Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory
polyarthritis characterized by synovial heterogeneity (1). Fibroblast-like synoviocytes (FLS)
serve a role in RA, a disease characterized by a continuous
inflammatory response in the synovial membrane, leading to
cartilage and joint damage (2).
FLS, together with other immune cells, including macrophages and
neutrophils, constitute an inflammatory environment in the synovium
and further recruit numerous immune cells, contributing to joint
damage (3). Following stimulation
of inflammatory cytokines, including tumor necrosis factor (TNF-α),
pro-inflammatory signaling is activated in FLS via release of a
number of pro-inflammatory molecules, including interleukin (IL)-6
and IL-1β, which further aggravate the inflammatory processes
(4). Consequently, elucidation of
the mechanisms underlying the inflammatory process in FLS is
necessary for identification of novel targets for treatment of
RA.
The competing endogenous RNA (ceRNA) theory suggests
that transcripts having common miRNA binding sites may modulate the
other's expression by competing for the same miRNAs (5). The ceRNA network has been
demonstrated to serve a role in tumors, pulmonary arterial
hypertension and autophagy (6–9).
However, the role of the ceRNA network in RA remains to be
elucidated.
A number of previous studies demonstrated a role of
miR-155 in RA (10–13). miR-155 has been identified as a
proinflammatory regulator in clinical and experimental arthritis
(10). Transcription factor PU.1
(PU.1), which is upregulated in miR-155 deficient B cells, is a
target of miR-155 (11).
miR-155/PU.1 pathway serves a role in RA, in addition to other
diseases (11–13). However, to the best of the authors
knowledge, the ceRNA network mediated by miR-155 in RA has not been
previously investigated.
Therefore, the present study aimed to determine
whether there is a miR-155-mediated ceRNA network that serves a
role in the inflammatory process and the TNF-α-induced
proliferation and cytokine release of FLS.
Materials and methods
Cells and reagents
Human rheumatoid arthritis FLS (RA-FLS) MH7A cells,
human FLS (HFLS) and 293 cells were purchased from the Cell Bank of
the Chinese Academy of Sciences of China (Shanghai, China). All
cells were cultured in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented
with 15% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in a humidified atmosphere in an environment containing 5%
CO2. TNF-α was purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
To measure the expression level of miR-155, total
RNA extracted from the MH7A and HFLS cells with TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was
reverse-transcribed using miScript II RT kit (Qiagen GmbH, Hilden,
Germany). For determination of PU.1 and FOXO3 mRNA expression,
iScript™ Advanced cDNA Synthesis kit (cat. no. 1725037; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) for RT-PCR was used with the
Prime PCR™ Assay Validation Report primer reagent (cat. no.
1723894; Bio-Rad Laboratories, Inc.). The RT-qPCR experiment was
performed using ABI Prism 7500 Sequence Detector (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The 2−∆∆Cq
method was used to determine the expression of each transcript
(14). U6 small nuclear RNA
(Guangzhou RiboBio Co., Ltd., Guangzhou, China) and GAPDH were used
as control for miR-155 and genes, respectively. Primers are
presented in Table I.
 | Table I.Primer sequences used for polymerase
chain reaction. |
Table I.
Primer sequences used for polymerase
chain reaction.
|
| Sequence (5′→3′) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| Cyclin D1 |
GCCCGCAGCCCCCGCCGGGCCCCGC |
GCGGGGCCCGGCGGGGGCTGCGGGC |
| c-Myc |
TTTTGAGAGGTGGAGAAAGAGATG |
AAAGAAGTGCAGAAATAATGAGCG |
| FOXO3 |
AATAAAGCAGTCAAAAAGAAGTCC |
ACAGCAAGAAAAATAAAACCAGAG |
| Dicer1 |
ACTGTGGAAGTAGTAGGAAAGGGG |
ACAAAGCAGAAGTGAGGAAAGAAG |
| GAPDH |
AGGTCGGTGTGAACGGATTTG |
GGGGTCGTTGATGGCAACA |
| PU.1-CDS |
CTAGACTAGTATGTTACAGGC |
CTAGTCTAGATCAGTGGGGCG |
|
| GTGCAAAATGGAAG | GGTGGCGCCGCTCG |
| PU.1 3′UTR |
CTAGACTAGTCCAGGCCT |
CTAGTCTAGAGATGGATT |
|
|
CCCCGCTGGCCATAGCA |
GAGAATAACTTTACTTG |
| miR-155 |
TTAATGCTAATGTGTAGGGGT |
ACCCCTACACATTAGCATTAA |
Plasmid construction and cell
transfection
Lentiviral small hairpin (sh)RNA against human Dicer
1 (Lenti-shDicer), FOXO3 (Lenti-shFOXO3), PU.1 (Lenti-shPU.1) and a
scrambled non-targeting shRNA were purchased from Sigma-Aldrich
(Merck KGaA), and cloned into pLKO.1 vector (Sigma-Aldrich, Merck
KGaA). PU.1 3′UTR or coding sequences of PU.1 were amplified by
polymerase chain reaction (PCR) and PU.1 3′UTR with mutant-type of
miR-155 binding site (mut) was synthesized using Fast Multi Site
Mutagenesis System (Beijing Transgen Biotech Co., Ltd., Beijing,
China). PCR-amplified sequences of PU.1 3′UTR, PU.1 3′UTR with
mutant-type of miR-155 binding site or coding sequences of PU.1
were cloned using SpeI and Xbal sites of
pLVX-IRES-ZsGreen1 vector (Hunan China Sun Medical Devices, Co.,
Ltd., Hunan, China), referred to as Lenti-PU.1-3′UTR,
Lenti-PU.1-3′UTR-mut and Lenti-PU.1-coding sequence (CDS). The
temperature protocol for PCR was as follows: Initial degeneration
at 95°C for 5 min, followed by 35 cycles of degeneration at 95°C
for 40 sec, annealing at 58°C for 30 sec, extension at 72°C for 2
min and a final extension step at 72°C for 10 min. The primers used
for PCR using 2×Taq PCR MasterMix (cat. no. PC1120; Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) are
presented in Table I. The cDNA
reverse transcribed from total RNA, according to the aforementioned
protocol, was used as the template. Lentiviral particles were
packaged in 293 cells, as previously described (15).
miR-155 mimic/inhibitor (mimics,
5′-UUAAUGCUAAUGUGUAGGGGU-3′; inhibitor, 5′-AAUUACGAUUAUACAUCCUC-3′)
and negative control (NC; 5′-GGCGCUAGCGUCACGUUUA-3′) were
synthesized by Guangzhou RiboBio Co., Ltd..
Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to transfect miR-155 mimic, inhibitor
and NC (all 5 nM).
Cell viability assay
To determine the effects of PU.1 3′UTR on RA-FLS
proliferation, MH7A cells were transfected with Lenti-PU.1-3′UTR
and subsequently treated with 50 ng/ml of TNF-α for 1 h (5
wells/group). Cells that were untreated served as the control
group. A total of 24, 48 and 72 h following Lenti-PU.1-3′UTR
transfection, proliferation of MH7A was detected using an MTT assay
kit (Beyotime Institute of Biotechnology, Haimen, China) following
the manufacturer's protocol. Acetic acid (30%) was used to dissolve
the purple formazan and a 570 nm wavelength was used to measure
formazan.
ELISA
To determine the effect of PU.1 3′UTR on
TNF-α-induced inflammation of MH7A cells, MH7A cells were
transfected with Lenti-PU.1-3′UTR and subsequently treated with 50
ng/ml of TNF-α for 1 h (n=5/group). A total of 48 h following
transfection with Lenti-PU.1-3′UTR, the concentration of IL-6 (cat.
no. ab46042) and IL-1β (cat. no. ab214025; both Abcam, Cambridge,
UK) in cell culture supernatant (obtained by centrifugation at
13,500 × g for 5 min at room temperature) was performed using IL-6
and IL-1β ELISA kits according to the manufacturer's protocol.
Transwell migration and invasion
assays
To determine the effects of PU.1 3′UTR on RA-FLS
MH7A migration and invasion ability, Transwell migration and
invasion assays were carried out using 24-well Millicell Hanging
Cell Culture inserts, Polyethylene Terephthalate 8 µm (Merck KGaA,
Darmstadt, Germany), which were coated without or with Matrigel
matrix gel (BD Biosciences, Franklin Lakes, NJ, USA). The detailed
procedure was performed as previously described (16).
Adhesion assay
To determine the effect of PU.1 3′UTR on RA-FLS MH7A
adhesion ability, the colorimetric MTT assay was used to determine
the number of adherent cells. Acetic acid (30%) was used to
dissolve purple formazan and 570 nm wavelength was used to measure
formazan. Detailed procedure was conducted as previously described
(17).
Luciferase reporter assay
To determine whether PU.1 3′UTR mediated its effect
through the 3′UTR, pMIR-Report vector (Promega Corporation,
Madison, WI, USA) was used to clone the fragments of PU.1 3′UTR and
FOXO3 3′UTR, named Luc-PU.1-3′UTR and Luc-FOXO3-3′UTR,
respectively. To investigate whether PU.1-mediated regulation of
FOXO3 depends on the 3′UTR, MH7A cells overexpressing PU.1 3′UTR or
PU.1 shRNA were co-transfected with Luc-FOXO3-3′UTR or an empty
vector and β-gal expression vector (Hunan China Sun Medical
Devices, Co., Ltd.) using Lipofectamine® 2000 according
to the manufacturer's protocols. Transfection efficiency was
determined using a Luciferase Reporter Assay kit (cat. no.
K801-200; BioVision, Inc., Milpitas, CA, USA). β-gal activity was
determined using a β-Galactosidase Enzyme Assay System with
Reporter Lysis Buffer (cat. no. E2000; Promega Corporation) for
normalization, according to the manufacturer's protocols.
RNA immune co-precipitation (RIP)
assays
The RIP assays were performed as previously
described (16). Total RNA in the
RIP assay product was determined via the aforementioned RT-qPCR
protocol.
Western blot analysis
Detailed procedure was previously described
(7). Antibodies against PU.1 (cat.
no. ab76543; 1:5,000), FOXO3 (cat. no. ab12162; 1:4,000), Cyclin D1
(cat. no. ab134175; 1:3,000), c-Myc (cat. no. ab32072; 1:5,000) and
Dicer (cat. no. ab14601; 1:2,000) were all purchased from Abcam.
β-actin (cat. no. AM1021B; 1:5,000) was purchased from OriGene
Technologies, Inc. (Rockville, MD, USA). Membranes were blocked
with 5% non-fat milk and incubated with the primary antibodies in
5% bovine serum albumin (cat. no. A8020; Beijing Solarbio Science
& Technology Co., Ltd.) at 4°C overnight, followed by
incubation with the following secondary antibodies: Horseradish
peroxidase (HRP)-conjugated goat-anti-rabbit (cat. no. TA130003;
1:5,000) and HRP-conjugated goat-anti-mouse (cat. no. THRPA140003;
1:5,000; both OriGene Technologies, Inc.). Blots were washed 3
times for 15 min each, and the signals were detected using enhanced
chemiluminescence detection kit (Thermo Fisher Scientific, Inc.)
followed by exposure and densitometric analysis was performed using
the Tanon 5200 automatic chemiluminescence image analysis system
(Tanon Science & Technology Co., Ltd., Shanghai, China).
β-actin was used as an internal control.
Statistical analysis
All data were obtained from at least three
independent experiments, and presented as the mean ± standard
deviation. Datasets with only two groups were analyzed using a
Student's t-test. Differences between multiple groups were analyzed
using one-way analysis of variance with the Tukey-Kramer post-hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
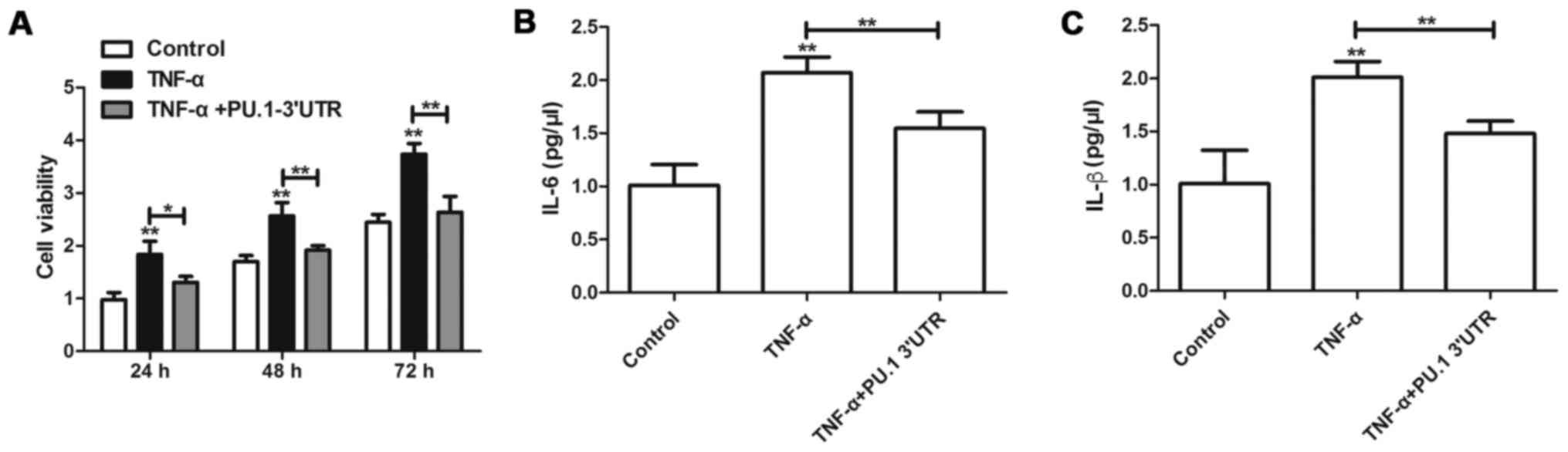
PU.1 3′UTR inhibits TNF-α-induced
proliferation and inflammation of MH7A cells
To determine the effects of PU.1 3′UTR on
RA-FLSMH7Aproliferation, MH7A cells were transfected with
Lenti-PU.1-3′UTR for 6 h and subsequently treated with 50 ng/ml of
TNF-α for 1 h. A total of 24, 48 and 72 h following transfection
with Lenti-PU.1-3′UTR, proliferation of MH7A was assessed using an
MTT assay. The results indicated that, compared with the control
group, TNF-α induced the cell viability of MH7A cells (Fig. 1A). However, transfection with
Lenti-PU.1-3′UTR inhibited the proliferation induced by TNF-α. To
determine the effects of PU.1 3′UTR on inflammation, MH7A cells
were transfected with Lenti-PU.1-3′UTR and subsequently treated
with TNF-α for 48 h. ELISA analyses were used to detect the
concentration of IL-6 and IL-1β in cell culture supernatant.
Overexpression of PU.1 3′UTR significantly attenuated TNF-α-induced
production of IL-6 and IL-1β (Fig. 1B
and C).
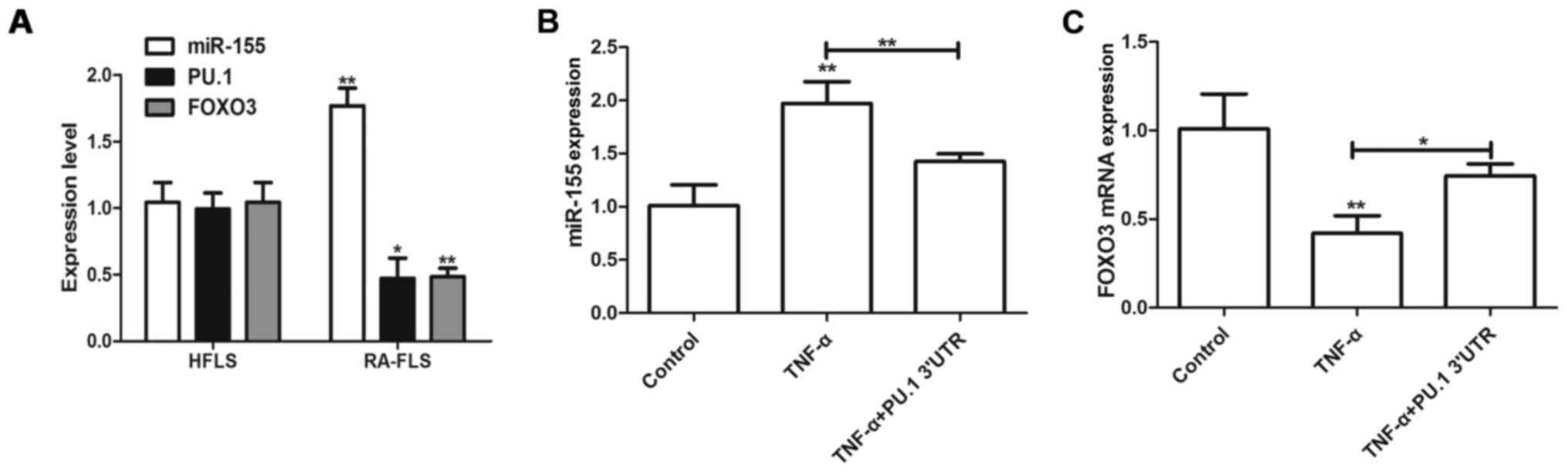
PU.1 3′UTR inhibits TNF-α-induced
miR-155 expression
miR-155/PU.1 and miR-155/FOXO3 signaling pathways
have been previously demonstrated to serve roles in RA and TNF-α
was demonstrated to induce miR-155 expression in RA-FLS (2). The present study aimed to detect
expression levels of miR-155 and FOXO3 in RA-FLS and HFLS. miR-155
level was upregulated in RA-FLS, however PU.1 and FOXO3 expression
decreased, compared with the HFLS cells (Fig. 2A). Furthermore, transfection with
Lenti-PU.1-3′UTR inhibited the TNF-α-mediated upregulation of
miR-155 (Fig. 2B). Furthermore,
transfection with Lenti-PU.1-3′UTR reversed the TNF-α-mediated
inhibition of FOXO3 (Fig. 2C).
Therefore, the results of the present study demonstrated that PU.1
3′UTR may regulate RA and be associated with miR-155 and FOXO3
activities.
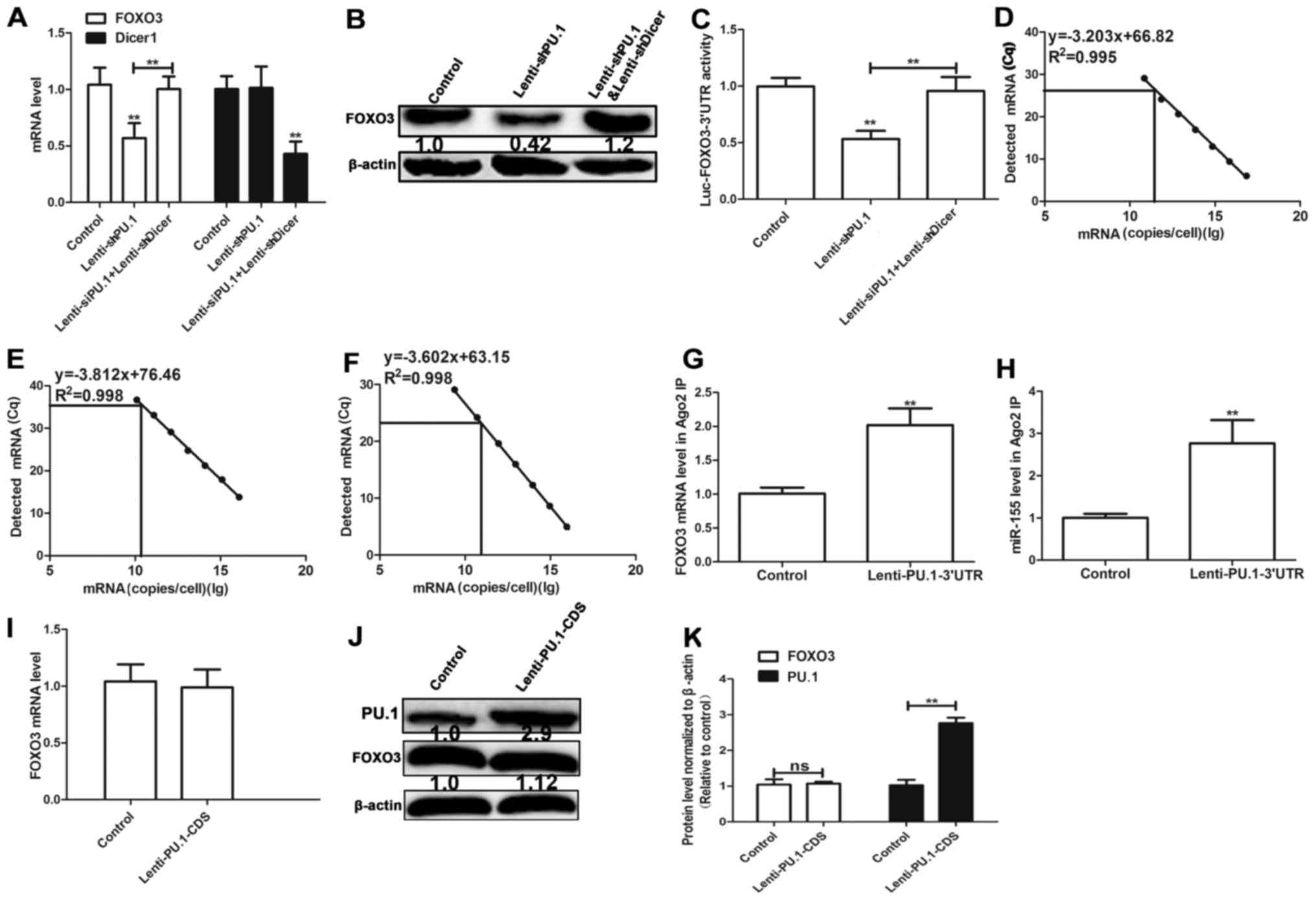
PU.1 3′UTR promotes FOXO3 expression
by acting as a ceRNA for FOXO3
Since PU.1 3′UTR and FOXO3 3′UTR both bind miR-155,
PU.1 3′UTR may serve a role of a ceRNA for FOXO3 in MH7A cells. It
was hypothesized in the present study that the effect of PU.1 3′UTR
on FOXO3 expression is mediated by miRNAs. Cell lines with Dicer 1
knockdown, a ribonuclease critical for miRNA biogenesis, were used
in the present study. Dicer 1 deficiency significantly reduces
levels of mature miRNAs (6).
Transfection with Lenti-shDicer1 demonstrated 60% knockdown
efficiency and knockdown of PU.1 decreased FOXO3 expression in
Dicer 1-proficient cells, however not that in isogenic Dicer 1
knockdown cells (Fig. 3A and B).
In order to determine whether PU.1 3′UTR mediated its effect
through its 3′UTR, FOXO3 3′UTR-luciferase reporter
(Luc-FOXO3-3′UTR) was co-expressed in Dicer 1-proficient and
-deficient MH7A cells with PU.1 knockdown. Knockdown of PU.1
decreased the activity of Luc-FOXO3-3′UTR, which was reversed in
Dicer 1-knockdown cells (Fig. 3C).
The absolute number of PU.1, FOXO3 and miR-155 transcripts was
determined by RT-qPCR combined with an internal standard curve to
determine their abundance (18).
The results revealed that miR-155, PU.1 and FOXO3 were expressed at
8.66×1010, 1.16×1010 and 5.22×1010
copies/cell multiplicity of infection in MH7A cells, respectively
(Fig. 3D-F). RIP assay
demonstrated that PU.1 3′UTR overexpression facilitated the
association between protein argonaute-2 (ago2) and FOXO3 mRNA
(Fig. 3G) and the association
between ago2 and miR-155 (Fig.
3H). The results indicate that miR-155 directly binds PU.1 and
FOXO3. Additionally, ectopic expression of PU.1-coding sequence
(CDS) increased PU.1 protein level, however did not affect the
protein level of FOXO3 (Fig.
3I-K). The aforementioned results suggest that PU.1 3′UTR
promotes FOXO3 expression by acting as a ceRNA for FOXO3.
PU.1 3′UTR inhibition of TNF-α-induced
proliferation and inflammation of MH7A cells is dependent on FOXO3
expression
Following elucidation of the interaction between
PU.1 and FOXO3, potential relevance of this phenotype for
TNF-α-induced proliferation and inflammation was investigated in
RA-FLS MH7A cells. MH7A cells were transfected with
Lenti-PU.1-3′UTR and Lenti-shFOXO3 for 6 h, and treated with 50
ng/ml TNF-α for 1 h. Following 24, 48 and 72 h of treatment,
proliferation of MH7A was assessed using an MTT assay. The results
indicated that knockdown of FOXO3 attenuated the inhibitory effect
of Lenti-PU.1-3′UTR on TNF-α induced proliferation of MH7A cells
(Fig. 4A). Expression levels of
cell proliferation markers cyclin D1 and c-Myc were significantly
inhibited in TNF-α activated MH7A cells following PU.1 3′UTR
overexpression (Fig. 4B-D).
However, these effects were reversed in MH7A cells co-transfected
with Lenti-shFOXO3. Similarly, knockdown of FOXO3 reversed the
anti-inflammatory effects of PU.1 3′UTR and resulted in
upregulation of IL-6 and IL-β levels, compared with the
TNF-α+Lenti-PU.1-3′UTR group (Fig. 4E
and F).
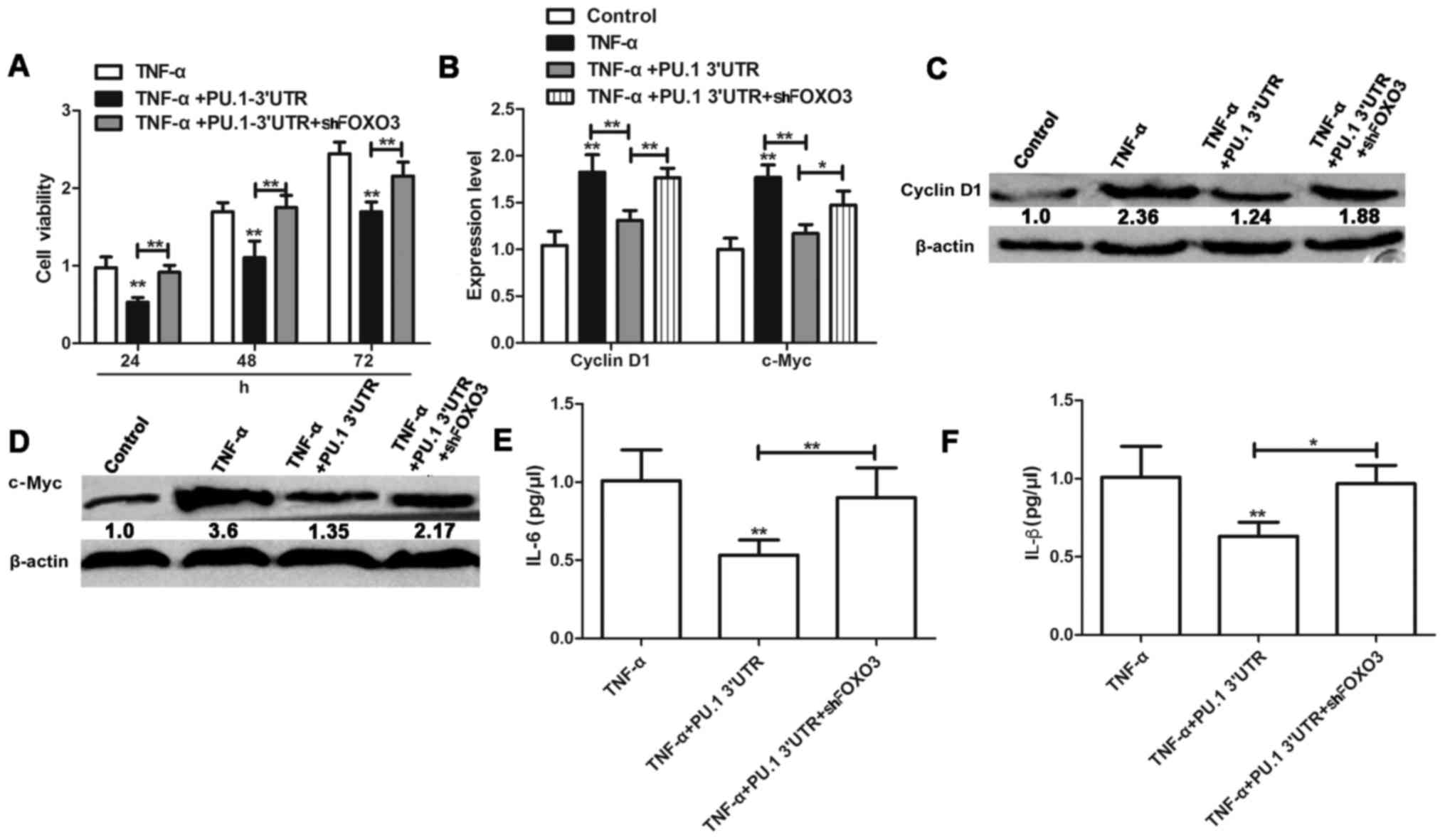 | Figure 4.PU.1 3′UTR inhibits TNF-α-induced
proliferation and inflammation of MH7A cells dependent on FOXO3
expression. (A) Viability of MH7A cells transfected with
Lenti-PU.1-3′UTR and Lenti-shFOXO3 or untreated prior to incubation
with TNF-α, was evaluated by MTT assay. (B) mRNA expression level
of cyclin D1 and c-Myc. Expression of (C) cyclin D1, (D) c-Myc, (E)
IL-6 and (F) IL-1β were detected in cells transfected or not
transfected with Lenti-PU.1-3′UTR+Lenti-shFOXO3 prior to TNF-α
treatment. Data are presented as the mean ± standard deviation.
n=3. *P<0.05 and **P<0.01 vs. the respective control group.
PU.1, transcription factor PU.1; UTR, untranslated region; TNF-α,
tumor necrosis factor-α; FOXO3, forkhead box protein O3; sh, small
hairpin RNA; c-Myc, Myc proto-oncogene protein; IL,
interleukin. |
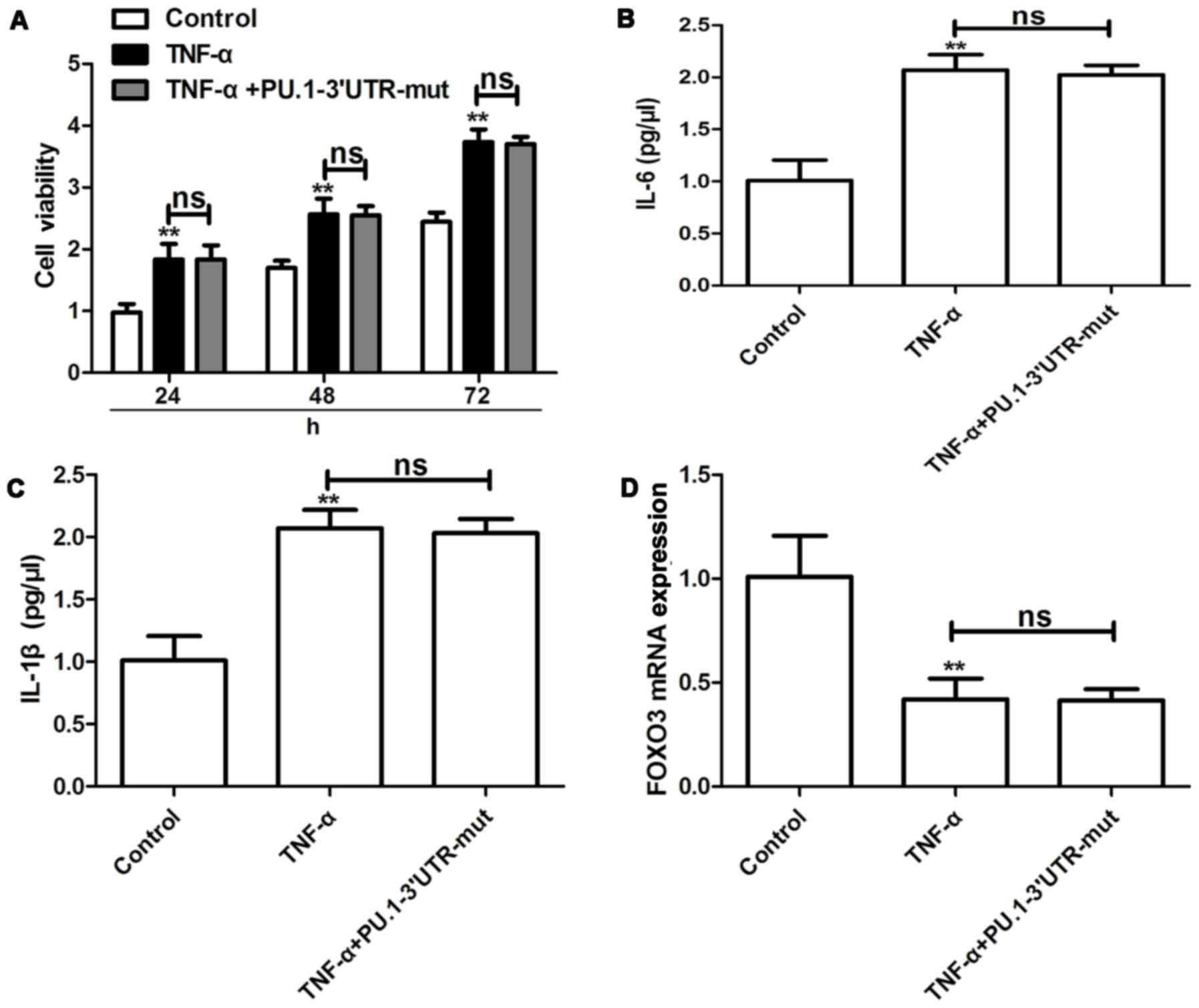
Effects of PU.1 3′UTR with a mutant type of miR-155
binding site on TNF-α-induced proliferation, cytokine release and
FOXO3 expression were investigated in RA-FLSMH7A cells. It was
determined that PU.1 3′UTR-mut exhibited no effect on TNF-α-induced
proliferation (Fig. 5A), cytokine
release (Fig. 5B and C) and FOXO3
expression (Fig. 5D) in RA-FLSMH7A
cells. The aforementioned results suggest that PU.1 3′UTR inhibits
TNF-α-induced RA-FLS proliferation by upregulation of FOXO3 via
competitive binding of miR-155.
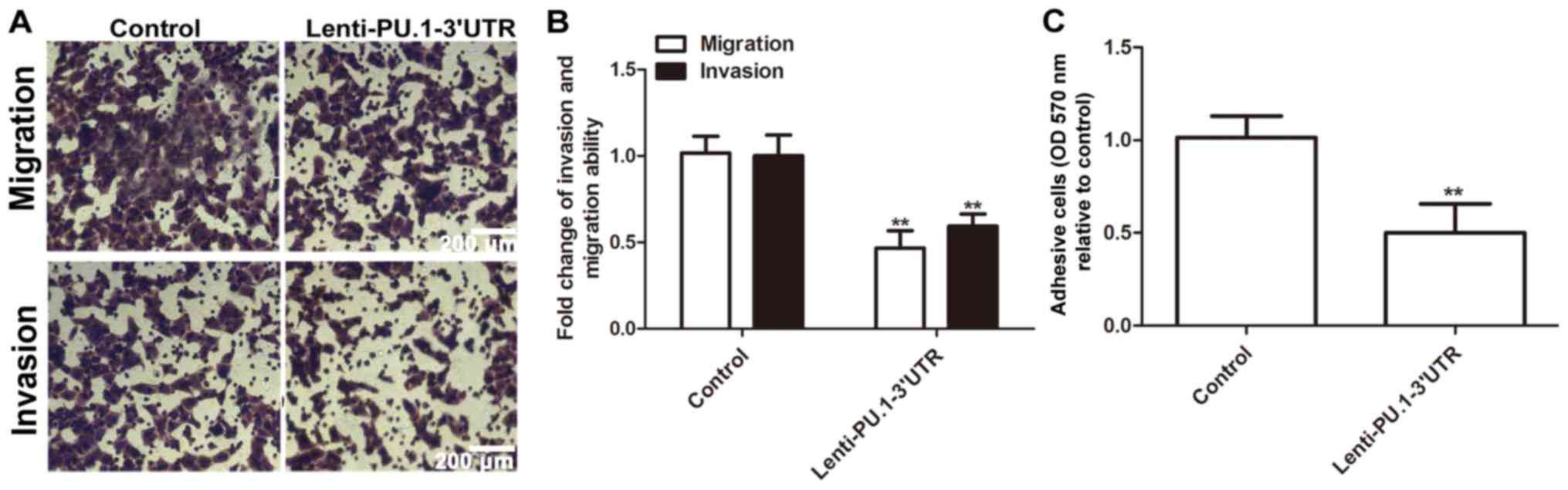
PU.1 3′UTR inhibits MH7A cell
migration, invasion and adhesion
Following determination that RA-FLS exhibit an
enhanced invasion phenotype, the present study further investigated
whether PU.1 3′UTR modulated RA-FLSMH7A migration, invasion and
adhesion abilities. Infection with Lenti-PU.1-3′UTR markedly
reduced migration and invasion abilities of MH7A cells (Fig. 6A and B). Additionally,
overexpression of PU.1 3′UTR significantly attenuated the adhesive
ability of MH7A cells (Fig. 6C).
Therefore, the results of the present study demonstrated that
ectopic expression of PU.1 3′UTR significantly inhibited migration,
invasion and adhesion of MH7A cells.
Discussion
RA-FLS cells serve a role in RA and have been
hypothesized to be associated with inflammation and joint damage in
RA (1,3). Currently, although the immune system
is the target for treatment of RA, developing novel drugs is
necessary (19). Therefore, RA-FLS
maybe targeted in novel methods of treatment of RA.
In the present study, the effects of PU.1 3′UTR on
TNF-α-induced proliferation of RA-FLS and the associated release of
pro-inflammatory cytokines were investigated. It was determined
that PU.1 3′UTR protected against TNF-α-induced RA-FLS
proliferation, decreased the expression of cell proliferation
markers cyclin D1 and c-Myc, and reduced the levels of inflammatory
factors IL-6 and IL-1β. ceRNA network between RNA transcripts
mediated by shared miRNAs represents a novel method of gene
regulation, which confers novel functions to non-coding RNAs
(ncRNAs) or non-coding parts of mRNAs, for example 3′UTRs (6). ceRNA network has been demonstrated to
serve a role in development (20).
However, the roles of the ceRNA network in RA remain to be
elucidated.
To determine whether PU.1 3′UTR exerts its functions
by acting as a ceRNA in RA, FOXO3 has been investigated in the
present study based on the observation that PU.1 and FOXO3 are
potential targets of miR-155 in inflammation (11–13).
miRNA-155 is a proinflammatory regulator in clinical and
experimental arthritis (10),
these results are consistent with the fact that ceRNAs and the
shared miRNAs primarily hold the opposite effects (7). The present study demonstrated that
PU.1 3′UTR-mediated upregulation of FOXO3 expression was dependent
on the activity of miRNA. The aforementioned observation indicated
that a ceRNA network is involved in the development of RA. However,
multiple miRNAs may interact with a single 3′UTR. Future studies
should therefore aim to investigate other ceRNAs, including ncRNAs,
pseudogenes and circular RNAs, which maybe associated with the
ceRNA network and therefore modulate RA pathogenesis.
In conclusion, the present study demonstrated that
ceRNA interactions may mediate the crosstalk between different
signaling pathways during RA development. Further analysis of ceRNA
interactions between PU.1 and FOXO3 during RA development may
provide insights into gene regulatory networks and their clinical
implications for RA.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
KW and BD designed the research included in the
present study; ZX, YQ and PS analyzed the data; ZX, YQ, PS and BW
performed the research; and ZX and YQ wrote the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dennis G Jr, Holweg CT, Kummerfeld SK,
Choy DF, Setiadi AF, Hackney JA, Haverty PM, Gilbert H, Lin WY,
Diehl L, et al: Synovial phenotypes in rheumatoid arthritis
correlate with response to biologic therapeutics. Arthritis Res
Ther. 16:R902014. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu N, Feng X, Wang W, Zhao X and Li X:
Paeonol protects against TNF-α-induced proliferation and cytokine
release of rheumatoid arthritis fibroblast-like synoviocytes by
upregulating FOXO3 through inhibition of miR-155 expression.
Inflamm Res. 66:603–610. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chang SK, Gu Z and Brenner MB:
Fibroblast-like synoviocytes in inflammatory arthritis pathology:
the emerging role of cadherin-11. Immunol Rev. 233:256–266. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartok B and Firestein GS: Fibroblast-like
synoviocytes: Key effector cells in rheumatoid arthritis. Immunol
Rev. 233:233–255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li X, Zheng L, Zhang F, Hu J, Chou J, Liu
Y, Xing Y and Xi T: STARD13-correlated ceRNA network inhibits EMT
and metastasis of breast cancer. Oncotarget. 7:23197–23211.
2016.PubMed/NCBI
|
|
7
|
Zheng L, Li X, Gu Y, Lv X and Xi T: The
3′UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in
breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res
Treat. 150:105–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma K, Zhao Q and Li S: Competing
endogenous RNA network in pulmonary arterial hypertension. Int J
Cardiol. 172:e527–e528. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ge D, Han L, Huang S, Peng N, Wang P,
Jiang Z, Zhao J, Su L, Zhang S, Zhang Y, et al: Identification of a
novel MTOR activator and discovery of a competing endogenous RNA
regulating autophagy in vascular endothelial cells. Autophagy.
10:957–971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kurowska-Stolarska M, Alivernini S,
Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna
M, Fraser AR, Stolarski B, et al: MicroRNA-155 as a proinflammatory
regulator in clinical and experimental arthritis. Proc Natl Acad
Sci USA. 108:11193–11198. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alivernini S, Kurowska-Stolarska M,
Tolusso B, Benvenuto R, Elmesmari A, Canestri S, Petricca L,
Mangoni A, Fedele AL, Di Mario C, et al: MicroRNA-155 influences
B-cell function through PU.1 in rheumatoid arthritis. Nat Commun.
7:129702016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu D, Nakagawa R, Lazzaro S, Staudacher P,
Abreu-Goodger C, Henley T, Boiani S, Leyland R, Galloway A, Andrews
S, et al: The miR-155-PU.1 axis acts on Pax5 to enable efficient
terminal B cell differentiation. J Exp Med. 211:2183–2198. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huskova H, Korecka K, Karban J, Vargova J,
Vargova K, Dusilkova N, Trneny M and Stopka T: Oncogenic
microRNA-155 and its target PU.1: An integrative gene expression
study in six of the most prevalent lymphomas. Int J Hematol.
102:441–450. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang H, Wang F and Hu Y: STARD13 promotes
hepatocellular carcinoma apoptosis by acting as a ceRNA for Fas.
Biotechnol Lett. 39:207–217. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y and Lin G: TP53INP1 3′-UTR
functions as a ceRNA in repressing the metastasis of glioma cells
by regulating miRNA activity. Biotechnol Lett. 38:1699–1707. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang J, Li T, Gao C, Lv X, Liu K, Song H,
Xing Y and Xi T: FOXO1 3′UTR functions as a ceRNA in repressing the
metastases of breast cancer cells via regulating miRNA activity.
FEBS Lett. 588:3218–3224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Denzler R, Agarwal V, Stefano J, Bartel DP
and Stoffel M: Assessing the ceRNA hypothesis with quantitative
measurements of miRNA and target abundance. Mol Cell. 54:766–776.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu J, Wang H, Chen J and Wei W:
Inhibition of plasma kallikrein-kinin system to alleviate renal
injury and arthritis symptoms in rats with adjuvant-induced
arthritis. Immunopharmacol Immunotoxicol. 40:134–148. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Feng L, Han Z, Li Y, Wu A, Shao T,
Ding N, Li L, Deng W, Di X, et al: Extensive ceRNA-ceRNA
interaction networks mediated by miRNAs regulate development in
multiple rhesus tissues. Nucleic Acids Res. 44:9438–9451.
2016.PubMed/NCBI
|