Introduction
Although osteosarcoma is a relatively rare type of
cancer, it is considered to be one of the major causes of
cancer-associated mortality among children and young adults
(1). In the USA, ~1,000 new cases
of osteosarcoma are reported each year (2). Young patients are typically diagnosed
with primary conventional osteosarcoma, while secondary
osteosarcoma is more common among elderly patients (2). Osteosarcoma primarily affects the
long bones, but other bones in the body can also be affected
(3). Similar to other tumors,
development of osteosarcoma is a complex process with various
signaling pathways involved (4).
Therefore, in-depth analyses of signal transduction involved
regulation in tumorigenesis may aid in elucidation of molecular
mechanisms underlying the onset, development and progression of
osteosarcoma.
The role of noncoding RNAs in the development of
various pathophysiological conditions has been well characterized
(5). Long non-coding RNAs (lncRNA)
are a type of noncoding RNAs of >200 nucleotides in length
(6). lncRNAs have roles in the
onset and development of different human cancer types, including
breast cancer, non-small-cell lung cancer and colorectal cancer
(7–10). Expression of lncRNA H19 has been
reported to be positively associated with progression of various
malignant human tumors (11,12).
It has been reported that H19 is frequently overexpressed during
the development of osteosarcoma (13) and that H19 can promote migration
and invasion of osteosarcoma cells (14). However, the molecular mechanism
underlying the function of H19 in osteosarcoma remains to be
elucidated.
In the present study, expression of H19 in
osteosarcoma tumor tissue and adjacent healthy tissue was detected.
Effects of H19 on migration and invasion of multiple osteosarcoma
cells lines were determined. Association between H19 and the NF-κB
signaling pathway was investigated. In addition, prognostic value
of expression of H19 for patients with osteosarcoma was also
evaluated.
Materials and methods
Patients
A total of 40 patients with osteosarcoma were
selected in Tianjin Medical University Cancer Institute and
Hospital from January 2010 to January 2013. All patients were
diagnosed based on pathological and imaging examinations. Inclusion
criteria were: i) Patient pathologically diagnosed as osteosarcoma
and ii) patients willing to cooperate with researchers. Exclusion
criteria were: i) Patient with other malignancies; ii) patients
with metal disorders; iii) patients who had received treatment
prior to admission. Surgical resection was performed for all
patients. The patients included 18 males and 22 females (age range
8–66 years; mean age of 31±8.1 years). Distant metastasis was
observed in 22 patients. During surgical operations, cancer tissues
and normal tissues within the region 0.5 cm around tumor were
collected for subsequent experiments. The present study was
approved by the Ethics Committee of Tianjin Medical University
Cancer Institute and Hospital (Tianjin, China), and all patients
signed informed consent. A follow-up study was performed for 48
months to monitor the survival.
Cell lines and cell culture
Human MG-63, U2OS and SAOS-2 osteosarcoma cell
lines, and hFOB normal bone cell line, were purchased from American
Type Culture Collection (Manassas, VA, USA). MG-63 and SAOS-2 cells
were cultured with Eagle's minimum Essential medium (cat. no.
30–2003; American Type Culture Collection) containing 10%
heat-inactivated fetal bovine serum (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). U2OS and SAOS-2 cells were cultured with
McCoy's 5a modified medium (cat. no. 30-2007; American Type Culture
Collection) containing 10% fetal bovine serum (Thermo Fisher
Scientific, Inc.). All cells were cultured at 37°C and 5%
CO2. Cells were harvested during logarithmic growth
phase for subsequent experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from tissues and
cells. Concentration and quality of RNA samples were determined
using a NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific,
Inc.). Only RNA samples with a ratio of A260/A280 between 1.8 and
2.0 were used for reverse transcription to synthesize cDNA using
SuperScript III Reverse Transcriptase (Thermo Fisher Scientific,
Inc.). Reaction conditions for reverse transcription were: 25°C for
5 min, 55°C for 20 min and 80°C for 10 min. SYBR®-Green
Real-Time PCR Master Mix (Thermo Fisher Scientific, Inc.) was used
for qPCR. The following primers were used in PCR reactions:
5′-TGAGCTCTCAGGAGGGAGGATGG-3′ (forward) and
5′-TTGTCACGTCCACCGGACCTG-3′ (reverse) for H19;
5′-GACCTCTATGCCAACACAGT-3′ (forward) and 5′-AGTACTTGCGCTCAGGAGGA-3′
(reverse) for β-actin. PCR reactions were performed using CFX96
Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The following thermocycling conditions were
used for the PCR: Initial denaturation at 95°C for 40 sec; 40
cycles of 95°C for 15 sec and 60°C for 45 sec. Relative expression
was calculated using 2−ΔΔCq method (15). Relative expression level of H19 was
normalized to endogenous control β-actin.
Establishment of H19-silenced cell
lines
H19 small interfering RNA (siRNA; cat. no. 1299001;
Thermo Fisher Scientific, Inc.) and Silencer™ Select
Negative Control No. 1 siRNA (cat. no. 4390843; Thermo Fisher
Scientific, Inc.) were used. Cells were cultured overnight to reach
80–90% confluence prior to transfection. Transfection (50 nM siRNA)
with Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was performed according to the
manufacturer's protocol. Cells were cultured for another 6 h before
subsequent experiments.
Cell migration and invasion assay
Transwell cell migration assay (BD Biosciences,
Franklin Lakes, NJ, USA) was performed to measure cell migration
ability. Briefly, 5×104 cells in 100 µl serum-free
Dulbecco's modified Eagle medium (Thermo Fisher Scientific) were
transferred to the upper chamber, while the lower chamber was
filled with RPMI-1640 medium (Thermo Fisher Scientific, Inc.)
containing 20% fetal calf serum (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Membranes were collected 24 h later and
stained with 0.5% crystal violet at room temperature
(Sigma-Aldrich; Merck KGaA) for 20 min. Stained cells were counted
under a light microscope (Olympus Corporation, Tokyo, Japan).
Invasion assay was performed using the same method with the upper
chamber was pre-coated with Matrigel (EMD Millipore, Billerica, MA,
USA).
Western blot analysis
Total protein extraction was performed using RIPA
buffer (Thermo Fisher Scientific, Inc.), and protein concentration
was measured using THE bicinchoninic acid method. Subsequently, 30
µg of protein was subjected to 10% SDS-PAGE gel electrophoresis,
and transferred to polyvinylidene fluoride membrane at 20 V for 1
h. Following blocking with 5% skimmed milk at room temperature for
1 h, membranes were washed with TBST (0.1% Tween 20) 3 times for 15
min each time. Membranes were subsequently incubated with primary
antibodies, including rabbit anti-p-PI3K (1:2,000; cat. no.
ab182651), anti-PI3K (1:2,000; cat. no. ab5451; Abcam), anti-p-AKT
(1:2,000; cat. no. ab18206; Abcam), anti-AKT (1:2,000; cat. no.
ab126811; Abcam), anti-NF-κB inhibitor α (IκBα; 1:1,000; cat. no.
ab76429; Abcam) and anti-GAPDH (1:1,000; ab8245; all Abcam,
Cambridge, UK) overnight at 4°C. Subsequently, membranes were
washed three times with TBST, 15 min each time. Membranes were
incubated with anti-rabbit immunoglobulin G-horseradish peroxidase
conjugated secondary antibody (1:1,000; cat. no. MBS435036;
MyBioSource, Inc., San Diego, CA, USA) at room temperature for 2 h.
Following washing twice with TBST, 15 min each time, Enhanced
Chemiluminescence detection reagent (Sigma-Aldrich; Merck KGaA) was
used to detect the signals. Images were analyzed using Image J
software version 1.8.0 (National Institutes of Health, Bethesda,
MD, USA) to calculate relative expression level of each protein
relative to endogenous control β-actin.
Statistical analysis
SPSS software (version 19.0; IBM Corp., Armonk, NY,
USA) was used for all statistical analysis. Normal distribution
data are presented as the mean ± standard deviation. Comparisons
between two groups were performed using t test and comparisons
among multiple groups were performed using one-way analysis of
variance and least significant difference test. Data that were not
normally distributed were analyzed using non-parametric
Mann-Whitney U test. In multivariate logistic regression analysis,
mortality was set as dependent variable, and gender (being a male),
age (>40), distant metastasis and high H19 expression level were
independent variables. Survival curves were plotted using
Kaplan-Meier method and compared using log-rank test. With the
median expression level of H19 in cancer tissues as the cutoff
score, 40 patients with osteosarcoma were divided into high
expression level group (n=20) and low expression level group
(n=20), and the association between the expression level of H19 and
clinicopathological features was analyzed by χ2 test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Comparison of expression levels of H19
in cancer tissues and adjacent healthy tissues
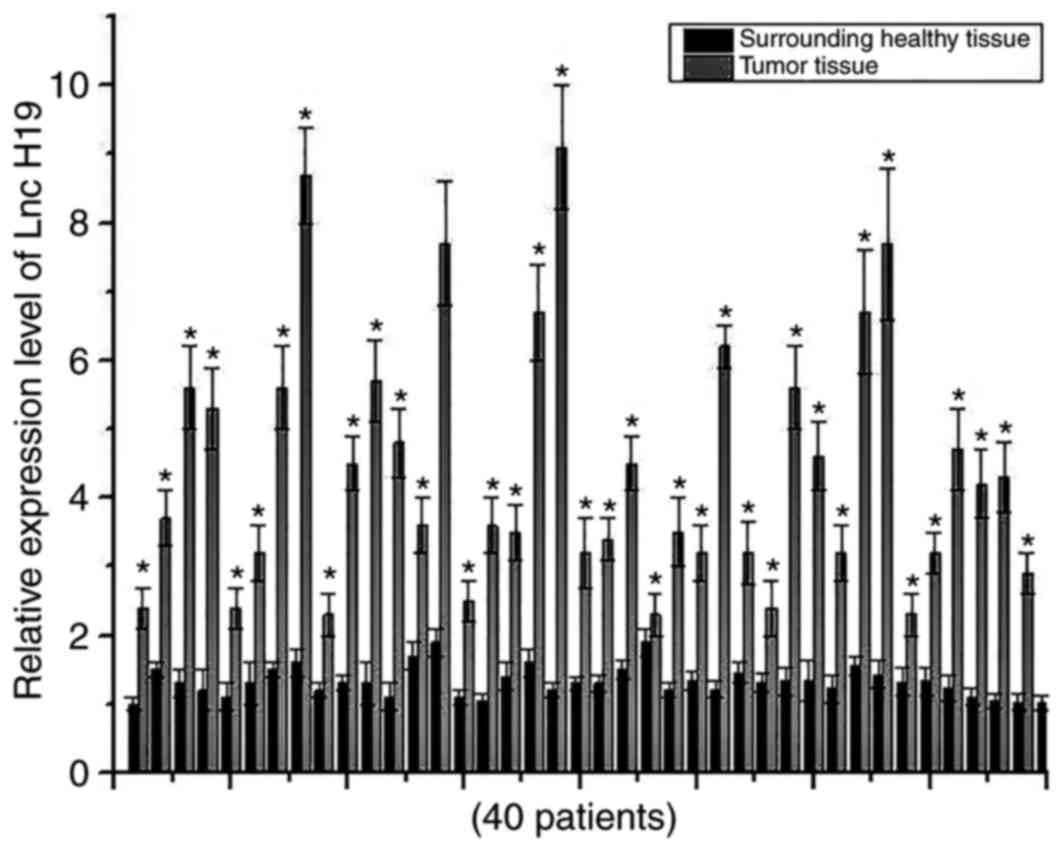
Expression levels of H19 in cancer tissues and
adjacent healthy tissues of 40 patients with osteosarcoma were
detected using RT-qPCR. Expression levels of H19 were significantly
increased in cancer tissues of all 40 patients compared with the
respective adjacent healthy tissues, indicating the potential role
of H19 in development of osteosarcoma (Fig. 1).
Association between expression of H19
and clinicopathological features, and the prognostic values
Univariate analysis demonstrated that the expression
level of H19 was positively associated with distant metastasis
(P<0.01), not with gender and age (Table I). Multivariate regression analysis
demonstrated that aging, distant metastasis and high expression
level of H19 were associated with mortality of patients with
osteosarcoma (P<0.05; Table
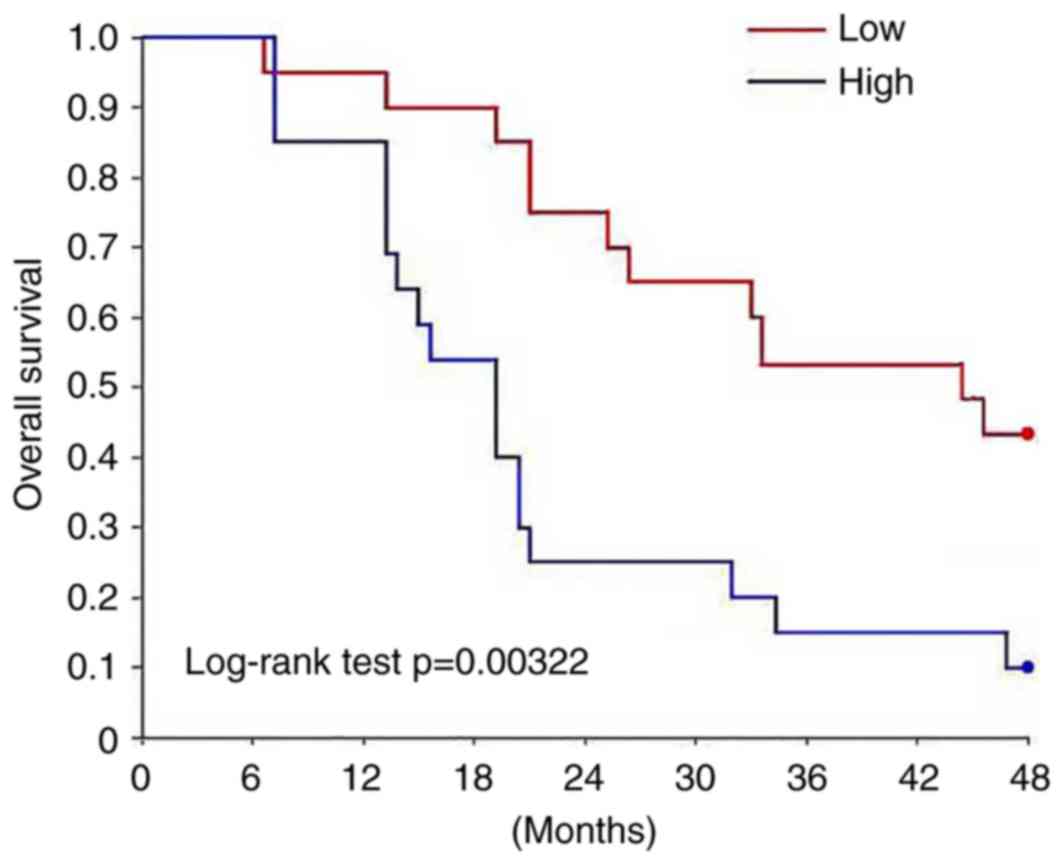
II). Survival curves were plotted using the Kaplan-Meier method
to evaluate the prognostic value of H19 for osteosarcoma. Survival
curves were compared using log-rank test. The overall survival rate
of patients with high expression level of H19 was significantly
lower compared with patients with low expression level of H19
(P<0.05; Fig. 2).
 | Table I.Correlation between the expression
level of H19 and clinicopathological features. |
Table I.
Correlation between the expression
level of H19 and clinicopathological features.
|
|
|
| H19 expression |
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
feature | Group | Total no. | High | Low | P-value |
|---|
| Sex | Male | 18 | 8 | 10 | 0.79 |
|
| Female | 22 | 12 | 10 |
|
| Age (years) | >25 | 13 | 6 | 7 | 0.72 |
|
| ≤30 | 27 | 11 | 16 |
|
| Distant
metastasis | Yes | 22 | 17 | 5 | <0.01 |
|
| No | 18 | 4 | 14 |
|
 | Table II.Multivariate regression analysis of
factors associated with mortality of patients with
osteosarcoma. |
Table II.
Multivariate regression analysis of
factors associated with mortality of patients with
osteosarcoma.
| Clinicopathological
feature | Risk ratio | 95% CI | P-value |
|---|
| Gender (male) | 1.02 | 0.61–5.52 | 0.33 |
| Age (>40) | 3.83 | 2.12–7.73 | 0.04 |
| Distant
metastasis | 6.92 | 2.85–12.82 | 0.01 |
| High H19 expression
level | 6.53 | 2.27–9.58 | 0.01 |
Effects of H19 knockdown on
osteosarcoma cell migration and invasion
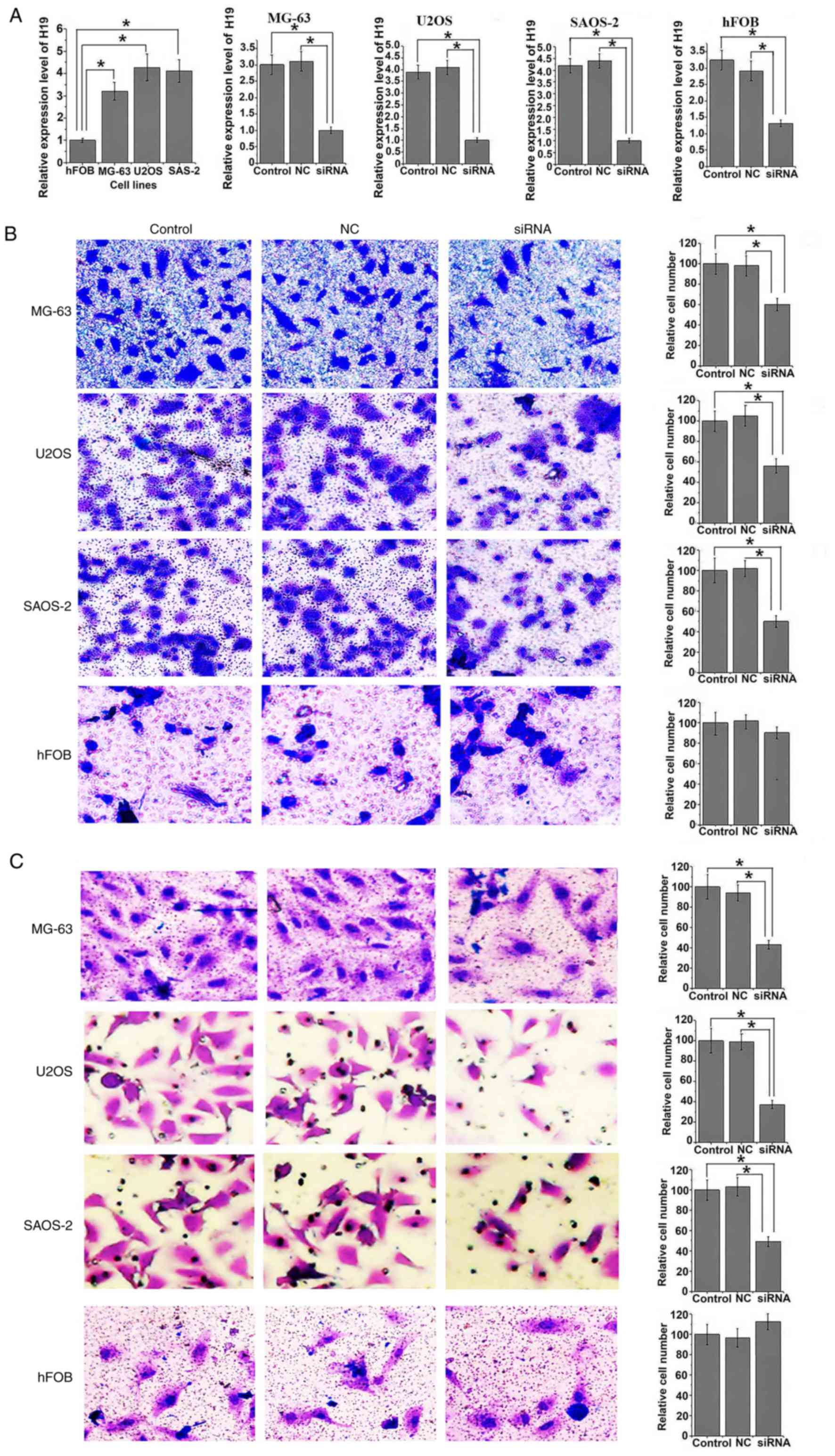
Expression level of H19 was significantly increased
in human osteosarcoma cell lines MG-63, U2OS and SAOS-2 compared
with normal bone cell line hFOB (all P<0.05; Fig. 3A). Following siRNA transfection,
expression level of H19 was significantly reduced in all three
osteosarcoma cell lines and in the normal hFOB cell line compared
with the controls (all P<0.05). Following siRNA H19
transfection, migration ability was significantly reduced in all
osteosarcoma cell lines compared with control cells (all
P<0.05). Similar results were observed in the invasion assay,
where invasion ability of all osteosarcoma cell lines was
significantly reduced following siRNA transfection (all P<0.05).
However, no significant differences in migration and invasion
abilities were identified in hFOB cells with and without H19
knockdown (Fig. 3B and C).
Effects of H19 knockdown on the NF-κB
signaling pathway
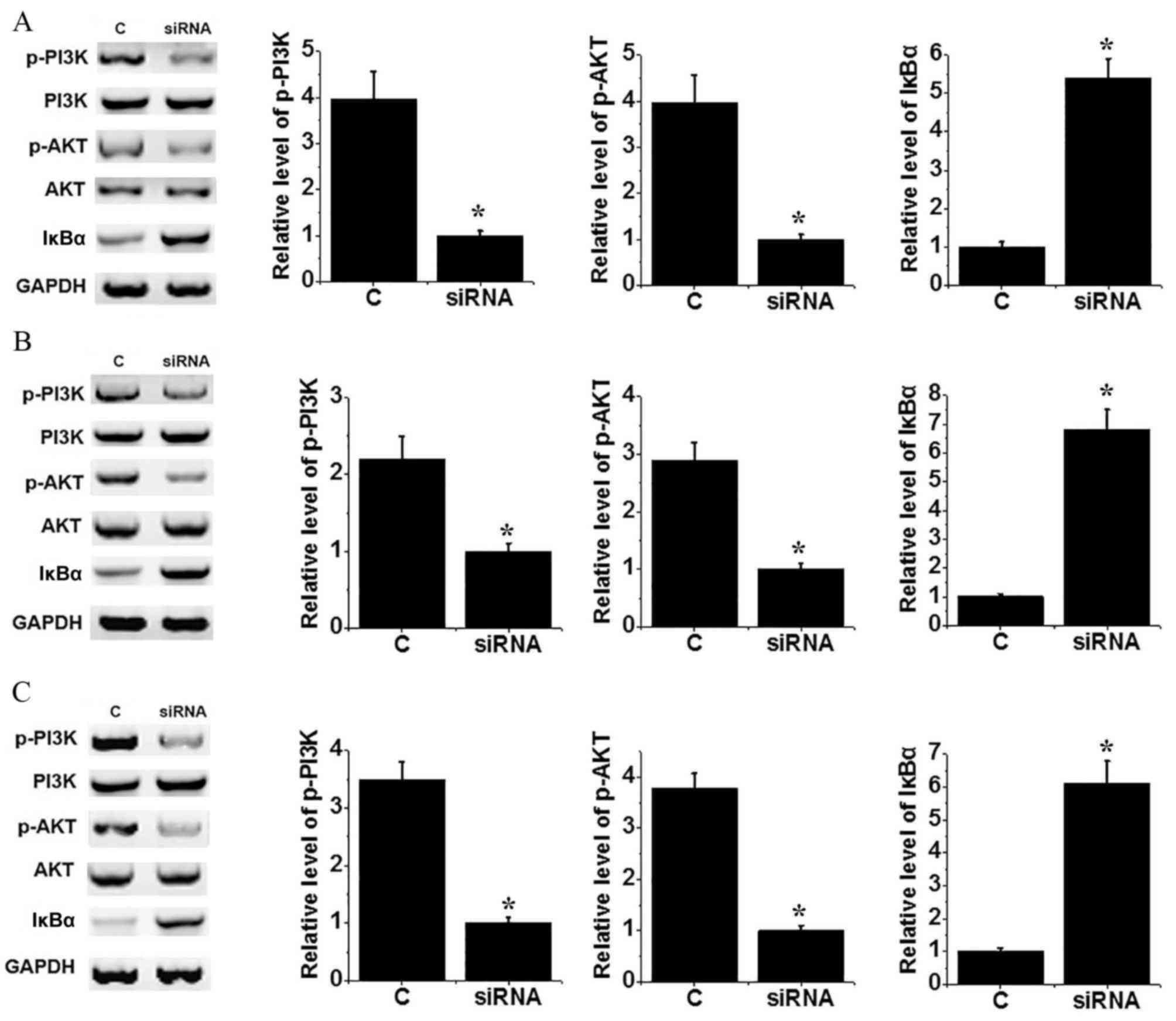
The PI3K/AKT signaling pathway can activate the
NF-κB pathway by regulating degradation of IκBα. No significant
differences in levels of PI3K and AKT were identified between the
control and H19 knockdown groups of MG-64 (Fig. 4A), U2OS (Fig. 4B) and SAOS-2 (Fig. 4C) cells. Levels of p-PI3K and p-AKT
were decreased significantly in cells transfected with siRNA
compared with control cells (P<0.05), suggesting inactivation of
the PI3K/AKT signaling pathway following H19 knockdown. In
addition, the level of IκBα protein increased significantly in
cells transfected with siRNA compared with control cells (all
P<0.05). The above data suggest that H19 knockdown can
inactivate NF-κB pathway, potentially through interaction with the
PI3K/AKT signaling pathway.
Discussion
The role of lncRNA in the development of
osteosarcoma has been demonstrated by numerous studies (14,16).
Sun et al (14) reported
that hepatocellular carcinoma up-regulated long non-coding RNA
(HULC) was upregulated in osteosarcoma tissues compared with
adjacent healthy tissues, and increased expression level of HULC
was associated with a shorter overall survival of patients with
osteosarcoma. By contrast, downregulation of HULC significantly
reduced proliferation, migration and invasion of in vitro
cultured osteosarcoma cells (14).
lncRNA taurine up-regulated 1 (TUG1) was also demonstrated to
inhibit apoptosis and promote proliferation of osteosarcoma cells,
and TUG1 may be a novel target for treatment of osteosarcoma
(16). H19 is an oncogenic lncRNA
and was demonstrated to be involved in the progression of various
types of human cancer (11,12).
A previous study reported that activation of the Hedgehog signaling
pathway upregulated the expression of H19, which in turn promoted
tumorigenesis (13). Consistent
with previous studies, in the present study, the expression level
of H19 in 40 osteosarcoma patients was increased markedly in tumor
tissue compared with adjacent healthy tissue. The results suggest
that H19 may be involved in development of osteosarcoma. In
addition, the expression level of H19 was positively associated
with the occurrence of distant metastasis of osteosarcoma, but not
with gender or age, and the overall survival of patients with
osteosarcoma exhibiting high H19 expression significantly shorter
compared with patients with low expression H19. Thus, the results
of the present study suggest that increased expression of H19 may
have a prognostic role for patients with distant metastasis of
osteosarcoma.
lncRNA H19 serves a role in the progression of
different types of cancer primarily by promoting tumor metastasis.
Shi et al (17) reported
that miR-675 derived from H19 can promote invasion of glioma cells.
In another study, H19 promoted migration and invasion of human
hepatocellular carcinoma cells by activating with the
AKT/GSK-3β/Cdc25A signal transduction pathway (18). In the present study, H19 knockdown
reduced migration and invasion abilities of three osteosarcoma cell
lines. The results of the present study suggest that expression
level of H19 is positively associated with invasion and migration
abilities of osteosarcoma cells.
NF-κB signaling has roles in numerous aspects of
initiation and progression of tumorigenesis (19). Previous studies demonstrated that
H19 can interact with the NF-κB pathway to perform its biological
functions (20). It has also been
demonstrated that the NF-κB pathway can be activated by the
PI3K/AKT signaling pathway via the regulation of degradation of
IκBα (21,22). In the present study,
phosphorylation levels of PI3K and AKT decreased, and expression
level of IκBα increased following knockdown of H19, while no
significant differences in expression levels of PI3K and AKT were
identified between osteosarcoma cells with and without H19
knockdown. The above results suggest that knockdown of H19 can
mediate inactivation of the NF-κB pathway by inhibiting the
activation PI3K/AKT pathway.
In conclusion, expression level of H19 increased in
osteosarcoma tissues compared with adjacent healthy tissues.
Expression level of H19 was positively associated with the
occurrence of distant metastasis, and elevated expression level of
H19 indicated poor prognosis of patients with osteosarcoma.
Expression of H19 affected migration and invasion of osteosarcoma
by activating the NF-κB pathway. Future studies should focus on
identifying targets of H19 to further elucidate the underlying
mechanism of H19 in osteosarcoma. The present study was limited by
small sample size. Further studies with larger sample sizes are
necessary to confirm the conclusions of the present study.
Acknowledgements
Not applicable.
Funding
The present study was supported by Major Science and
Technology Projects of Anhui Province (grant no. 17030801014).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MST designed the experiments. ZJ performed the
experiments. MST and ZJ analysed the data. MST wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Tianjin Medical University Cancer Institute and
Hospital (Tianjin, China), and all patients signed informed
consent.
Consent for publication
All participants signed informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Botter SM, Neri D and Fuchs B: Recent
advances in osteosarcoma. Curr Opin Pharmacol. 16:15–23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin PP and Patel S: OsteosarcomaBone
Sarcoma. Springer; New York, NY: pp. 75–97. 2013, View Article : Google Scholar
|
|
3
|
Moore DD and Luu HH:
OsteosarcomaOrthopaedic Oncology. Springer; New York, NY: pp.
65–92. 2014
|
|
4
|
Akiyama T, Dass CR and Choong PF: Novel
therapeutic strategy for osteosarcoma targeting osteoclast
differentiation, bone-resorbing activity, and apoptosis pathway.
Mol Cancer Ther. 7:3461–3469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Perkel JM: Visiting ‘noncodarnia’.
Biotechniques. 54:3012013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ning S, Zhang J, Wang P, Zhi H, Wang J,
Liu Y, Gao Y, Guo M, Yue M, Wang L and Li X: Lnc2Cancer: A manually
curated database of experimentally supported lncRNAs associated
with various human cancers. Nucleic Acids Res. 44:D980–D985. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Y, Li H, Hou S, Hu B, Liu J and Wang
J: The noncoding RNA expression profile and the effect of lncRNA
AK126698 on cisplatin resistance in non-small-cell lung cancer
cell. PLoS One. 8:e653092013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
11
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chan LH, Wang W, Yeung W, Deng Y, Yuan P
and Mak KK: Hedgehog signaling induces osteosarcoma development
through Yap1 and H19 overexpression. Oncogene. 33:4857–4866. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun XH, Yang LB, Geng XL, Wang R and Zhang
ZC: Increased expression of lncRNA HULC indicates a poor prognosis
and promotes cell metastasis in osteosarcoma. Int J Clin Exp
Pathol. 8:2994–3000. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Q, Geng PL, Yin P, Wang XL, Jia JP
and Yao J: Down-regulation of long non-coding RNA TUG1 inhibits
osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J
Cancer Prev. 14:2311–2315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lv J, Ma L, Chen XL, Huang XH and Wang Q:
Downregulation of LncRNAH19 and MiR-675 promotes migration and
invasion of human hepatocellular carcinoma cells through
AKT/GSK-3β/Cdc25A signaling pathway. J Huazhong Univ Sci Technolog
Med Sci. 34:363–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hoesel B and Schmid JA: The complexity of
NF-κB signaling in inflammation and cancer. Mol Cancer. 12:862013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan JX: LncRNA H19 promotes
atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur
Rev Med Pharmacol Sci. 21:322–328. 2017.PubMed/NCBI
|
|
21
|
Hyam SR, Lee IA, Gu W, Kim KA, Jeong JJ,
Jang SE, Han MJ and Kim DH: Arctigenin ameliorates inflammation in
vitro and in vivo by inhibiting the PI3K/AKT pathway and polarizing
M1 macrophages to M2-like macrophages. Eur J Pharmacol. 708:21–29.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han W, Xiong Y, Li Y, Fang W, Ma Y, Liu L,
Li F and Zhu X: Anti-arthritic effects of clematichinenoside (AR-6)
on PI3K/Akt signaling pathway and TNF-α associated with
collagen-induced arthritis. Pharm Biol. 51:13–22. 2013. View Article : Google Scholar : PubMed/NCBI
|