Introduction
Tristetraprolin (TTP) is a RNA-binding protein with
two zinc fingers, which are necessary for TTP to bind to the
AU-rich elements (AREs) in the 3′-untraslated region (3′-UTR) of
target mRNA (1), and to
subsequently promote mRNA deadenylation and decay (2,3).
TTP-mediated mRNA decay is a typical model of posttranscriptional
regulation in inflammatory mediator expression (4). However, TTP does not have deadenylase
activity and other proteins that have deadenylase activity are
required to cooperate with TTP to mediate mRNA decay.
The carbon catabolite repressor protein 4
(CCR4)-negative on TATA (NOT) complex consists of multiple subunits
that are involved in the regulation of gene expression at different
levels (5). One of the enzymatic
activities of the CCR4-NOT complex is deadenylation (6). In yeast, the CCR4 subunit provides
deadenylase activity, while in other eukaryotes the deadenylase
activity is provided by CCR4 and CCR4-associated factor 1 [CAF1; as
known as CCR4-NOT complex subunit 7 (CNOT7) in human cells]
(7,8). Schwede et al (9) reported that CNOT7, rather than CCR4,
is necessary for the decay of an ARE-containing reporter mRNA in
human cells. In humans, CNOT7 is one of the subunits of the
CCR4-NOT complex and interacts with CNOT1, the scaffold protein of
the CCR4-NOT complex (10).
Another previous study demonstrated that CNOT1 may directly bind to
TTP (11). These results implied
that CNOT1 could interact with CNOT7 and TTP. The authors' previous
studies have demonstrated that CNOT1, CNOT7 and TTP were
coimmunoprecipitated in human pulmonary microvascular endothelial
cells (HPMECs); TTP could bind to the AREs of intercellular
adhesion molecule-1 (ICAM-1) and interleukin-8 (IL-8) mRNA, and
CNOT7 was involved in ICAM-1 and IL-8 regulation by TTP (12,13).
On the basis of these results, together with the results of other
studies (11,14), it was hypothesized that CNOT1 may
provide a platform to recruit CNOT7 and TTP with ARE-bearing mRNA,
and be involved in TTP-mediated ICAM-1 and IL-8 mRNA decay.
The present study in HPMECs reported that CNOT1
knockdown increased ICAM-1 and IL-8 mRNA stabilization, and protein
production following stimulation with tumor necrosis factor
(TNF)-α. The immunofluorescence results demonstrated that CNOT1,
CNOT7 and TTP co-localized in the cytoplasm. In addition, CNOT1
silencing abolished CNOT7 and TTP coimmunoprecipitation (Co-IP).
However, CNOT7 silencing did not influence CNOT1 and TTP Co-IP, and
TTP silencing additionally did not influence CNOT1 and CNOT7 Co-IP.
In conclusion, these results together with the results of our
previous study, revealed that CNOT1 may provide a platform to
recruit TTP and CNOT7, and may be involved in TTP-mediated ICAM-1
and IL-8 mRNA decay in HPMECs.
Materials and methods
Materials
The materials used in the present study were as
follows: HPMECs and Endothelial Cell Medium (ECM; ScienCell
Research Laboratories, Inc., San Diego, CA, USA). TNF-α (cat. no.
300-01A; PeproTech, Rocky Hill, NJ, USA). Lipofectamine 2000™ (cat.
no. 11668-027), Opti-Minimum Essential Medium I reduced serum
medium and TRIzol® reagent (all Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Actinomycin D (ActD;
cat. no. A1410; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Immunoprecipitation kit (IP; cat. no. K286-25; BioVision, Inc.,
Milpitas, CA, USA). The Radioimmunoprecipitation Assay Lysis Buffer
(cat. no. P0013C; Beyotime Institute of Biotechnology, Haimen,
China). Protease inhibitors (cat. no. R1321) and RevertAid First
Strand cDNA Synthesis kit (cat. no. K1622; Fermentas; Thermo Fisher
Scientific, Inc.). Ribolock RNase Inhibitors (cat. no. E00381;
Thermo Fisher Scientific, Inc.). Protein assay reagent (cat. no.
KGPBCA; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). SYBR
Green PCR Master Mix (cat. no. 04913850001; Roche Diagnostics GmbH,
Mannheim, Germany). IL-8 ELISA kit (cat. no. 431507; BioLegend,
Inc., San Diego, CA, USA). CNOT1, CNOT7 and TTP small interfering
RNAs (siRNAs) and the control siRNAs (Shanghai GenePharma Co.,
Ltd., Shanghai, China). The following antibodies were used: Rabbit
anti-TTP antibody (cat. no. ABE285; Merck KGaA), mouse anti-CNOT7
antibody (cat. no. sc-101009), mouse anti-TTP antibody (cat. no.
sc374305; all Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
rabbit anti-CNOT1 antibody (cat. no. 14276-1-AP; Wuhan Sanying
Biotechnology, Wuhan, China), rabbit anti-ICAM-1 antibody (cat. no.
4915; Cell Signaling Technology, Inc., Danvers, MA, USA), mouse
anti-β-tubulin antibody (cat. no. M20005; Abmart, Shanghai, China),
mouse anti-GAPDH antibody (cat. no. AT0002; CMCTAG, Inc.,
Milwaukee, WI, USA), rabbit immunoglobulin G (IgG; cat. no. A7016)
and mouse IgG (cat. no. A7028; Beyotime Institute of
Biotechnology).
Cell culture, treatment and
transfection
HPMECs were cultured in ECM at 37°C in an incubator
supplied with 5% CO2. Cells were subcultured to passages
5 to 8 for all experiments and activated with TNF-α (10 ng/ml) in
the subsequent experiments. siRNAs were used to silence CNOT1,
CNOT7 and TTP. The siRNA transfection experiment was performed as
previously described (13) using
Lipofectamine 2000™ and the final concentration of CNOT1 siRNA, TTP
siRNA and CNOT7 siRNA was 100, 80 and 100 nM, respectively. The
cells were incubated for 24 h, transfection medium was replaced
with complete endothelial cell medium, and the incubation was
continued for an additional 24 h prior to using the cells. The
target sequences of siRNAs are as follows: CNOT1 siRNA 1 sense,
5′-CUUCACGUCGUGAAUACCUCATT-3′ and antisense,
5′-UGAGGUAUUCACGACGUGAAGTT-3′; CNOT1 siRNA 2 sense,
5′-CAAUUCGCCAACUUAUCAUGCTT-3′ and antisense,
5′-GCAUGAUAAGUUGGCGAAUUGTT-3′, negative control siRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. TTP siRNAs, CNOT7 siRNAs, and their
control siRNAs were as previously described (12,13).
Western immunoblotting
experiments
The western immunoblotting experiments were
performed as previously described (13). The primary antibodies (TTP, CNOT1,
CNOT7, ICAM-1, β-tubulin and GAPDH antibodies) were diluted at
1:1,000 with 0.1% PBST containing 5% bovine serum albumin (BSA;
cat. no. ST023; Beyotime Institute of Biotechnology). The
horseradish peroxidase-conjugated secondary antibodies [goat
anti-rabbit immunoglobulin G (IgG; cat. no. A0208; Beyotime
Institute of Biotechnology) and goat anti-mouse IgG (cat. no.
A0216; Beyotime Institute of Biotechnology)] were diluted at
1:5,000 with 0.1% PBST containing 5% BSA. The target bands were
developed using an enhanced chemiluminescent kit (cat. no. 32134;
Thermo Fisher Scientific, Inc.) and exposed on film. The band
images were obtained using the BenQ Scanner (5560C) and analyzed
with ImageJ software 1.8.0 (National Institutes of Health,
Bethesda, MD, USA; rsb.info.nih.gov/ij/).
ELISA
An IL-8 ELISA kit was employed to detect IL-8 levels
in the HPMEC culture supernatant. An IL-8 standard was used to
construct standard curves. Absorbance at 450 nm was read using a
microplate reader and IL-8 concentrations were calculated,
according to the standard curves.
RNA isolation, cDNA synthesis and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Following cell transfection and activation using
TNF-α (10 ng/ml), total RNA was extracted at 0, 2, 4, 8 and 12 h
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and synthesized and analyzed as previously
described (13). The GAPDH, ICAM-1
and IL-8 mRNA primers were the same as those previously described
(12). The 2−Δ∆Cq
method was used for RNA quantification (15).
Analysis of mRNA stability using
RT-qPCR
Cells were transfected using siRNAs for 48 h, as
aforementioned, and subsequently activated using TNF-α (10 ng/ml)
for 4 h at 37°C, then ActD (5 µg/ml) was added to block
transcription at 37°C for 60, 120, 180 and 240 min. RNA was
extracted at 0, 60, 120, 180 and 240 min following the addition of
ActD, synthesized and analyzed as previously described (12).
Immunofluorescence experiments
HPMECs were stimulated using TNF-α (10 ng/ml) for 4
h and fixed with 4% formaldehyde at room temperature for 30 min,
permeabilized with 0.2% Triton X-100 and incubated with antibodies
(1:100, diluted with 0.1% PBST containing 5% BSA) specific to
CNOT1, TTP and CNOT7. Following incubation with these antibodies
for 2 h at room temperature, the cells were washed with PBS and
incubated with Alexa Fluor 488 (cat. no. A11001) or Alexa Fluor 568
(cat. no. A11011)-conjugated secondary antibodies (both Thermo
Fisher Scientific, Inc.; 1:200, diluted with 0.1% PBST containing
2% BSA). DAPI was used to stain the cell nuclei for 5 min at room
temperature at a concentration of 1 µg/ml. The cells were observed
using a Leica DM2500 fluorescence microscope (Leica Microsystems
GmbH, Wetzlar, Germany; magnification, ×630).
Co-IP experiments
The Co-IP experiments were performed as previously
described (12). HPMECs were
transfected with CNOT1, CNOT7 or TTP siRNA for 48 h, as
aforementioned, and stimulated with TNF-α (10 ng/ml) for 4 h, then
~2×107 cells were lysed in 500 µl non-Denaturing Lysis
buffer, mixed on a rotary mixer for 30 min at 4°C, centrifuged at
10,000 × g for 10 min at 4°C and the cell extract was susbequently
transferred to chilled fresh tubes. A total of 10 µl supernatant
was collected for use as the input. Following antibody [8 µg CNOT1
antibody (cat. no. 14276-1-AP), 10 µg TTP antibody (cat. no.
ABE285) and 10 µg CNOT7 antibody (cat. no. sc-101009)] and Protein
A/G Sepharose beads (from the IP kit) binding for 4 h at 4°C on a
rotary mixer, the 2X SDS-PAGE loading buffer was added to the
washed beads and boiled. The eluent was saved by centrifugation at
2,000 × g for 2 min at 4°C for western immunoblotting
experiments.
Statistical analysis
SPSS 19.0 was used for statistical analysis (IBM
Corp., Armonk, NY, USA). The data are expressed as the mean ±
standard error of the mean. The experiments were repeated three
times. Student's t-test and analysis of variance with Bonferroni's
post hoc test were used to analyze the differences between the
groups and compare the differences between the different time
points within the same treatment group, respectively. P<0.05 was
considered to indicate a statistically significant difference.
Results
CNOT1 knockdown promotes the
TNF-α-induced expression of ICAM-1 and IL-8 in HPMECs
In previous studies it was demonstrated that CNOT1,
CNOT7 and TTP were coimmunoprecipitated, TTP was able to bind to
the ARE sequences of ICAM-1 and IL-8 mRNAs, and CNOT7 was involved
in ICAM-1 and IL-8 regulation by TTP (12,13).
As CNOT1, CNOT7 and TTP were coimmunoprecipitated and TTP and CNOT7
were involved in the regulation of ICAM-1 and IL-8 expression,
whether CNOT1 affected ICAM-1 and IL-8 expression was examined
further in the present study.
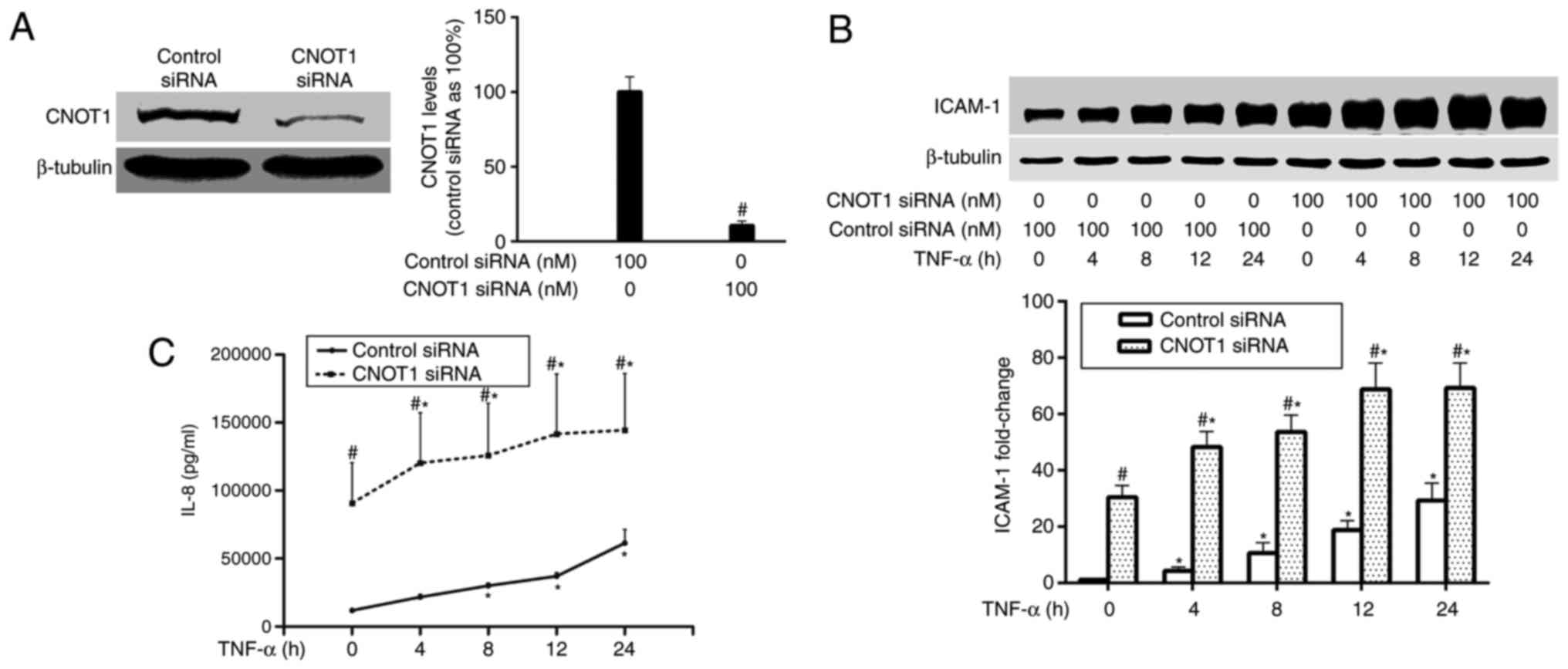
Following transfection, HPMECs were stimulated using
TNF-α (10 ng/ml). Total protein and the supernatant were collected
at 0, 4, 8, 12 and 24 h following TNF-α stimulation. Western
immunoblotting results indicated that CNOT1 siRNA (100 nM)
significantly reduced CNOT1 to ~11% of the control (P<0.05;
Fig. 1A). Fig. 1B demonstrated that TNF-α
stimulation significantly increased ICAM-1 levels, which in CNOT1
knocked-down HPMECs were increased when compared with the control
siRNA treated cells (P<0.05). In addition, the ELISA results
demonstrated that CNOT1 knockdown significantly promoted IL-8
expression compared with the control group and the peak level of
IL-8 was observed at 24 h following TNF-α activation (P<0.05;
Fig. 1C). These results indicated
that CNOT1 may be involved in the TNF-α-induced expression of
ICAM-1 and IL-8 in HPMECs.
CNOT1 silencing increases ICAM-1 and
IL-8 mRNA stability in HPMECs
As CNOT1 silencing increased ICAM-1 and IL-8 levels
in HPMECs, whether CNOT1 effected the expression and stability of
ICAM-1 and IL-8 mRNA with AREs at the 3′-UTR was investigated.
Cells were transfected with CNOT1 siRNA or control siRNA for 48 h,
and then activated using TNF-α (10 ng/ml). Total RNA was extracted
at 0, 2, 4, 8 and 12 h and analyzed using RT-qPCR. Fig. 2A demonstrated that the ICAM-1 mRNA
expression levels reached the peak level at 4 h following TNF-α
activation in control HPMECs and subsequently reduced to baseline
following 8 h. CNOT1-silenced HPMECs expressed ~2.7-fold more
ICAM-1 mRNA when compared with the control at 4 h and the ICAM-1
mRNA levels at 12 h of TNF-α activation remained significantly
elevated compared with the baseline (P<0.05). Similar results
were observed for IL-8 mRNA (P<0.05; Fig. 2B). These results demonstrated that
CNOT1 knockdown promoted ICAM-1 and IL-8 mRNA expression in HPMECs
following TNF-α activation.
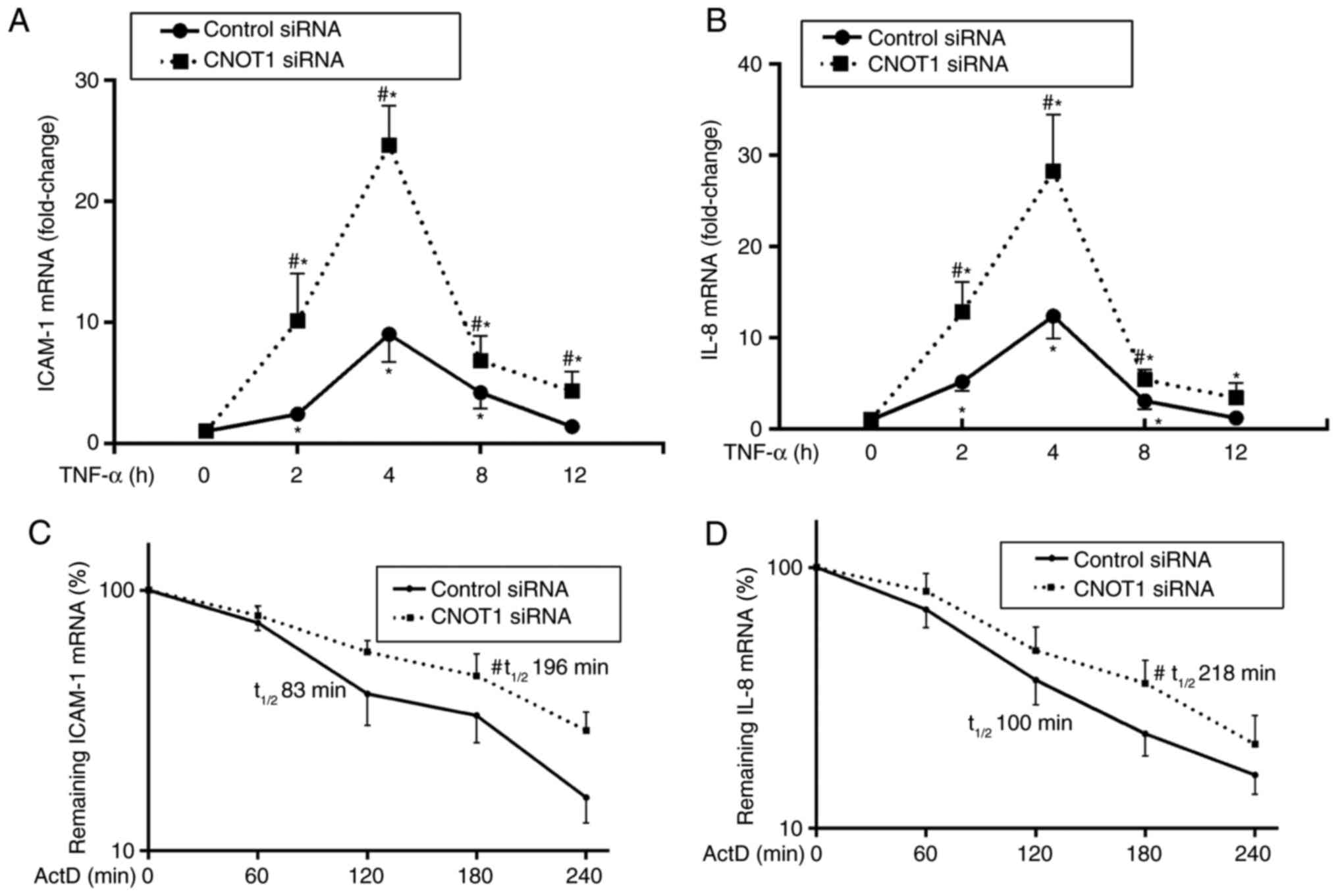 | Figure 2.CNOT1 silencing improves ICAM-1 and
IL-8 mRNA stability. (A) ICAM-1 and (B) IL-8 mRNA expression
significantly increased in CNOT1-silenced HPMECs following TNF-α
stimulation. (C) CNOT1 knockdown improved ICAM-1 and IL-8 mRNA
stabilization, and the average t1/2 of ICAM-1 mRNA was
196 and 83 min in the CNOT1-silenced HPMECs and the control,
respectively. (D) The t1/2 was 294 and 131 min for IL-8
mRNA in the CNOT1-silenced HPMECs and the control, respectively.
The data are expressed as the mean ± standard error of the mean
(n=3). #P<0.05 vs. control siRNA-transfected cells;
*P<0.05 vs. cells at TNF-α 0 h. IL, interleukin; siRNA, small
interfering RNA; HPMECs, human pulmonary microvascular endothelial
cells; CNOT1, subunit 1 of the carbon catabolite repressor protein
4-negative on TATA complex; TNF, tumor necrosis factor; ICAM-1,
intercellular adhesion molecule 1; t1/2, half life;
ActD, Actinomycin D. |
Furthermore, whether CNOT1 influenced ICAM-1 and
IL-8 mRNA expression by altering mRNA stability was analyzed.
CNOT1-silenced and control HPMECs were activated using TNF-α for 4
h, and ActD was added to block transcription. Total RNA was
isolated at 0, 60, 120, 180 and 240 min following the addition of
ActD and analyzed with qPCR. As demonstrated in Fig. 2C and D, CNOT1 knockdown
significantly stabilized ICAM-1 and IL-8 mRNA when compared with
the control, and the average half-life of ICAM-1 mRNA was 196 min
in CNOT1 knockdown HPMECs and 83 min in the control (P<0.05).
For IL-8 mRNA, the average half-life was 218 min in CNOT1 knockdown
HPMECs and 100 min in the control (P<0.05). These results
implied that CNOT1 reduced ICAM-1 and IL-8 mRNA levels by
decreasing the stability of the two mRNAs.
CNOT7, CNOT1 and TTP co-localize in
the cytoplasm of HPMECs
The results of the authors' previous studies
together with the above results indicated that CNOT7, CNOT1 and TTP
were coimmunoprecipitated in HPMECs, and all of them may be
involved in the regulation of ICAM-1 and IL-8 expression (12,13).
The cellular localization of CNOT7, CNOT1 and TTP was subsequently
investigated in HPMECs by immunofluorescence experiments in the
present study. Fig. 3A
demonstrated that CNOT7 and CNOT1 were detected in the cytoplasm
and nuclei. CNOT7 was localized in the cytoplasm with a small
fraction localized in the nuclei, while CNOT1 was detected in the
nuclei and a small fraction located in the cytoplasm. Fig. 3B indicated that CNOT7 and TTP were
primarily localized in the cytoplasm and a small fraction of the
two were located in the nuclei. As illustrated in Fig. 3C, TTP and CNOT1 were detected in
the cytoplasm and nuclei. These results demonstrated that CNOT7,
CNOT1 and TTP were detected in the cytoplasm and nuclei, which
implied that they were co-localized in the cytoplasm and nuclei
following TNF-α stimulation.
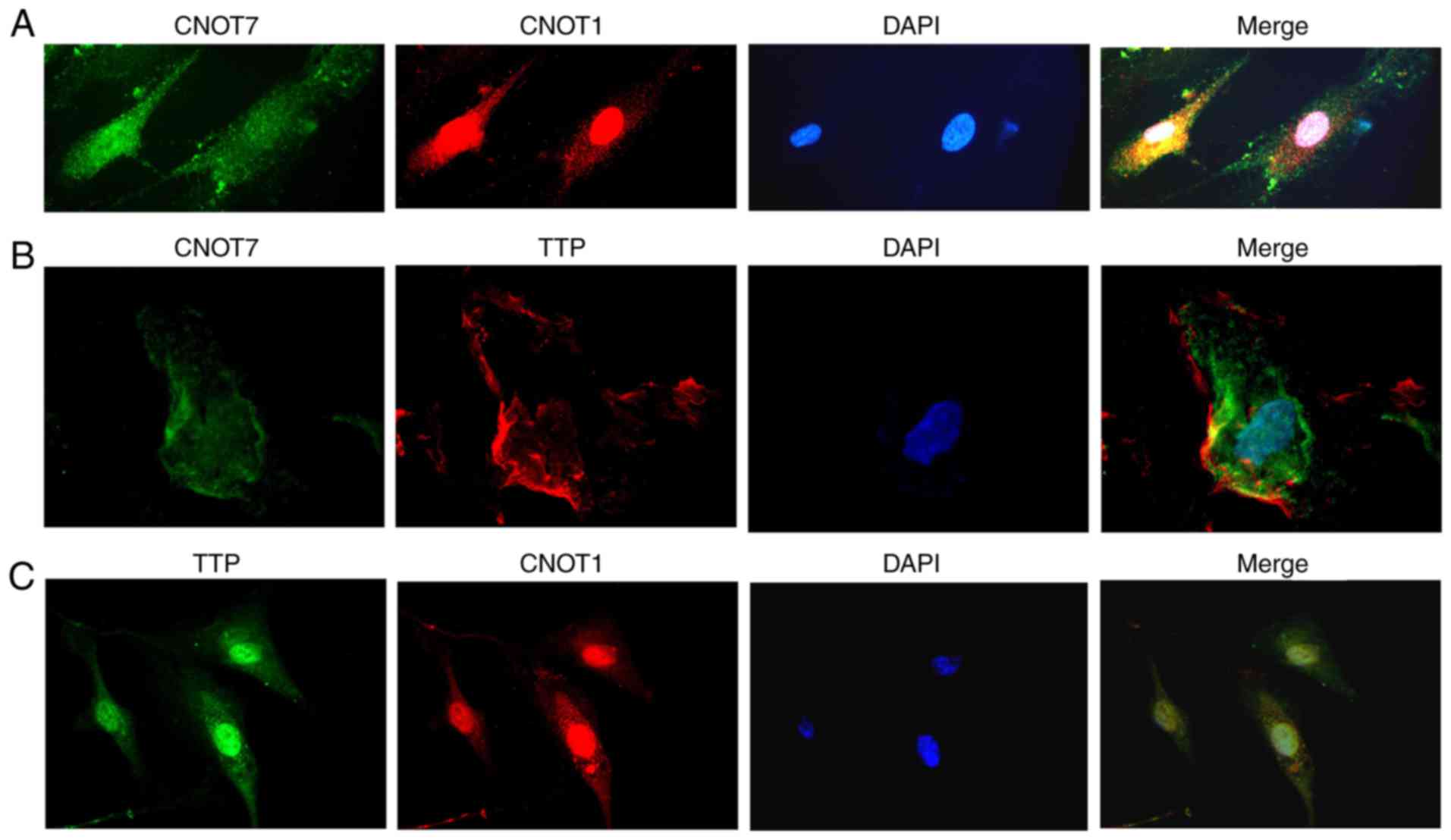 | Figure 3.CNOT7, CNOT1 and TTP co-localize in
the cytoplasm of HPMECs. HPMECs were activated using tumor necrosis
factor-α (10 ng/ml) and DAPI was used to stain the cell nuclei.
CNOT7 and CNOT1 were detected in the cytoplasm and nuclei
(magnification, ×630); (A) CNOT7 was primarily located in the
cytoplasm and a small fraction was localized in the nuclei, while
CNOT1 was primarily detected in the nuclei and a small fraction was
localized in cytoplasm. (B) CNOT7 and TTP were mainly located in
the cytoplasm, though a small fraction was localized in the nuclei.
(C) TTP and CNOT1 were detected in the cytoplasm and nuclei. siRNA,
small interfering RNA; HPMECs, human pulmonary microvascular
endothelial cells; CCR4-NOT, carbon catabolite repressor protein
4-negative on TATA; CNOT1, subunit 1 of the CCR4-NOT complex;
CNOT7, subunit 7 of the CCR4-NOT complex; TTP, tristetraprolin. |
Association of CNOT1 with TTP and
CNOT7
As CNOT1, CNOT7 and TTP were co-localized in the
cytoplasm and were coimmunoprecipitated in HPMECs (12), together with the results published
by other studies that CNOT1 could interact with CNOT7 and TTP
(10,11), it was hypothesized that CNOT1 may
serve as a platform that can recruit CNOT7 and TTP with
ARE-containing mRNA, leading to mRNA decay. CNOT1, CNOT7 and TTP
siRNA were used to silence the corresponding protein to investigate
the effect of knocking-down one protein on the Co-IP of the other
two proteins. The western immunoblotting results indicated that
CNOT7 siRNA (100 nM) and TTP siRNA (80 nM) reduced the CNOT7 and
TTP levels to ~10 and ~15% of the controls, respectively (Fig. 4A and B). The efficiency of CNOT1
silencing was investigated as described above. The Co-IP
experiments were performed as described above and the results
demonstrated that CNOT1, TTP and CNOT7 were immunoprecipitated
using the CNOT1, TTP or CNOT7 antibody, whereas no CNOT1, TTP or
CNOT7 band was detected when using the control antibody in the
control cells (Fig. 4C-E).
Notably, TTP was undetectable when using CNOT7 antibody in
CNOT1-silenced cells, and CNOT7 was not detectable when using TTP
antibody (Fig. 4C). However, TTP
knockdown did not impact CNOT1-CNOT7 Co-IP (Fig. 4D); neither did CNOT7 knockdown
affect CNOT1-TTP Co-IP (Fig. 4E).
These results indicated that CNOT1, CNOT7 and TTP were
coimmunoprecipitated in HPMECs, and CNOT1 could serve as a
platform, which recruits CNOT7 and TTP, while TTP and CNOT7 could
not directly combine.
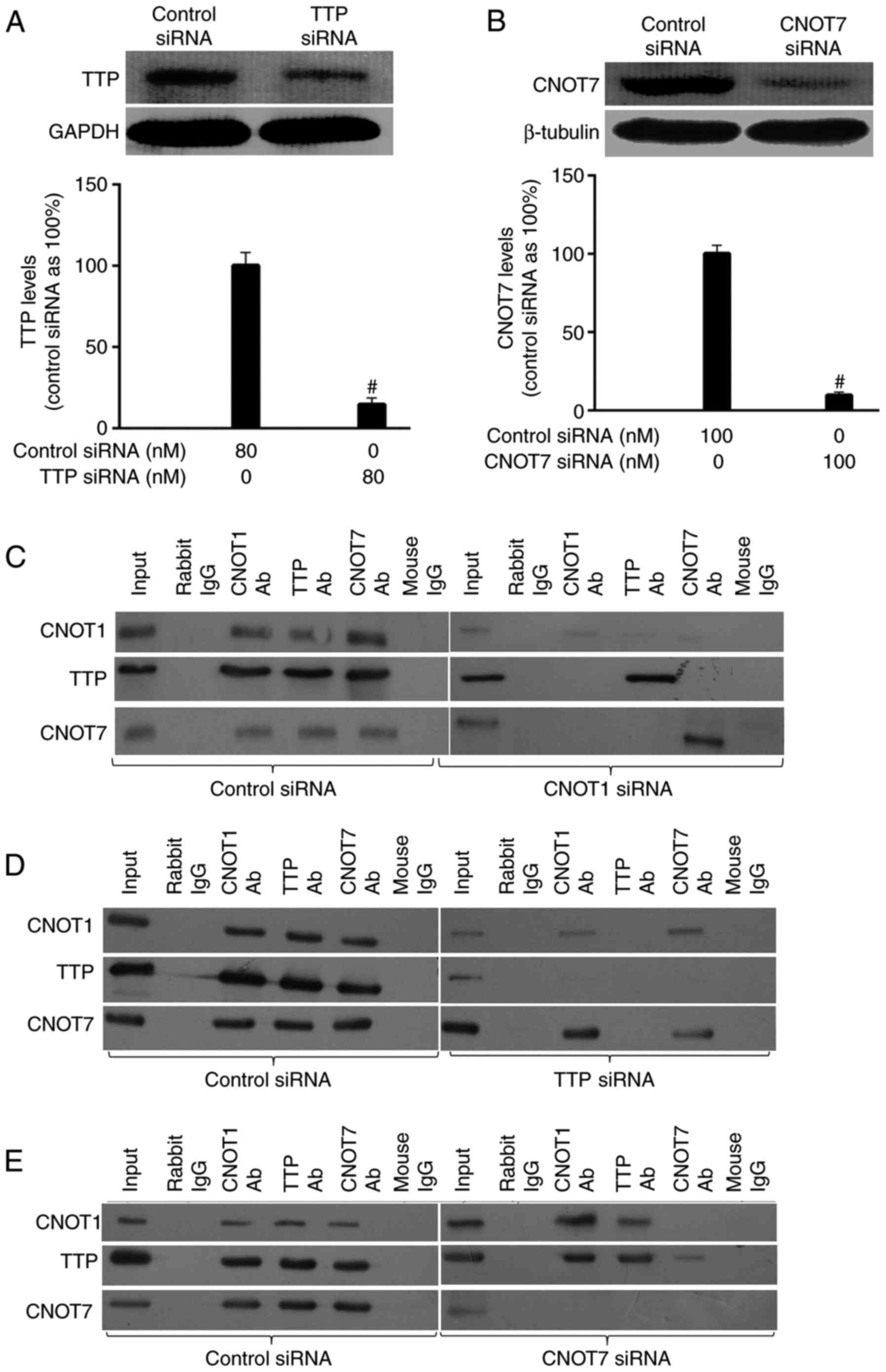 | Figure 4.Association of CNOT1 with TTP and
CNOT7. Western immunoblotting results indicated that CNOT7 siRNA
(100 nM) and TTP siRNA (80 nM) reduced (A) TTP and (B) CNOT7 levels
to 15 and ~10% of the controls, respectively. Following
transfection, human pulmonary microvascular endothelial cells were
activated using tumor necrosis factor-α (10 ng/ml) for 4 h, the
Co-IP experiments were subsequently performed using rabbit or mouse
IgG as the control. The Co-IP results demonstrated that CNOT1, TTP
and CNOT7 were immunoprecipitated by the CNOT1, TTP or CNOT7 Ab,
whereas no CNOT1, TTP or CNOT7 bands were detectable when using the
control antibody in the control cells. (C) TTP was undetectable
when using the CNOT7 antibody in CNOT1-silenced cells, neither was
CNOT7 detectable when using the TTP antibody. (D) However, TTP
knockdown did not impact CNOT1-CNOT7 Co-IP; nor did (E) CNOT7
knockdown affect CNOT1-TTP Co-IP. The data are expressed as the
mean ± standard error of the mean (n=3). #P<0.05 vs.
control siRNA-transfected cells. siRNA, small interfering RNA;
CCR4-NOT, carbon catabolite repressor protein 4-negative on TATA;
CNOT1, subunit 1 of the CCR4-NOT complex; CNOT7, subunit 7 of the
CCR4-NOT complex; ICAM-1, intercellular adhesion molecule 1; Co-IP,
coimmunoprecipitation; TTP, tristetraprolin; IgG, immunoglobulin G;
Ab, antibody. |
Discussion
Inflammatory mediator expression is regulated at the
transcriptional and/or posttranscriptional levels. ARE-mediated
mRNA decay (AMD) is an important process during posttranscriptional
regulation (16). A number of
RNA-binding proteins including TTP (12,13),
Human antigen R (17) and
poly(A)-binding protein (18) are
involved in AMD. TTP is known to serve a critical role in AMD. TTP
may bind to the ARE motifs on the 3′-UTR of target mRNA and cause
mRNA rapid deadenylation and decay (2,3). It
was demonstrated in our previous studies that TTP bound to the AREs
of ICAM-1 and IL-8 mRNA, and destabilized the two mRNAs (12,13).
TTP and CNOT7 are involved in the regulation of ICAM-1 and IL-8
expression (12,13). In addition, TTP, CNOT7 and CNOT1
were coimmunoprecipitated in HPMECs (12,13).
In the present study, CNOT1 silencing increased ICAM-1 and IL-8
mRNA stability. CNOT1, CNOT7 and TTP co-localized in the cytoplasm,
and CNOT1 silencing abolished CNOT7 and TTP Co-IP. However, CNOT7
silencing did not influence CNOT1 and TTP Co-IP, and TTP silencing
additionally did not influence CNOT1 and CNOT7 Co-IP. These results
implied that CNOT1 may serve as a platform to recruit TTP and
CNOT7, and be involved in TTP-mediated mRNA decay in HPMECs.
TTP has been proven to mediate ARE-bearing mRNA
rapid deadenylation and degradation (2,3);
however, TTP itself does not have any deadenylase activity.
Previous studies have investigated the proteins that are involved
in TTP-mediated mRNA decay. Yamashita et al (19) reported that the poly(A)-specific
nuclease 2 (PAN2)-PAN3 complex and the CCR4-NOT complex serve
critical roles in deadenylation. The PAN2-PAN3 complex hydrolyses
poly(A) in a distributive manner (individual and not continuous),
whereas the CCR4-NOT complex hydrolyses poly(A) in a processive
(continuous) manner and may promote rapid deadenylation (19). Previous studies have additionally
demonstrated that CCR4 is involved in the regulation of mRNAs
without a dedicated destabilizing motif (19,20)
and CAF1 (CNOT7 in human cells), rather than CCR4, is necessary for
the degradation of ARE-containing mRNA (9). These results demonstrated that CNOT7
may serve a critical role in ARE-bearing mRNA decay and the
authors' previous study confirmed this hypothesis (12). Although TTP, CNOT7 and CNOT1 were
coimmunoprecipitated in HPMECs (12), the associations between TTP and
CNOT7 have not been completely identified.
CNOT7 is a subunit protein of the CCR4-NOT complex,
while CNOT1 is the scaffold protein of the CCR4-NOT complex
(10). Fabian et al
(11) reported that CNOT1 may
directly bind to the C terminus of TTP, which provides critical
evidence for investigating the model of TTP-mediated mRNA decay. As
TTP and CNOT7 interact with CNOT1 (11,21),
together with the results of our previous study that revealed that
TTP, CNOT7 and CNOT1 were coimmunoprecipitated in HPMECs, it was
hypothesized that CNOT1 may serve as a platform that recruits CNOT7
and TTP with ARE-containing mRNA, and they may collectively exert
post-transcriptional control of AMD. The authors' previous studies
confirmed that TTP and CNOT7 were involved in the regulation of the
expression of ICAM-1 and IL-8 (12,13).
Based on the above hypothesis, it was speculated that CNOT1 may
also be involved in the regulation of ICAM-1 and IL-8 expression,
and the results of the present study confirmed this supposition.
Furthermore, in the present study it was demonstrated that CNOT1,
CNOT7 and TTP co-localize in the cytoplasm, and CNOT1 silencing
abolished CNOT7 and TTP Co-IP; however, CNOT7 or TTP silencing did
not influence CNOT1-TTP or CNOT1-CNOT7 Co-IP, respectively. These
results implied that CNOT1 may bind to TTP and CNOT7, respectively,
while TTP and CNOT7 may not directly combine. These results,
together with those of the aforementioned previous studies, suggest
that CNOT1 may recruit CNOT7 and TTP with ARE-bearing mRNA and
together may promote AMD.
In conclusion, CNOT1 may directly bind to the C
terminus of TTP and recruit CNOT7 to deadenylate TTP-bound mRNA;
however, the mechanism by which TTP rapidly dissociates from CNOT1
and goes on to carry target mRNA remains unclear. In addition, the
upstream factors that regulate CNOT7 deadenylase activity are also
not well known. Further studies are required to address these
questions. A CNOT1 deficiency animal model is additionally required
to investigate the effect of CNOT1 on inflammatory mediator
expression and acute lung injury in vivo.
Acknowledgements
The authors would like to thank Mr. Jin Yang, Mr.
Wen-Xue Liang, Mrs. Huan-Huan Zhang, Mrs. Ting Zhang and Mr.
Shao-Lin Zhao (all from the First People's Hospital of Lianyungang
City, Lianyungang, China) for their technical help.
Funding
The present study was supported by the China
Postdoctoral Science Foundation (grant no. 2015M570420), the
Jiangsu Provincial Health and Family Planning Commission Science
Foundation (grant no. H201558), the National Natural Science
Foundation of China (grant nos. 81300052 and 81670073), the Natural
Science Foundation of Jiangsu Province (grant nos. BK20130402 and
BK20161383) and the Lianyungang Science and Technology Bureau
Foundation (grant no. SH1401).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
J-XS and RH analyzed and interpreted the data. X-CZ
and C-CL conducted the experiments. J-XS, J-SL and X-ML designed
the experiments. HW, YS and XS helped design the experiments and
analyzed the data. J-XS wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCR4
|
carbon catabolite repressor protein
4
|
|
NOT
|
Negative on TATA
|
|
CNOT1
|
subunit 1 of the CCR4-NOT complex
|
|
CNOT7
|
subunit 7 of the CCR4-NOT complex
|
|
TTP
|
tristetraprolin
|
|
ARE
|
AU-rich element
|
|
HPMECs
|
human pulmonary microvascular
endothelial cells
|
|
PAN
|
poly (A)-specific nuclease
|
References
|
1
|
Lai WS, Carballo E, Thorn JM, Kennington
EA and Blackshear PJ: Interactions of CCCH zinc finger proteins
with mRNA. Binding of tristetraprolin-related zinc finger proteins
to Au-rich elements and destabilization of mRNA. J Biol Chem.
275:17827–17837. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lai WS, Carballo E, Strum JR, Kennington
EA, Phillips RS and Blackshear PJ: Evidence that tristetraprolin
binds to AU-rich elements and promotes the deadenylation and
destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol.
19:4311–4323. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ogilvie RL, Sternjohn JR, Rattenbacher B,
Vlasova IA, Williams DA, Hau HH, Blackshear PJ and Bohjanen PR:
Tristetraprolin mediates interferon-gamma mRNA decay. J Biol Chem.
284:11216–11223. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vlasova-St Louis I and Bohjanen PR:
Post-transcriptional regulation of cytokine signaling by AU-rich
and GU-rich elements. J Interferon Cytokine Res. 34:233–241. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Collart MA and Timmers HT: The eukaryotic
Ccr4-not complex: A regulatory platform integrating mRNA metabolism
with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol.
77:289–322. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inada T and Makino S: Novel roles of the
multi-functional CCR4-NOT complex in post-transcriptional
regulation. Front Genet. 5:1352014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tucker M, Valencia-Sanchez MA, Staples RR,
Chen J, Denis CL and Parker R: The transcription factor associated
Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA
deadenylase in Saccharomyces cerevisiae. Cell. 104:377–386. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wahle E and Winkler GS: RNA decay
machines: Deadenylation by the Ccr4-not and Pan2-Pan3 complexes.
Biochim Biophys Acta. 1829:561–570. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwede A, Ellis L, Luther J, Carrington
M, Stoecklin G and Clayton C: A role for Caf1 in mRNA deadenylation
and decay in trypanosomes and human cells. Nucleic Acids Res.
36:3374–3388. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Petit AP, Wohlbold L, Bawankar P,
Huntzinger E, Schmidt S, Izaurralde E and Weichenrieder O: The
structural basis for the interaction between the CAF1 nuclease and
the NOT1 scaffold of the human CCR4-NOT deadenylase complex.
Nucleic Acids Res. 40:11058–11072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fabian MR, Frank F, Rouya C, Siddiqui N,
Lai WS, Karetnikov A, Blackshear PJ, Nagar B and Sonenberg N:
Structural basis for the recruitment of the human CCR4-NOT
deadenylase complex by tristetraprolin. Nat Struct Mol Biol.
20:735–739. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi JX, Li JS, Hu R, Shi Y, Su X, Li Q and
Zhang F: CNOT7/hCAF1 is involved in ICAM-1 and IL-8 regulation by
tristetraprolin. Cell Signal. 26:2390–2396. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi JX, Su X, Xu J, Zhang WY and Shi Y:
MK2 posttranscriptionally regulates TNF-alpha-induced expression of
ICAM-1 and IL-8 via tristetraprolin in human pulmonary
microvascular endothelial cells. Am J Physiol Lung Cell Mol
Physiol. 302:L793–L799. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sandler H, Kreth J, Timmers HT and
Stoecklin G: Not1 mediates recruitment of the deadenylase Caf1 to
mRNAs targeted for degradation by tristetraprolin. Nucleic Acids
Res. 39:4373–4386. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR
& the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan J, Heller NM, Gorospe M, Atasoy U and
Stellato C: The role of post-transcriptional regulation in
chemokine gene expression in inflammation and allergy. Eur Respir
J. 26:933–947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu T, Shi JX, Geng S, Zhou W, Shi Y and Su
X: The MK2/HuR signaling pathway regulates TNF-alpha-induced ICAM-1
expression by promoting the stabilization of ICAM-1 mRNA. BMC Pulm
Med. 16:842016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huntzinger E, Kuzuoglu-Ozturk D, Braun JE,
Eulalio A, Wohlbold L and Izaurralde E: The interactions of GW182
proteins with PABP and deadenylases are required for both
translational repression and degradation of miRNA targets. Nucleic
Acids Res. 41:978–994. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamashita A, Chang TC, Yamashita Y, Zhu W,
Zhong Z, Chen CY and Shyu AB: Concerted action of poly (A)
nucleases and decapping enzyme in mammalian mRNA turnover. Nat
Struct Mol Biol. 12:1054–1063. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mauxion F, Faux C and Seraphin B: The BTG2
protein is a general activator of mRNA deadenylation. EMBO J.
27:1039–1048. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Faraji F, Hu Y, Yang HH, Lee MP, Winkler
GS, Hafner M and Hunter KW: Post-transcriptional control of tumor
cell autonomous metastatic potential by CCR4-NOT deadenylase CNOT7.
PLoS Genet. 12:e10058202016. View Article : Google Scholar : PubMed/NCBI
|