice L, Rayar M, Desille M, Turlin B,
Fautrel A, Boucher E, Llamas-Gutierrez F, Meunier B, Boudjema K,
Clément B and Coulouarn C: Molecular profiling of stroma identifies
osteopontin as an independent predictor of poor prognosis in
intrahepatic cholangiocarcinoma. Hepatology. 58:1992–2000. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rahman M, Chan AP, Tang MJ and Tai IT:
Correction: A peptide of SPARC interferes with the interaction
between caspase8 and Bcl2 to resensitize chemoresistant tumors and
enhance their regression in vivo. PLoS One. 10:e01272262015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yang Z, Guo L, Liu D, Sun L, Chen H, Deng
Q, Liu Y, Yu M, Ma Y, Guo N and Shi M: Acquisition of resistance to
trastuzumab in gastric cancer cells is associated with activation
of IL-6/STAT3/Jagged-1/Notch positive feedback loop. Oncotarget.
6:5072–5087. 2015.PubMed/NCBI
|
|
68
|
Wang S, Huang J, Lyu H, Cai B, Yang X, Li
F, Tan J, Edgerton SM, Thor AD, Lee CK and Liu B: Therapeutic
targeting of erbB3 with MM-121/SAR256212 enhances antitumor
activity of paclitaxel against erbB2-overexpressing breast cancer.
Breast Cancer Res. 15:R1012013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Vora SR, Juric D, Kim N, Mino-Kenudson M,
Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M, Li X, et al: CDK
4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K
inhibitors. Cancer Cell. 26:136–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hudis C, Swanton C, Janjigian YY,
Mino-Kenudson M, Huynh T, Costa C, Lockerman EL, Pollack SF, Liu M,
Li X, et al: A phase 1 study evaluating the combination of an
allosteric AKT inhibitor (MK-2206) and trastuzumab in patients with
HER2-positive solid tumors. Breast Cancer Res. 15:R1102013.
View Article : Google Scholar : PubMed/NCBI
|