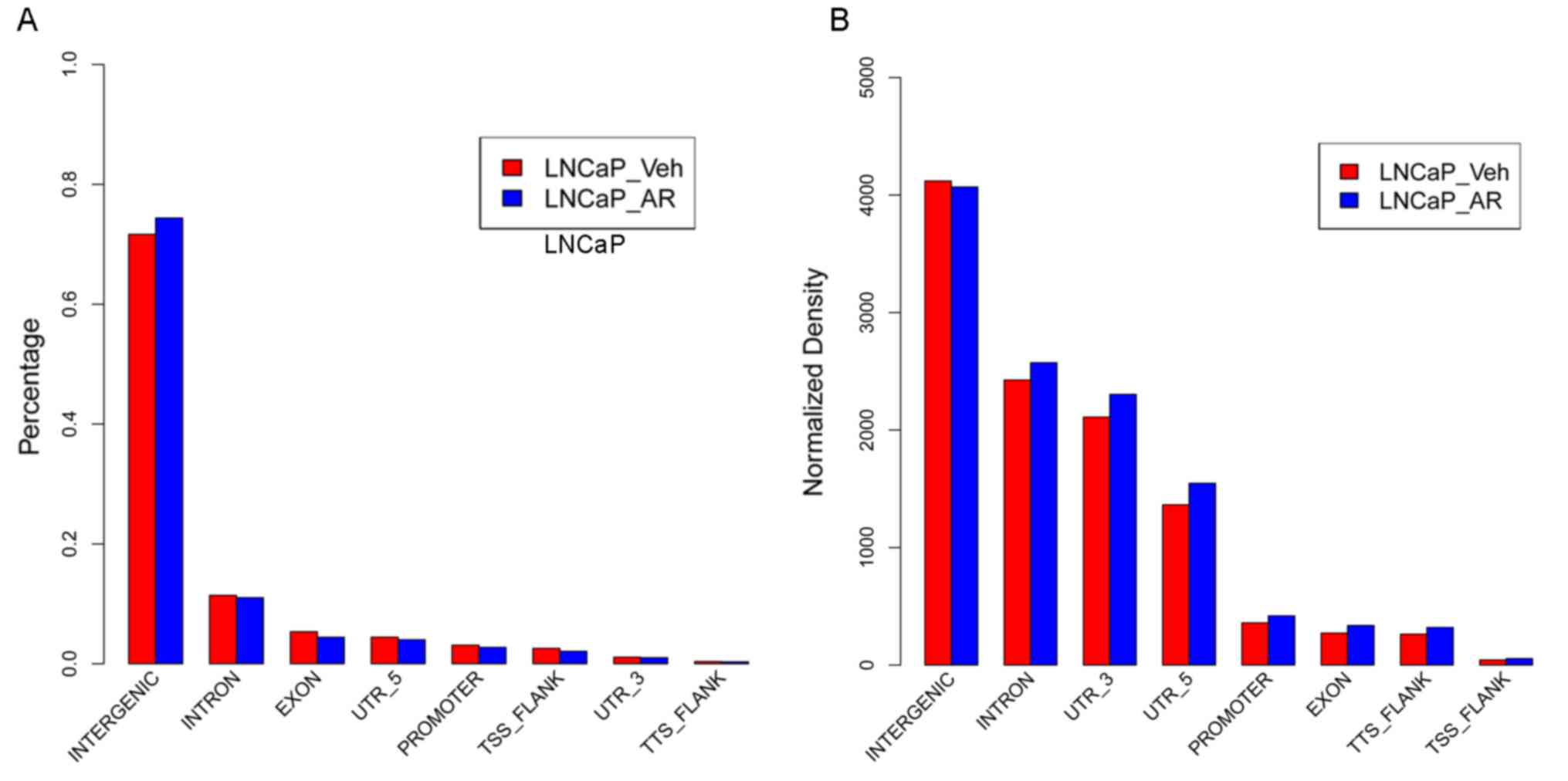
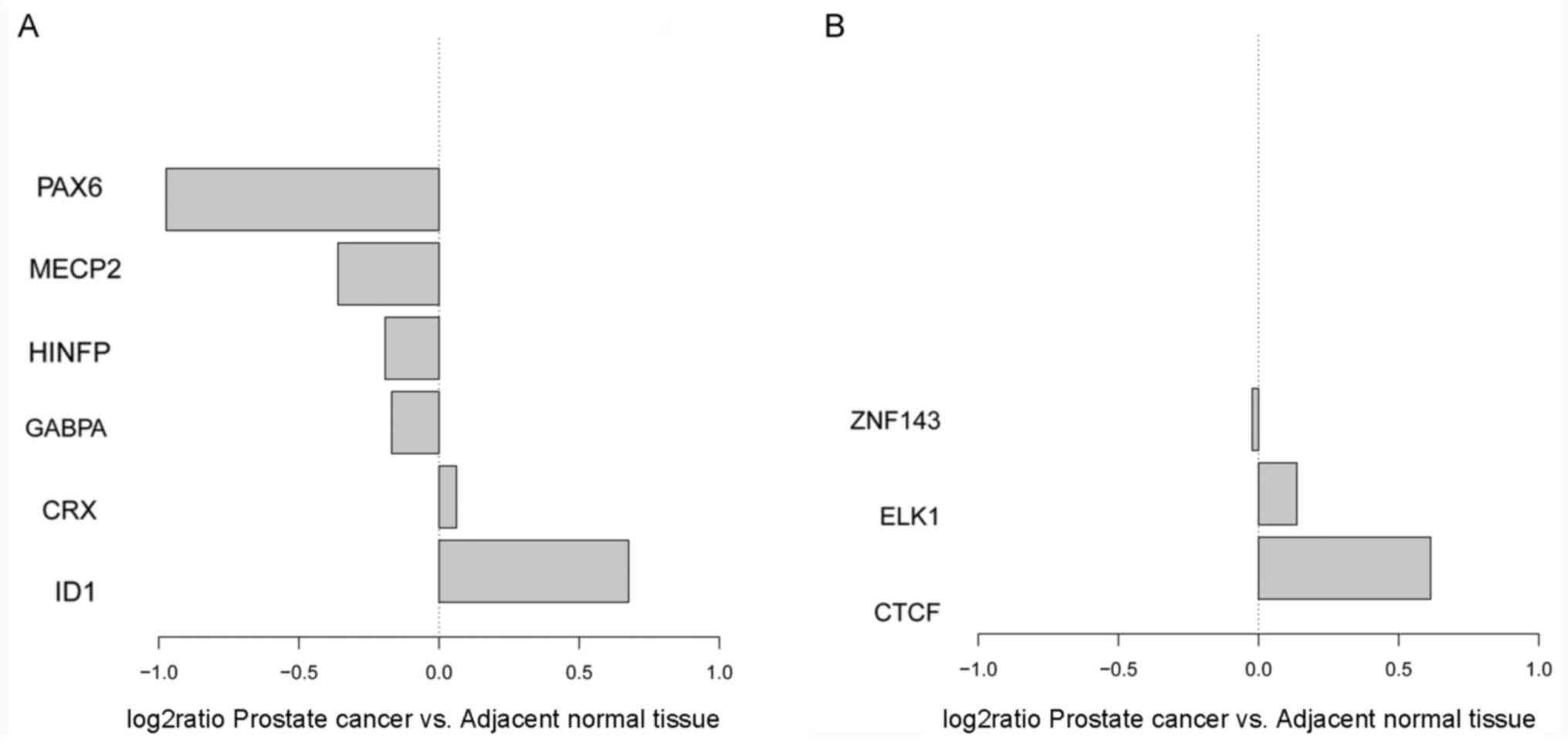
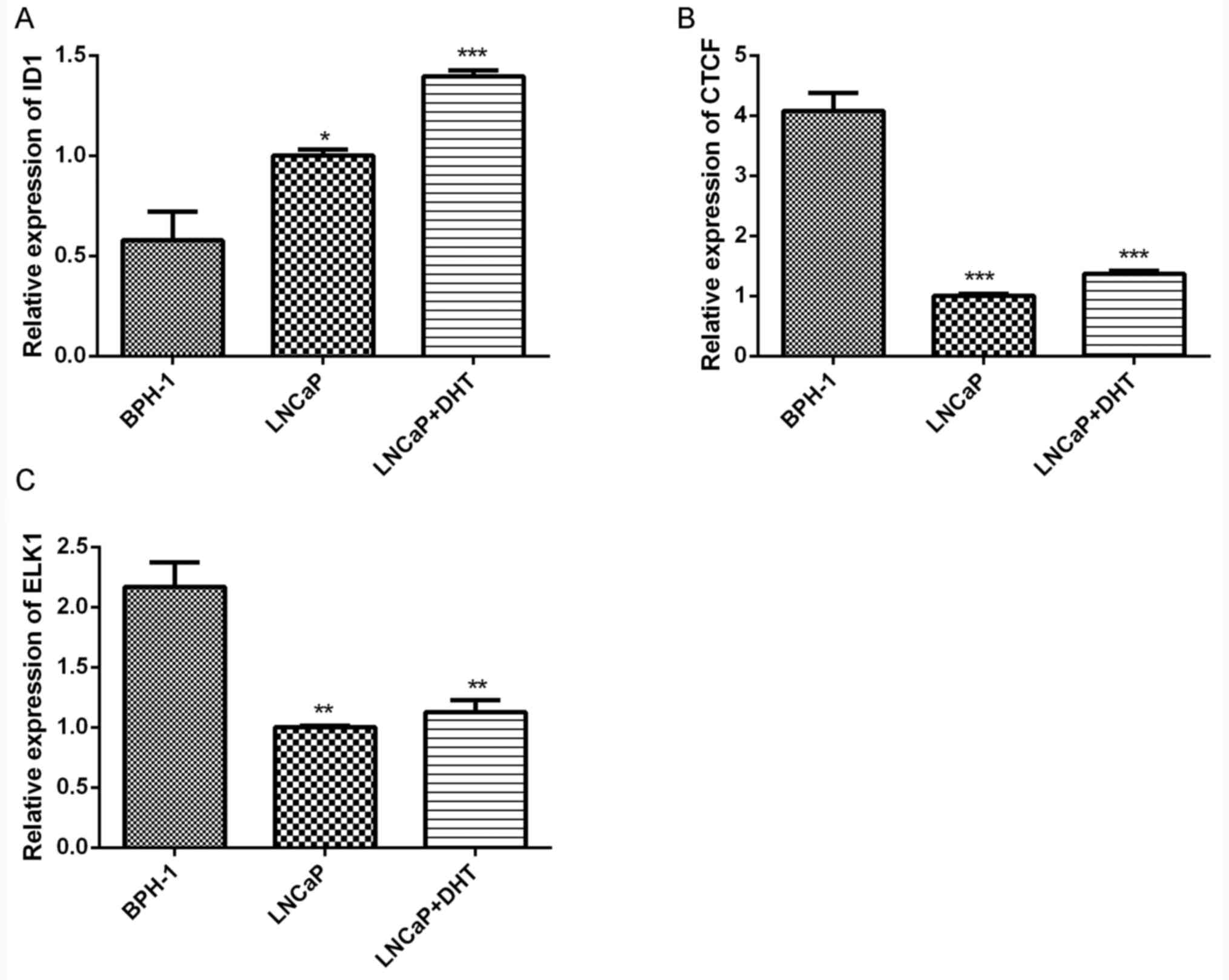
|
1
|
Heidenreich A, Bellmunt J, Bolla M, Joniau
S, Mason M, Matveev V, Mottet N, Schmid HP, van der Kwast T, Wiegel
T, et al: EAU guidelines on prostate cancer. Part 1: Screening,
diagnosis, and treatment of clinically localised disease. Eur Urol.
59:61–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Howlader N, Noone A, Krapcho M, Neyman N,
Aminou R, Waldron W, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z,
et al: SEER cancer statistics review, 1975–2008. Bethesda, MD:
National Cancer Institute; 2011, Also available online. Last
accessed. December 1–2011
|
|
3
|
Grignon DJ: Unusual subtypes of prostate
cancer. Mod Pathol. 17:316–327. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Culig Z, Hobisch A, Cronauer MV, Radmayr
C, Trapman J, Hittmair A, Bartsch G and Klocker H: Androgen
receptor activation in prostatic tumor cell lines by insulin-like
growth factor-I, keratinocyte growth factor, and epidermal growth
factor. Cancer Res. 54:5474–5478. 1994.PubMed/NCBI
|
|
7
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Heinlein CA and Chang C: Androgen receptor
(AR) coregulators: An overview. Endocr Rev. 23:175–200. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Edwards J, Krishna N, Grigor K and
Bartlett J: Androgen receptor gene amplification and protein
expression in hormone refractory prostate cancer. Br J Cancer.
89:552–556. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hong H, Kao C, Jeng MH, Eble JN, Koch MO,
Gardner TA, Zhang S, Li L, Pan CX, Hu Z, et al: Aberrant expression
of CARM1, a transcriptional coactivator of androgen receptor, in
the development of prostate carcinoma and androgen-independent
status. Cancer. 101:83–89. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin DL, Whitney MC, Yao Z and Keller ET:
Interleukin-6 induces androgen responsiveness in prostate cancer
cells through up-regulation of androgen receptor expression. Clin
Cancer Res. 7:1773–1781. 2001.PubMed/NCBI
|
|
12
|
Malinowska K, Neuwirt H, Cavarretta IT,
Bektic J, Steiner H, Dietrich H, Moser PL, Fuchs D, Hobisch A and
Culig Z: Interleukin-6 stimulation of growth of prostate cancer in
vitro and in vivo through activation of the androgen receptor.
Endocr Relat Cancer. 16:155–169. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pepke S, Wold B and Mortazavi A:
Computation for ChIP-seq and RNA-seq studies. Nat Methods. 6 11
Suppl:S22–S32. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He HH, Meyer CA, Chen MW, Jordan VC, Brown
M and Liu XS: Differential DNase I hypersensitivity reveals
factor-dependent chromatin dynamics. Genome Res. 22:1015–1025.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu FY, Zhao XM, Tang NL, Zhang Y and Chen
L: Comparative analysis of protein-coding genes and long non-coding
RNAs of prostate cancer between Caucasian and Chinese populations.
Systems Biology (ISB), 2012 IEEE 6th International Conference on
IEEE. 291–296. 2012. View Article : Google Scholar
|
|
16
|
Li H and Durbin R: Fast and accurate
long-read alignment with Burrows-Wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup: The sequence alignment/map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Liu T, Meyer CA, Eeckhoute J,
Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W and
Liu XS: Model-based analysis of ChIP-Seq (MACS). Genome Biol.
9:R1372008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ewert R: Broadband slat noise prediction
based on CAA and stochastic sound sources from a fast random
particle-mesh (RPM) method. Comput Fluids. 37:369–387. 2008.
View Article : Google Scholar
|
|
22
|
Tarazona S, García-Alcalde F, Dopazo J,
Ferrer A and Conesa A: Differential expression in RNA-seq: A matter
of depth. Genome Res. 21:2213–2223. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsu H and Lachenbruch PA: Paired t test.
Wiley Encyclopedia of Clinical Trials. 2008. View Article : Google Scholar
|
|
24
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao M, Sun J and Zhao Z: TSGene: A web
resource for tumor suppressor genes. Nucleic Acids Res.
41:D970–D976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He HH, Meyer CA, Shin H, Bailey ST, Wei G,
Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al: Nucleosome dynamics
define transcriptional enhancers. Nat Genet. 42:343–347. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schwarzenbach H, Chun FK, Isbarn H, Huland
H and Pantel K: Genomic profiling of cell-free DNA in blood and
bone marrow of prostate cancer patients. J Cancer Res Clin Oncol.
137:811–819. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Steinkamp MP, O'Mahony OA, Brogley M,
Rehman H, Lapensee EW, Dhanasekaran S, Hofer MD, Kuefer R,
Chinnaiyan A, Rubin MA, et al: Treatment-dependent androgen
receptor mutations in prostate cancer exploit multiple mechanisms
to evade therapy. Cancer Res. 69:4434–4442. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Costa VL, Henrique R and Jerónimo C:
Epigenetic markers for molecular detection of prostate cancer. Dis
Markers. 23:31–41. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Perk J, Iavarone A and Benezra R: Id
family of helix-loop-helix proteins in cancer. Nat Rev Cancer.
5:603–614. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Parrinello S, Lin CQ, Murata K, Itahana Y,
Singh J, Krtolica A, Campisi J and Desprez PY: Id-1, ITF-2, and
Id-2 comprise a network of helix-loop-helix proteins that regulate
mammary epithelial cell proliferation, differentiation, and
apoptosis. J Biol Chem. 276:39213–39219. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kebebew E, Treseler PA, Duh QY and Clark
OH: The helix-loop-helix transcription factor, Id-1, is
overexpressed in medullary thyroid cancer. Surgery. 128:952–957.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
O'Toole PJ, Inoue T, Emerson L, Morrison
IE, Mackie AR, Cherry RJ and Norton JD: Id proteins negatively
regulate basic helix-loop-helix transcription factor function by
disrupting subnuclear compartmentalization. J Biol Chem.
278:45770–45776. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ao J, Meng J, Zhu L, Nie H, Yang C, Li J,
Gu J, Lin Q, Long W, Dong X and Li C: Activation of androgen
receptor induces ID1 and promotes hepatocellular carcinoma cell
migration and invasion. Mol Onco. 6:507–515. 2012. View Article : Google Scholar
|
|
36
|
Xu B, Sun Y, Tang G, Xu C, Wang L, Zhang Y
and Ji J: Id-1 expression in androgen-dependent prostate cancer is
negatively regulated by androgen through androgen receptor. Cancer
Lett. 278:220–229. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ling MT, Wang X, Ouyang XS, Xu K, Tsao SW
and Wong YC: Id-1 expression promotes cell survival through
activation of NF-kappaB signalling pathway in prostate cancer
cells. Oncogene. 22:4498–4508. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li QJ, Yang SH, Maeda Y, Sladek FM,
Sharrocks AD and Martins-Green M: MAP kinase
phosphorylation-dependent activation of Elk-1 leads to activation
of the co-activator p300. EMBO J. 22:281–291. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen A, Xu J and Johnson A: Curcumin
inhibits human colon cancer cell growth by suppressing gene
expression of epidermal growth factor receptor through reducing the
activity of the transcription factor Egr-1. Oncogene. 25:278–287.
2005. View Article : Google Scholar
|
|
40
|
Liu T, Wu J and He F: Evolution of
cis-acting elements in 5′ flanking regions of vertebrate actin
genes. J Mol Evol. 50:22–30. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Patki M, Chari V, Sivakumaran S, Gonit M,
Trumbly R and Ratnam M: The ETS domain transcription factor ELK1
directs a critical component of growth signaling by the androgen
receptor in prostate cancer cells. J Biol Chem. 288:11047–11065.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kemp CJ, Moore JM, Moser R, Bernard B,
Teater M, Smith LE, Rabaia NA, Gurley KE, Guinney J, Busch SE, et
al: CTCF haploinsufficiency destabilizes DNA methylation and
predisposes to cancer. Cell Reports. 7:1020–1029. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ulaner GA, Vu TH, Li T, Hu JF, Yao XM,
Yang Y, Gorlick R, Meyers P, Healey J, Ladanyi M and Hoffman AR:
Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied
by reciprocal methylation changes of a CTCF-binding site. Hum Mol
Genet. 12:535–549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rakha EA, Pinder SE, Paish CE and Ellis
IO: Expression of the transcription factor CTCF in invasive breast
cancer: A candidate gene located at 16q22.1. Br J Cancer.
91:1591–1596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Paradowska A, Fenic I, Konrad L, Sturm K,
Wagenlehner F, Weidner W and Steger K: Aberrant epigenetic
modifications in the CTCF binding domain of the IGF2/H19 gene in
prostate cancer compared with benign prostate hyperplasia. Int J
Oncol. 35:87–96. 2009. View Article : Google Scholar : PubMed/NCBI
|