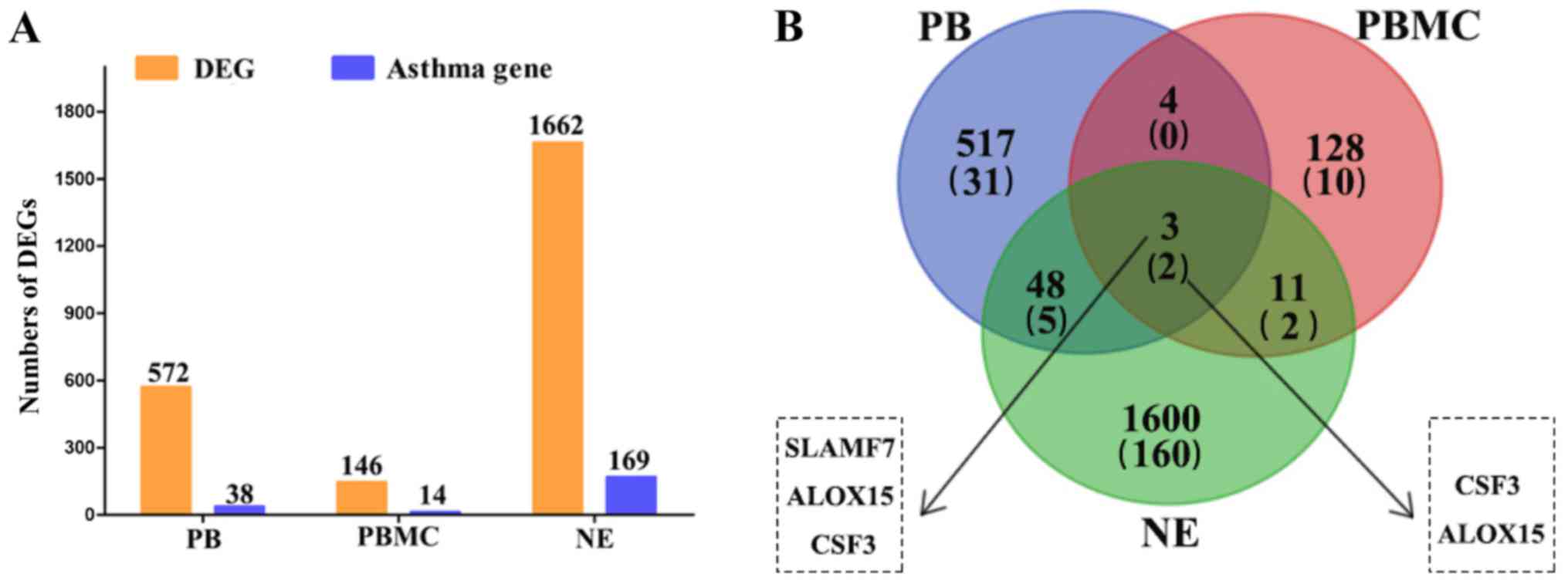
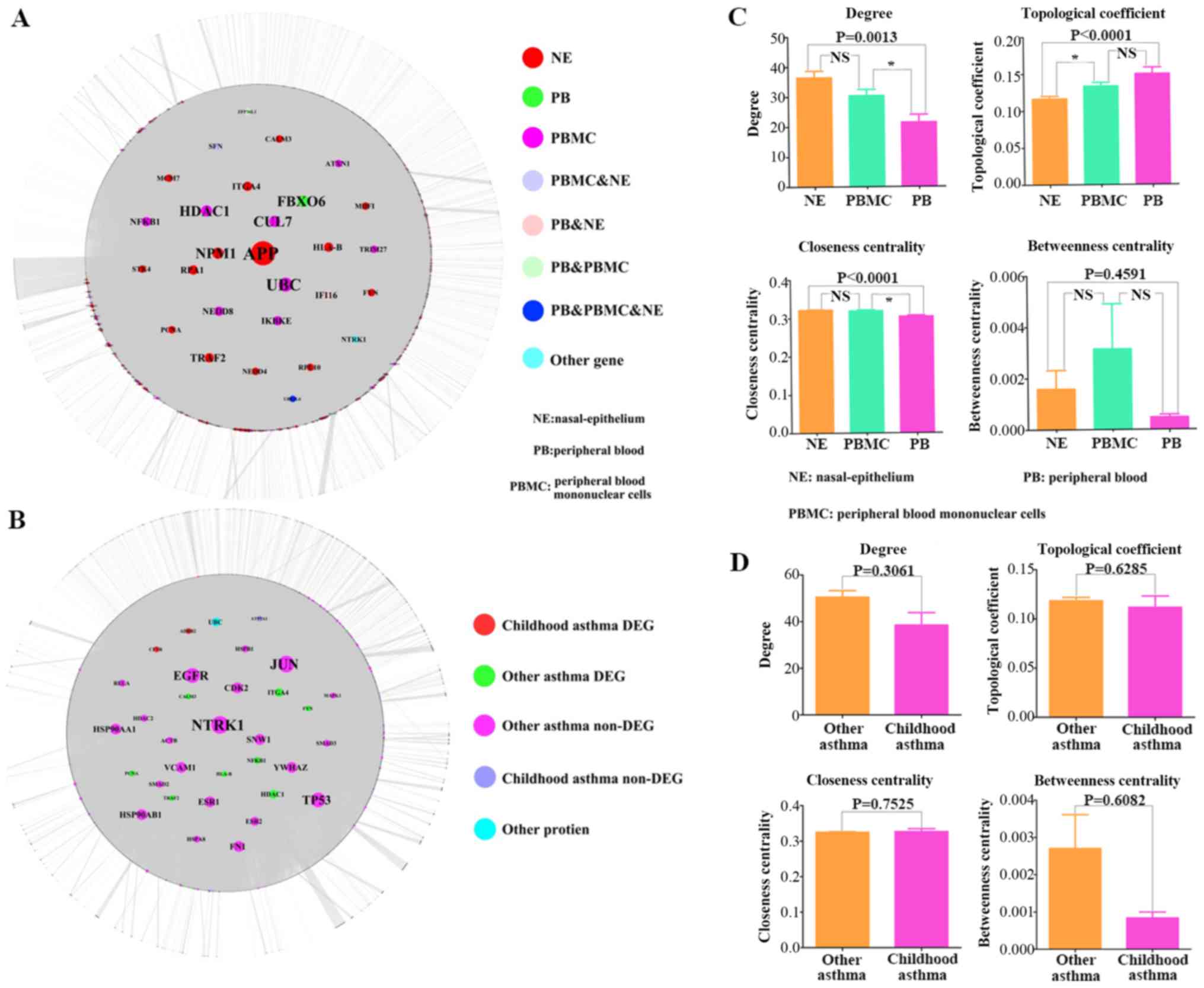
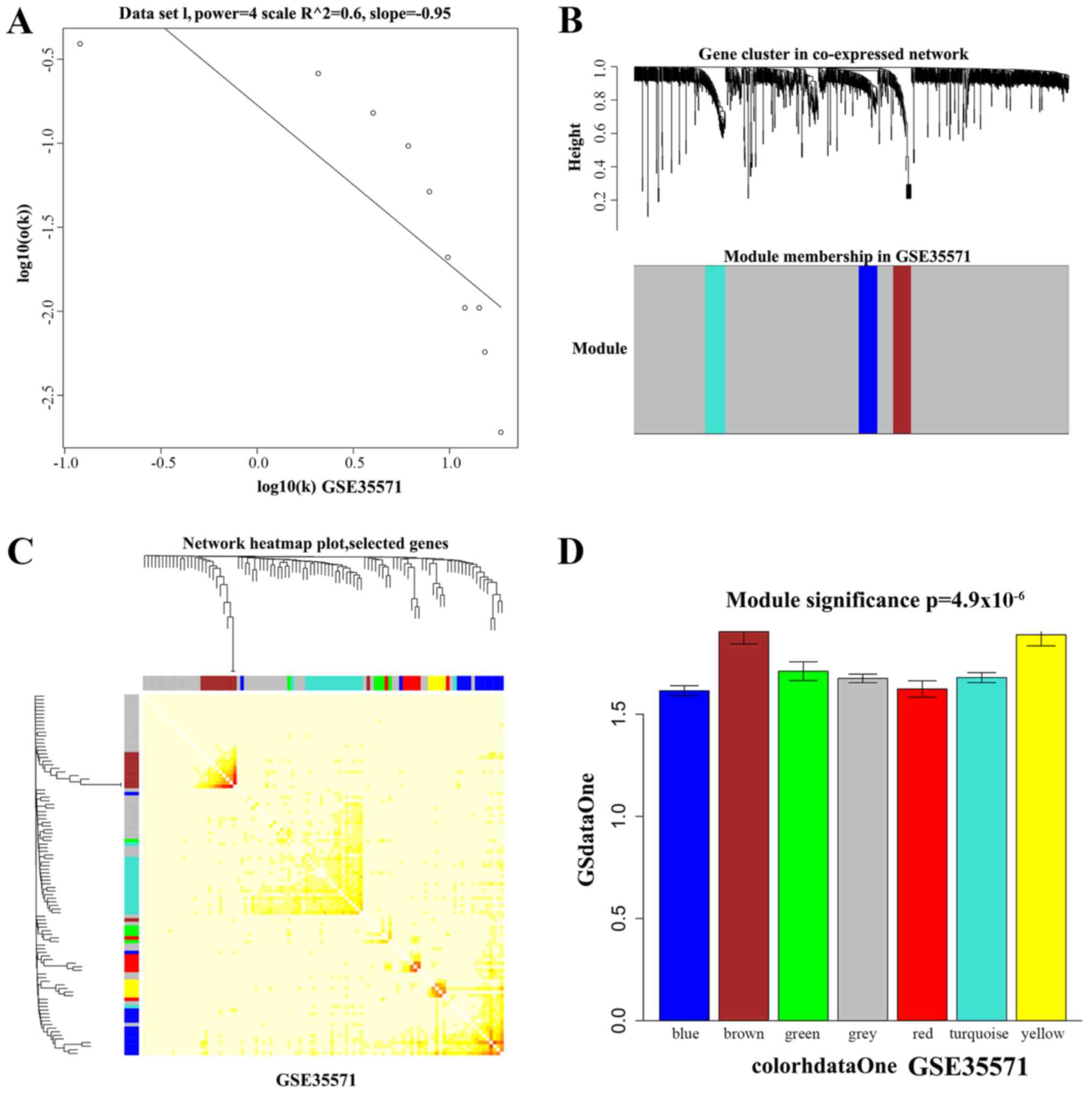
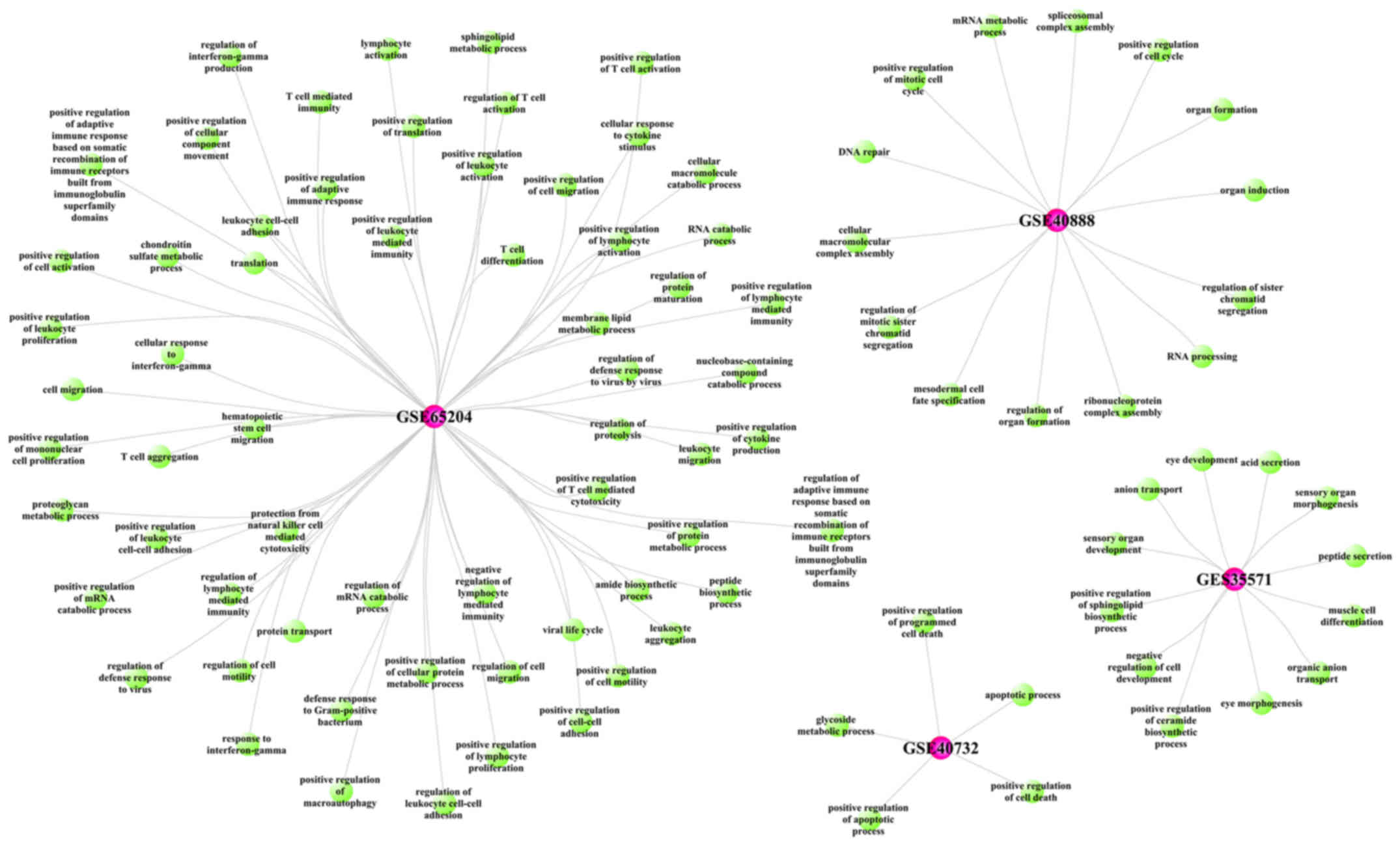
|
1
|
U.S. Department of Health and Human
Services, Centers for Disease Control and Prevention and National
Center for Health Statistics: Tables of Summary Health Statistics:
National Health Interview Survey. Table C-1b. 3–4. 2016.
|
|
2
|
James CV and Rosenbaum S: Paying for
quality care: Implications for racial and ethnic health disparities
in pediatric asthma. Pediatrics. 123 Suppl 3:S205–S210. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin HW and Lin SC: Environmental factors
association between asthma and acute bronchiolitis in young
children-a perspective cohort study. Eur J Pediatr. 171:1645–1650.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martinez FD: Genes, environments,
development and asthma: A reappraisal. Eur Respir J. 29:179–184.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goetghebuer T, Isles K, Moore C, Thomson
A, Kwiatkowski D and Hull J: Genetic predisposition to wheeze
following respiratory syncytial virus bronchiolitis. Clin Exp
Allergy. 34:801–803. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Finkelstein Y, Bournissen FG, Hutson JR
and Shannon M: Polymorphism of the ADRB2 gene and response to
inhaled beta-agonists in children with asthma: A meta-analysis. J
Asthma. 46:900–905. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wills-Karp M: Immunologic basis of
antigen-induced airway hyperresponsiveness. Annu Rev Immunol.
17:255–281. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang IV, Pedersen BS, Liu AH, O'Connor GT,
Pillai D, Kattan M, Misiak RT, Gruchalla R, Szefler SJ, Hershey
Khurana GK, et al: The nasal methylome and childhood atopic asthma.
J Allergy Clin Immunol. 139:1478–1488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang IV, Pedersen BS, Liu A, O'Connor GT,
Teach SJ, Kattan M, Misiak RT, Gruchalla R, Steinbach SF, Szefler
SJ, et al: DNA methylation and childhood asthma in the inner city.
J Allergy Clin Immunol. 136:69–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Raedler D, Ballenberger N, Klucker E, Böck
A, Otto R, da Costa Prazeres O, Holst O, Illig T, Buch T, von
Mutius E and Schaub B: Identification of novel immune phenotypes
for allergic and nonallergic childhood asthma. J Allergy Clin
Immunol. 135:81–91. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Williams-DeVane CR, Reif DM, Hubal EC,
Bushel PR, Hudgens EE, Gallagher JE and Edwards SW: Decision
tree-based method for integrating gene expression, demographic, and
clinical data to determine disease endotypes. BMC Syst Biol.
7:1192013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
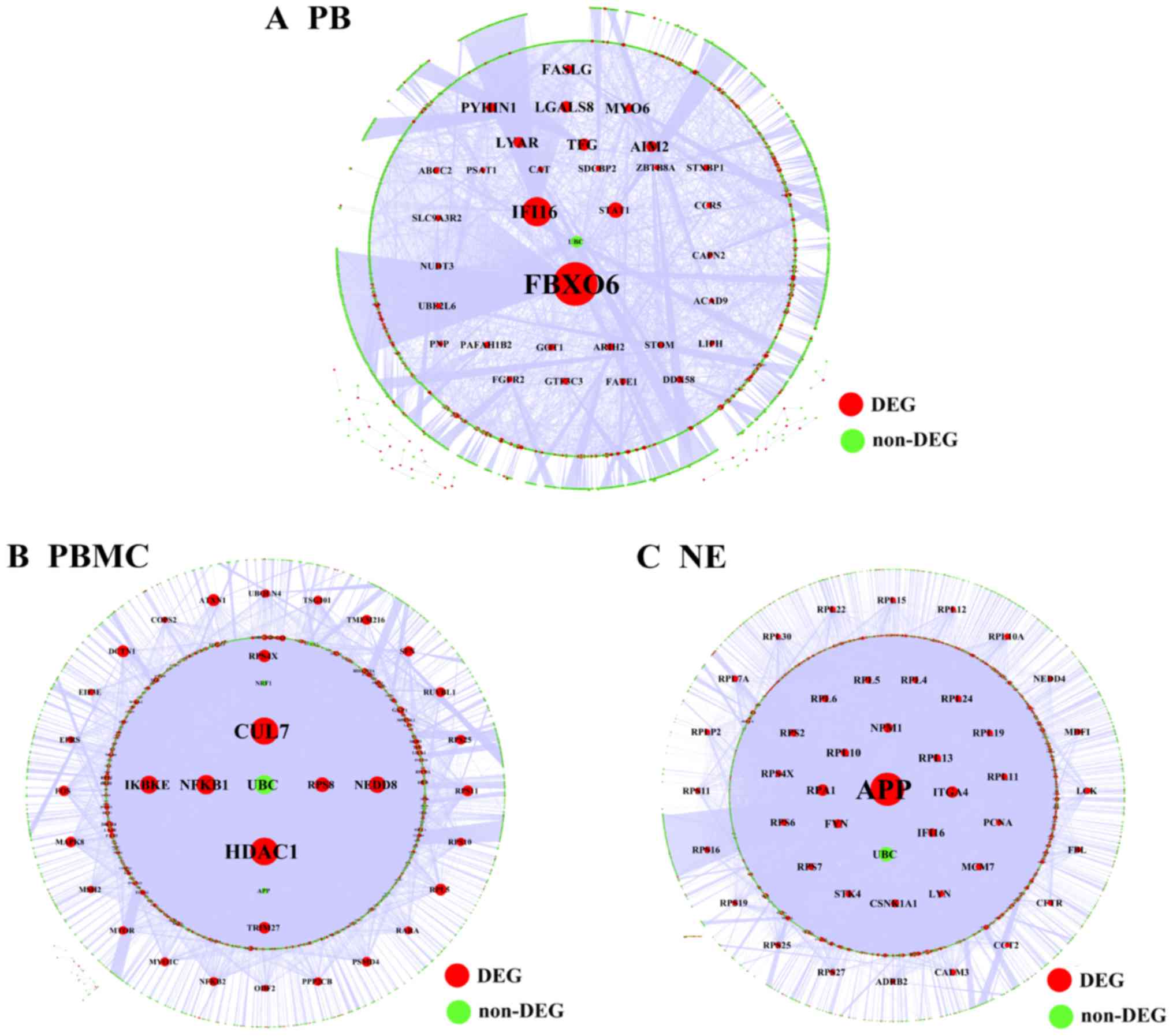
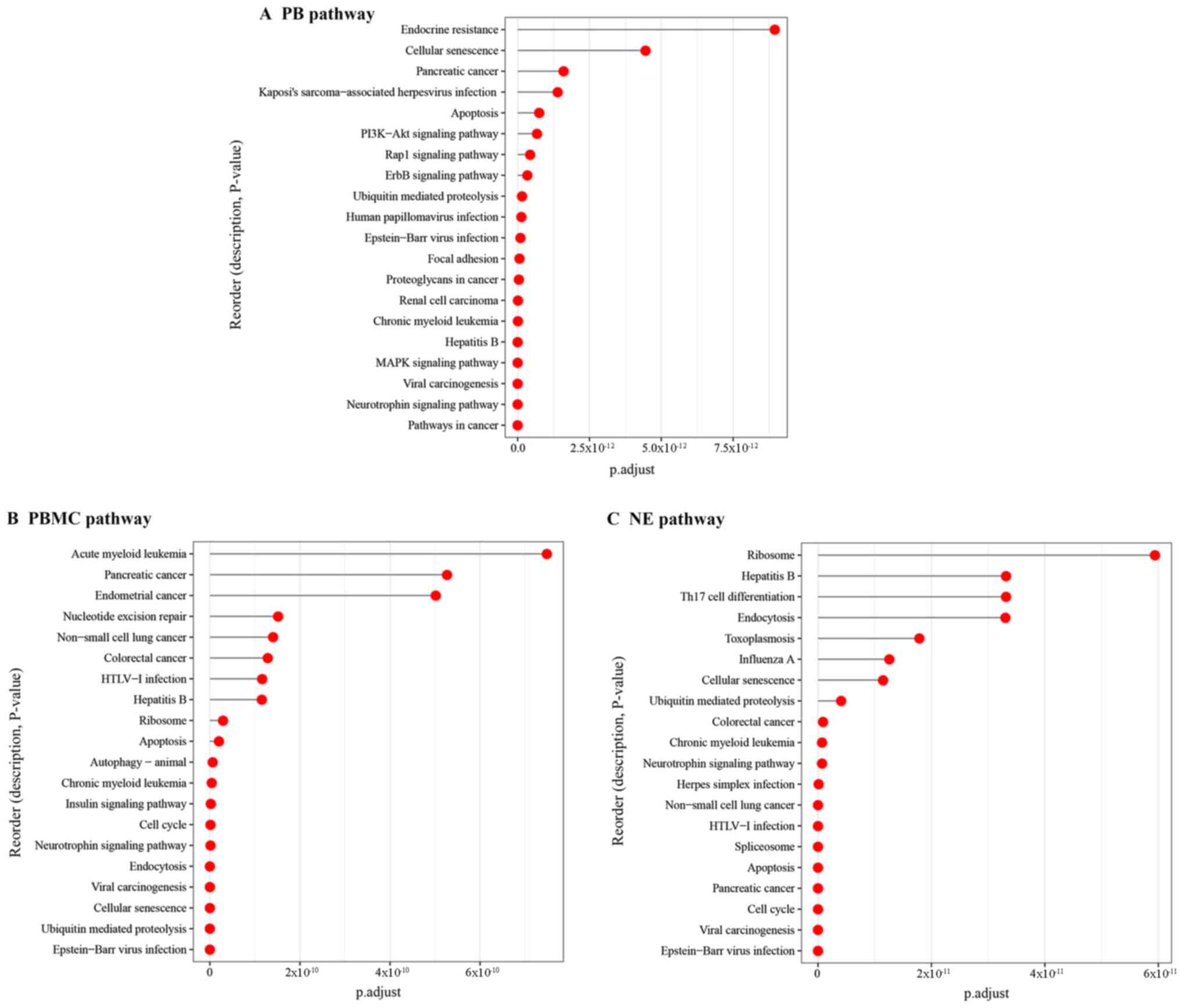
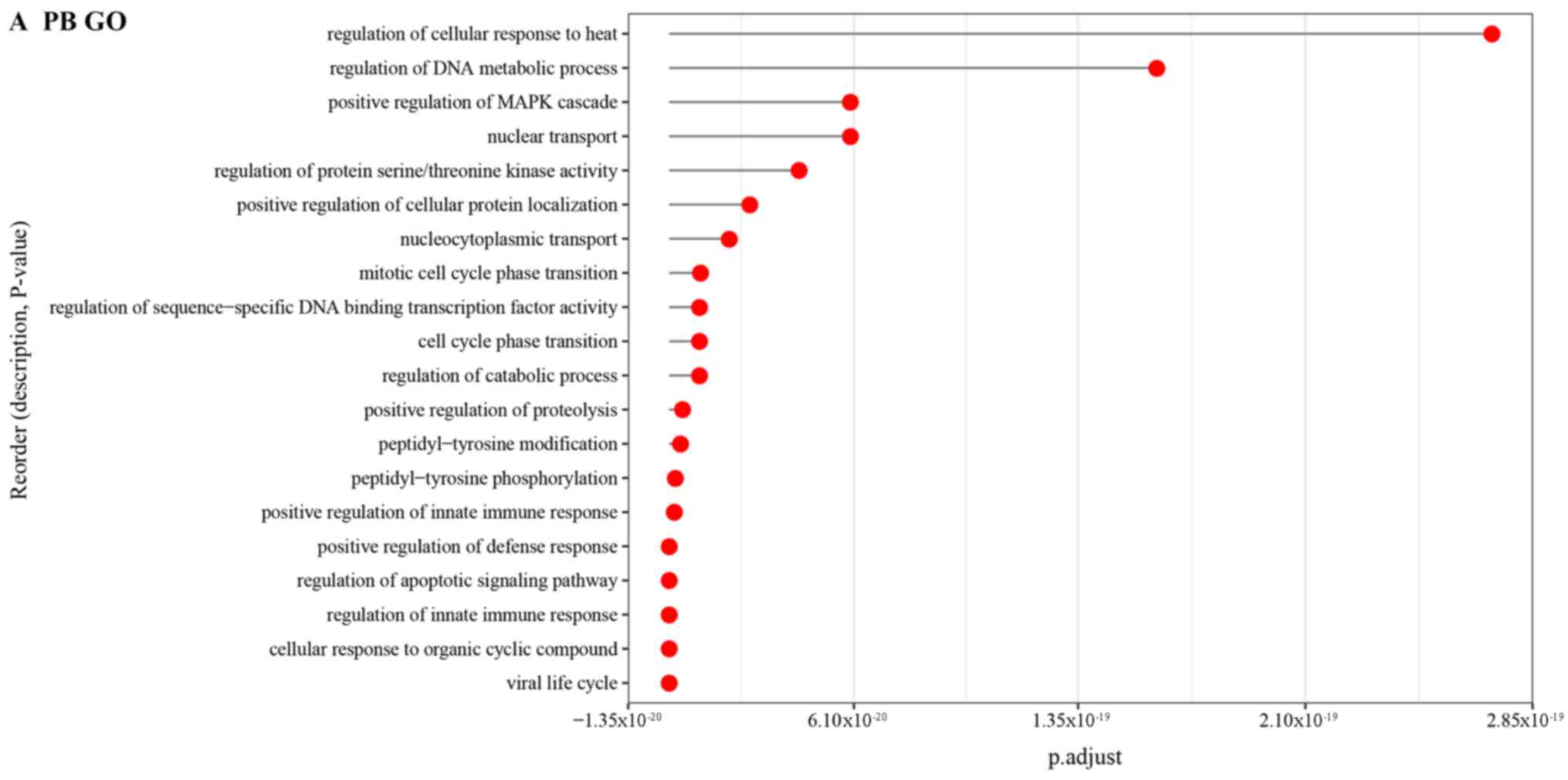
Chen X, Duan LH, Luo PC, Hu G, Yu X, Liu
J, Lu H and Liu B: FBXO6-mediated ubiquitination and degradation of
Ero1L inhibits endoplasmic reticulum stress-induced apoptosis. Cell
Physiol Biochem. 39:2501–2508. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao J, Wei J, Mialki RK, Mallampalli DF,
Chen BB, Coon T, Zou C, Mallampalli RK and Zhao Y: F-box protein
FBXL19-mediated ubiquitination and degradation of the receptor for
IL-33 limits pulmonary inflammation. Nat Immunol. 13:651–658. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cantley MD, Fairlie DP, Bartold PM, Marino
V, Gupta PK and Haynes DR: Inhibiting histone deacetylase 1
suppresses both inflammation and bone loss in arthritis.
Rheumatology (Oxford). 54:1713–1723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grausenburger R, Bilic I, Boucheron N,
Zupkovitz G, El-Housseiny L, Tschismarov R, Zhang Y, Rembold M,
Gaisberger M, Hartl A, et al: Conditional deletion of histone
deacetylase 1 in T cells leads to enhanced airway inflammation and
increased Th2 cytokine production. J Immunol. 185:3489–3497. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim MH, Kim SH, Kim YK, Hong SJ, Min KU,
Cho SH and Park HW: A polymorphism in the histone deacetylase 1
gene is associated with the response to corticosteroids in
asthmatics. Korean J Intern Med. 28:708–714. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chuang LS, Ian HI, Koh TW, Ng HH, Xu G and
Li BF: Human DNA-(cytosine-5) methyltransferase-PCNA complex as a
target for p21WAF1. Science. 277:1996–2000. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Milutinovic S, Zhuang Q and Szyf M:
Proliferating cell nuclear antigen associates with histone
deacetylase activity, integrating DNA replication and chromatin
modification. J Biol Chem. 277:20974–20978. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vignola AM, Chiappara G, Siena L, Bruno A,
Gagliardo R, Merendino AM, Polla BS, Arrigo AP, Bonsignore G,
Bousquet J and Chanez P: Proliferation and activation of bronchial
epithelial cells in corticosteroid-dependent asthma. J Allergy Clin
Immunol. 108:738–746. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maugeri N, Powell JE, t Hoen PA, de Geus
EJ, Willemsen G, Kattenberg M, Henders AK, Wallace L, Penninx B,
Hottenga JJ, et al: LPAR1 and ITGA4 regulate peripheral blood
monocyte counts. Hum Mutat. 32:873–876. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Herrenknecht K, Ozawa M, Eckerskorn C,
Lottspeich F, Lenter M and Kemler R: The uvomorulin-anchorage
protein alpha catenin is a vinculin homologue. Proc Natl Acad Sci
USA. 88:9156–9160. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu M, Subramanian V, Christie C, Castro M
and Mohanakumar T: Immune responses to self-antigens in asthma
patients: Clinical and immunopathological implications. Hum
Immunol. 73:511–516. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bernstein DI, Kashon M, Lummus ZL, Johnson
VJ, Fluharty K, Gautrin D, Malo JL, Cartier A, Boulet LP, Sastre J,
et al: CTNNA3 (α-catenin) gene variants are associated with
diisocyanate asthma: A replication study in a Caucasian worker
population. Toxicol Sci. 131:242–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wullschleger S, Loewith R and Hall MN: TOR
signaling in growth and metabolism. Cell. 124:471–484. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen H, Zhang L, Wang P, Su H, Wang W, Chu
Z, Zhang L, Zhang X and Zhao Y: mTORC2 controls Th9 polarization
and allergic airway inflammation. Allergy. 72:1510–1520. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Jing Y, Qiao J, Luan B, Wang X,
Wang L and Song Z: Activation of the mTOR signaling pathway is
required for asthma onset. Sci Rep. 7:45322017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meyer R, Hatada EN, Hohmann HP, Haiker M,
Bartsch C, Röthlisberger U, Lahm HW, Schlaeger EJ, van Loon AP and
Scheidereit C: Cloning of the DNA-binding subunit of human nuclear
factor kappa B: The level of its mRNA is strongly regulated by
phorbol ester or tumor necrosis factor alpha. Proc Natl Acad Sci
USA. 88:966–970. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shi HL, Liu JB and Lu AP: Expression
profiles of PI3K, NF-κB, and STAT1 in peripheral blood mononuclear
cells in children with bronchial asthma. Zhongguo Dang Dai Er Ke Za
Zhi. 18:614–617. 2016.(In Chinese). PubMed/NCBI
|
|
31
|
Li Z, Zheng J, Zhang N and Li C: Berberine
improves airway inflammation and inhibits NF-κB signaling pathway
in an ovalbumin-induced rat model of asthma. J Asthma. 53:999–1005.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Brossard M, Sarnowski C, Vaysse A,
Moffatt M, Margaritte-Jeannin P, Llinares-López F, Dizier MH,
Lathrop M, Cookson W, et al: Network-assisted analysis of GWAS data
identifies a functionally-relevant gene module for childhood-onset
asthma. Sci Rep. 7:9382017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vishweswaraiah S, Veerappa AM, Mahesh PA,
Jayaraju BS, Krishnarao CS and Ramachandra NB: Molecular
interaction network and pathway studies of ADAM33 potentially
relevant to asthma. Ann Allergy Asthma Immunol. 113:418–424.e1.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zou J, Zhu F, Liu J, Wang W, Zhang R,
Garlisi CG, Liu YH, Wang S, Shah H, Wan Y and Umland SP: Catalytic
activity of human ADAM33. J Biol Chem. 279:9818–9830. 2004.
View Article : Google Scholar : PubMed/NCBI
|