Introduction
Recent advances in surgical techniques and
developments in anticancer agents with improved therapeutic
efficacies have been made; however, gastric cancer (GC) remains to
be the most prevalent type of malignancy and is the second most
common cause of cancer-associated mortality worldwide (1,2).
Metastasis is one of the key molecular steps affecting the
prognosis of GC, but as a complex process, further investigation is
required (3). Identifying novel
diagnostic or prognostic biomarkers of GC may be considered a major
objective for the treatment of GC.
Epithelial-to-mesenchymal transition (EMT) is one of
the key molecular steps in the process associated with distant
metastasis (4), during which tumor
cells exhibiting epithelial characteristics acquire mesenchymal
characteristics by modulating cellular polarity and adhesion, and
via the loss of cell-cell junctions (5). Throughout the process of EMT, cancer
cells lose the expression of cellular adhesion proteins, such as
E-cadherin, which has been considered as a hallmark of EMT; cells
also acquire the expression of mesenchymal markers, including
vimentin (5,6). EMT permits the invasion and migration
in a variety of cancer types, including gastric and colon cancer
(5).
The transmembrane (TMEM) protein superfamily is a
group of transmembrane proteins that participate in particular
signaling pathways and tumor development associated with
established oncogenes, particularly TMEM16a (7,8),
TMEM17a and TMEM17b (9). The
associated proteins serve roles in colorectal, ovarian and bladder
cancer. TMEM41A, a member of the TMEM41 family, is a 264-amino acid
protein encoded by a gene mapped to human chromosome 3 (10,11).
Chromosome 3 consists of ~214 million bases, encoding <1,100
genes (11), and possesses various
human tumor-associated loci, as well as a chemokine receptor gene
cluster (12). Particular areas of
the chromosome 3 short arm tend to be lost in numerous types of
tumor cells (13,14).
At present, the exact roles of TMEM41A in GC have
not been determined. In the present study, the expression of
TMEM41A and the prognoses of 147 patients with GC, and the
functional role of TMEM41A in GC-associated metastasis were
evaluated to investigate the potential therapeutic role of TMEM41A
in the treatment of GC.
Materials and methods
Patient specimens and cell lines
Tumor tissues and matched adjacent normal gastric
tissues were obtained from 147 patients (31–84-years-old) who
underwent radical GC surgery between April 2007 and June 2010 at
the Qingdao Municipal Hospital (Qingdao, China). Patients that
received neoadjuvant chemotherapy or irregular therapy (standard
therapy treatments, followed by 6 months of chemotherapy) were
excluded from the present study. The adjacent normal tissues were
laterally resected at a distance of ≥5 cm from the tumor margin.
None of the subjects underwent neoadjuvant chemoradiotherapy. All
the resected samples were immediately frozen in liquid nitrogen and
maintained at −80°C until use.
Written informed consent was provided by all
patients prior to enrollment into the present study, and the study
protocols were approved by the Ethics Committee of Qingdao
University (Qingdao, China). The clinical data collected from the
subjects are summarized in Table
I, including patient age and sex, tumor location, size and
differentiation, lymph node involvement, distant metastasis, and
tumor, node and metastasis (TNM) classification. Postoperative
follow-up was performed every 3 months for a minimum of 5 years of
104 patients; 43 patients did not undergo follow-up due to
exclusion criteria that arose in the present study, including
patients withdrawal and failure to adhere to courses of standard
treatment.
 | Table I.Association between TMEM41A
expression and clinicopathologic features. |
Table I.
Association between TMEM41A
expression and clinicopathologic features.
| Variable | Low TMEM41A
expression | High TMEM41A
expression | P-value |
|---|
| Total | 39 | 65 |
|
| Sex |
|
| 0.545 |
|
Male | 21 | 35 |
|
|
Female | 18 | 30 |
|
| Age (years) |
|
| 0.491 |
|
≤50 | 15 | 26 |
|
|
>50 | 24 | 39 |
|
| Tumor size
(cm) |
|
| 0.566 |
| ≤5 | 25 | 40 |
|
|
>5 | 14 | 25 |
|
| Histology |
|
| 0.488 |
|
Adenocarcinoma | 34 | 57 |
|
|
Mucinous | 5 | 8 |
|
| Local tumor
invasion |
|
| 0.584 |
| Early
stage | 36 | 61 |
|
| Late
stage | 3 | 4 |
|
| Lymph node
metastasis |
|
| 0.0026 |
|
Negative | 25 | 22 |
|
|
Positive | 14 | 43 |
|
| Distant
metastasis |
|
| 0.0179 |
|
Negative | 34 | 43 |
|
|
Positive | 5 | 22 |
|
| TNM stage |
|
| 0.00238 |
|
I–II | 27 | 25 |
|
|
III–IV | 12 | 40 |
|
The human GC cell lines MKN-45, MGC-803, NCI-N87,
SNU-5, KATO III, HGC-27, BGC-823, SGC-7901 and AGS were obtained
from the American Type Culture Collection (Manassas, VA, USA), and
were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), McCoy's 5A complete
medium or RPMI-1640 (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), supplemented with 10% heat-inactivated fetal bovine serum
(FBS; Sigma-Aldrich; Merck KGaA). All cells were maintained in a
humidified atmosphere with 5% CO2 at 37°C.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was employed to isolate total RNA from cell lines
according to the manufacturer's protocol. cDNA was obtained from 2
µg RNA using the PrimeScript RT reagent kit (Takara Biotechnology
Co., Ltd., Dalian, China). The mRNA expression levels of TMEM41A
were determined by semi-qPCR with GoTaq polymerase (Promega
Corporation, Madison, WI, USA) following the manufacturer's
protocols. qPCR was followed by amplification with SYBR®
Premix Ex Taq (Takara Biotechnology Co., Ltd.) using an ABI 7500HT
system (ABI; Thermo Fisher Scientific, Inc.). The thermocycling
program was set as follows: Initial denaturation for 5 min at 95°C;
40 cycles at 95°C for 15 sec and 60°C for 30 sec. The primers used
were as follows: TMEM41A forward, 5′-CTGCTGTGCTGTGTGTTGAC-3′ and
reverse, 5′-GTGTCCCAGGAGAAAAGAGCA-3′; and β-actin forward,
5′-AGTGTGACGTGGACATCCGCAAAG-3′ and reverse,
5′-ATCCACATCTGCTGGAAGGTGGAC-3′. The 2−ΔΔCq method was
used to quantify the expression levels of RNA (15). For semi-qPCR, the DNA products were
run on a 1.5% agarose gel containing ethidium bromide. The bands
were quantified using a UV illuminating instrument (Bio-Rad
Laboratories, Inc., Hercules, CA, MA, USA).
Cell transfection
The small interfering (si)-negative control (NC;
cat. no. CON077) and si-TMEM41A (cat. no. 38422-2) oligonucleotides
were synthesized by Shanghai GenePharma Co., Ltd., (Shanghai,
China) and transfected at 20 nM. the oligonucleotides sequence of
the siRNA-TMEM41A (5′-3′) is:
Ccgg-(stem)gcGGAAGTAGCTTGCCTCACTCTCGAG
(loop)-AGTGAGGCAAGCTACTTCCGC(stem)-TTTTTg. The control
(pCMV6-NC) or TMEM41A overexpression (pCMV6-TMEM41A) vectors were
constructed by OriGene Technologies, Inc. (Rockville, MD, USA).
Oligonucleotide transfection was performed using 0.5–1.0 µl
siRNA/vector with Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol; successful transfection was confirmed via western
blotting as described below. Cells were collected at 48 h following
transfection.
Immunohistochemistry
Immunohistochemistry with the gastric tumor and
adjacent normal tissues was performed as previously described
(16). A TMEM41A rabbit polyclonal
antibody (1:50; sc-103285; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) was utilized as the primary antibody (overnight, 4°C); a
secondary antibody (1:1,000; GK600705; Gene Tech Biotechnology Co.,
Ltd. (Shanghai, China) was applied for 1 h at room temperature.
Subsequently, sections were counterstained with hematoxylin at room
temperature for 40 min and analyzed under a light microscope
(magnification, ×10). The sections were scored semi-quantitatively
by two observers independently, in a blinded manner and analyzed
under a light microscope. The scoring system was as follows: 0, 0%
immunoreactive cells; 1, <5% immunoreactive cells; 2, 5–50%
immunoreactive cells and 3, >50% immunoreactive cells. Scores of
0 or 1 were considered as low expression levels, and scores of 2 or
3 were considered as high expression levels.
Cell proliferation analysis
Cell proliferation assays of transfected cells were
performed using a Cell Counting kit-8 (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) according to the
manufacturer's protocols; absorbance measurements at 450 nm were
conducted to determine the cell proliferative ability in each well.
The cells were seeded in a dish at a density of 1×105
cells/dish and cultured for 24 h at 37°C prior to transfection;
detection was performed at 24, 48 and 72 h post-transfection.
Wound-healing assay
A wound-healing assay was employed to assess the
migration capacity of transfected cells in a defective monolayer.
In brief, cells were seeded in 6-well plates and a 100-µl pipette
tip was used to conduct scratches. The medium was discarded, the
cells were washed with PBS and fresh medium (RPMI-1640/DMEM and 10%
FBS) was added, followed by the analysis of scratch closure under a
light microscope at 18 h. Calculations were performed using the
following formula: (Sx time 0-Sx time
point/Sx time 0) ×100, where S indicated the
distance in µm.
Cell migration assays
A cell migration assay was performed using Costar
Transwell inserts (Thermo Fisher Scientific, Inc.) with a diameter
of 6.5 mm and a pore size of 8.0 µm. Briefly, cells
(1×104) resuspended in 300 µl serum-free medium were
seeded into the upper Transwell chamber, and the bottom chamber was
supplemented with 10% FBS. The cells were incubated for 24 h.
Subsequently, the upper chamber was stained with Diff-Quik stain
(Polysciences, Inc., Warrington, PA, USA) according to the
manufacturer's protocol. Cell numbers were counted in 10 fields of
view under an inverted microscope (magnification, ×40; DMI600B;
Leica Microsystems GmbH, Wetzlar, Germany), and the proportion of
migrating cells was determined after normalization to control
cells.
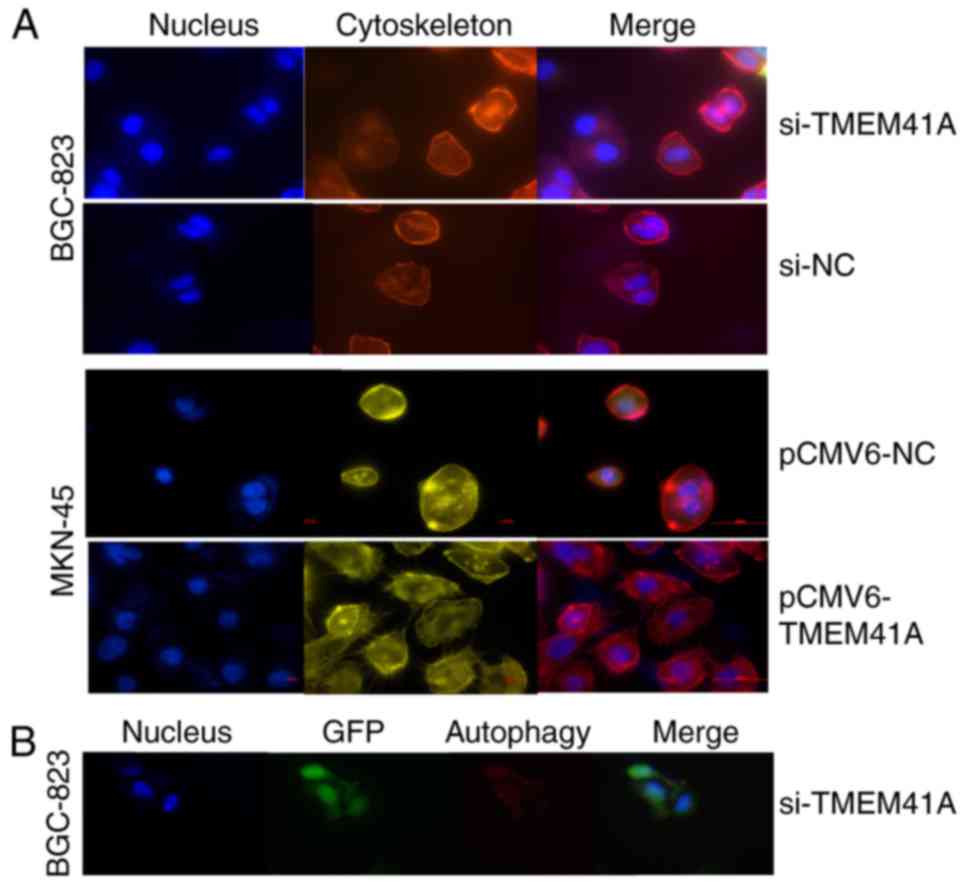
Cell immunofluorescence for
cytoskeleton stain and autophagy
Cells were seeded on coverslips when a single
monolayer was achieved from culture, fixed in 4% paraformaldehyde
for 10 min at room temperature, permeabilized in 0.1% Triton X-100
in PBS for 4 min. Subsequently, cells were blocked with 1% BSA for
20 min at room temperature, and then incubated at room temperature
for 30 min with rhodamine-conjugated phalloidin (Sigma-Aldrich;
Merck KGaA) at 1:200 in blocking solution (1% BSA in 0.1% Triton
X-100 in PBS). The nuclei were counterstained with DAPI for 20 min
at room temperature. Images were captured with a fluorescence
microscope.
Autophagosome formation was detected by red
fluorescent protein (RFP)-LC3 puncta incorporation into autophagic
vacuoles. Sterile coverslips were seeded with 5×105
cells into 6-well plates. Following adherence and washing twice
with PBS, cells were cultured in serum-free medium for 24 h.
Subsequently, the cells were transiently transfected with
RFP-LC3B-expressing GFP-vector (OriGene Technologies, Inc.) for 32
h with Lipofectamine® 2000, followed by washing with
cold-ice PBS and fixation with 4% formaldehyde in PBS at room
temperature for 20 min. The nuclei were counterstained with DAPI
for 20 min at room temperature. Images were captured with a
fluorescence microscope (magnification, ×100).
Tumor formation in nude mice
Stably siRNA-transfected BGC-823 cells selected via
incubation with 1–3 µg/l puromycin and were then suspended in 0.2
ml PBS and injected into the caudal vein of 12 5-week-old female
BALB/c nude mice (20–22 g) (5×106 cells/mouse). The mice
housed under a 12 h light/dark cycle with free access to food and
water at 20–24°C, 50% humidity; mice were monitored for 70 days and
then sacrificed. The tumor size was recorded and tumor volume was
calculated using the formula: (length × width2)/2.
Tumors formed in vivo were collected, sectioned and stained
with hematoxylin and eosin (H&E) at room temperature for 40 min
and analyzed under a light microscope.
Western blot analysis
Briefly, for western blotting, after washing with
cold-ice PBS, total protein of transfected cells was isolated by
incubation in radioimmunoprecipitation assay buffer (Cell Signaling
Technology, Inc., Danvers, MA, USA) containing complete protease
inhibitor cocktail (Roche Diagnostics, Basel, Switzerland) for 1 h
on ice. Following centrifugation (15,000 × g at 4°C for 20 min),
the supernatants were collected. Protein concentration was
determined using a bicinchoninic acid protein assay kit (Thermo
Fisher Scientific, Inc.). Samples (2–10 mg) were subjected to 8–12%
SDS-PAGE according to protein mass loaded and transferred to
polyvinylidene difluoride membranes activated with 100% methanol.
After blocking in 5% fat-free milk for 1 h, the membranes were
incubated with a primary antibody against TMEM41A (dilution, 1:200;
Santa Cruz Biotechnology, Inc.), E-cadherin (ab15148), N-cadherin
(ab18203), p62 (ab91526), LC3-I/II (ab51520), and β-actin (ab8227)
(dilution, 1:1,000; Abcam, Cambridge, MA, USA) overnight on ice and
incubated with the indicated secondary antibody (in-house; 1:1,000)
for 2 h at room temperature. The membrane was then washed with
tris-buffered saline with 1:1,000 Tween-20 in v/v. prior to
treatment with an enhanced chemiluminescent reagent and placed in a
dark room to allow the reaction to run to completion. β-actin was
used as a positive control.
Bioinformatic analysis
Investigation with The Cancer Genome Atlas (TCGA;
http://cancergenome.nih.gov/) dataset
was conducted to analyze the expression levels of TMEM41A in normal
gastric tissues; Protein Atlas (www.proteinatlas.org) also revealed that the mRNA and
protein expression levels of TMEM41A may be inconsistent.
Statistical analysis
Data were analyzed using SPSS version 18 (SPSS Inc.,
Chicago, IL, USA). The results are presented as the mean ± standard
error of the mean. Kaplan-Meier analysis was conducted, followed by
a log-rank test. All experiments were performed in triplicate. The
differences between groups were analyzed using one-way analysis of
variance followed by the Least Significant Difference t-test.
Categorical data was analyzed with a χ2 test. In all
cases, P<0.05 was considered to indicate a statistically
significant difference and P<0.01 was considered to indicate a
highly statistically significant difference.
Results
Expression of TMEM41A in human GC cell
lines
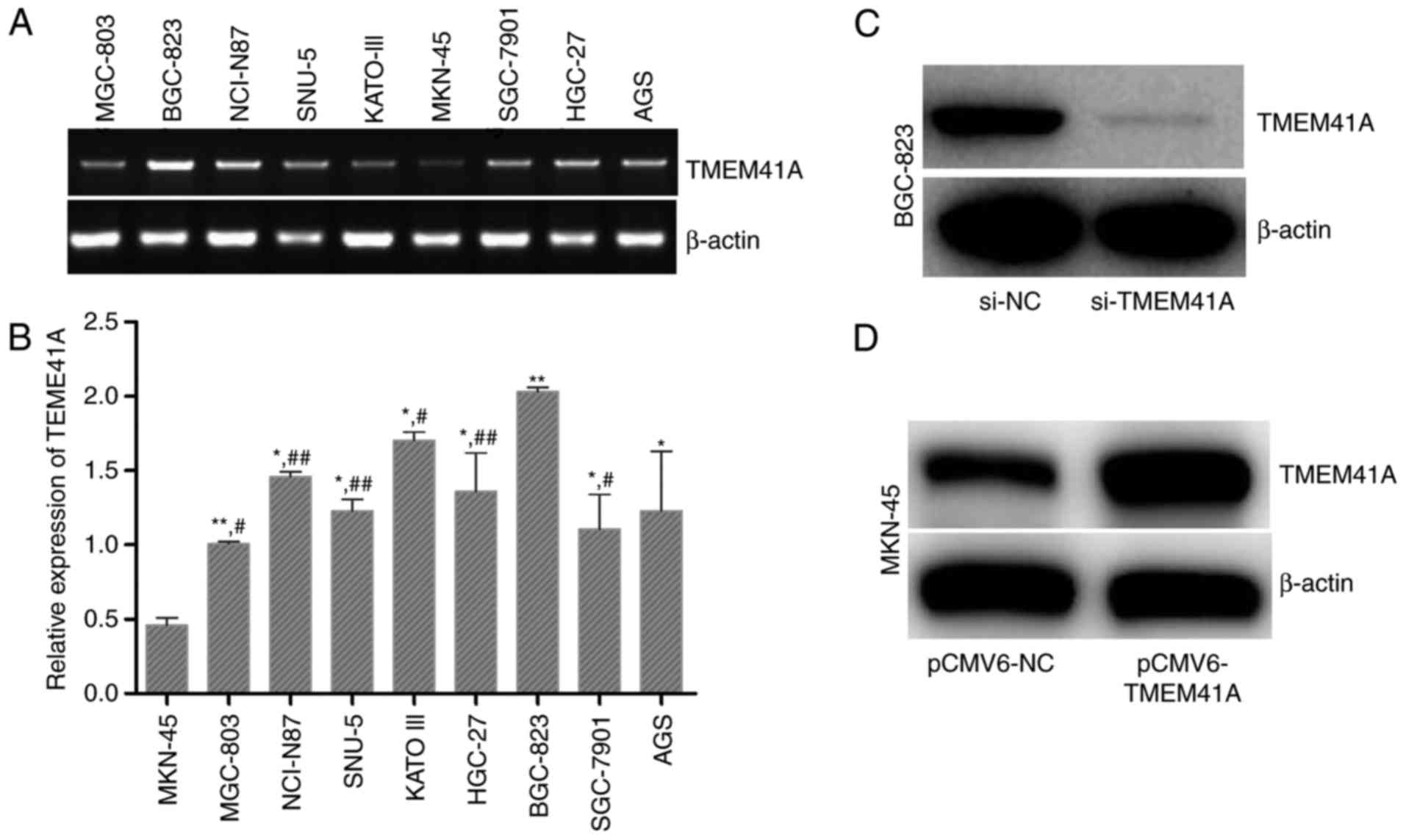
The expression levels of TMEM41A in the nine human
GC cell lines were determined. As presented in Fig. 1A and B
(F=5.381>F0.05; P<0.05), the expression levels of
TMEM41A were the highest in BGC-823 cells and the lowest in MKN-45
cells (Fig. 1B). Therefore,
BGC-823 cells were transfected with siRNA against TMEM41A, and
MKN-45 cells were transfected with a TMEM41A-overexpression plasmid
to investigate the effects of siRNA and plasmid transfection on the
expression levels of TMEM41A. Western blot analysis demonstrated
that si-TMEM41A markedly decreased the expression levels of TMEM41A
in BGC-823 cells, and that pCMV6-TMEM41A notably increased the
expression levels of TMEM41A in MKN-45 cells (Fig. 1C and D).
TMEM41A expression in human GC cells
and its association with clinicopathological characteristics
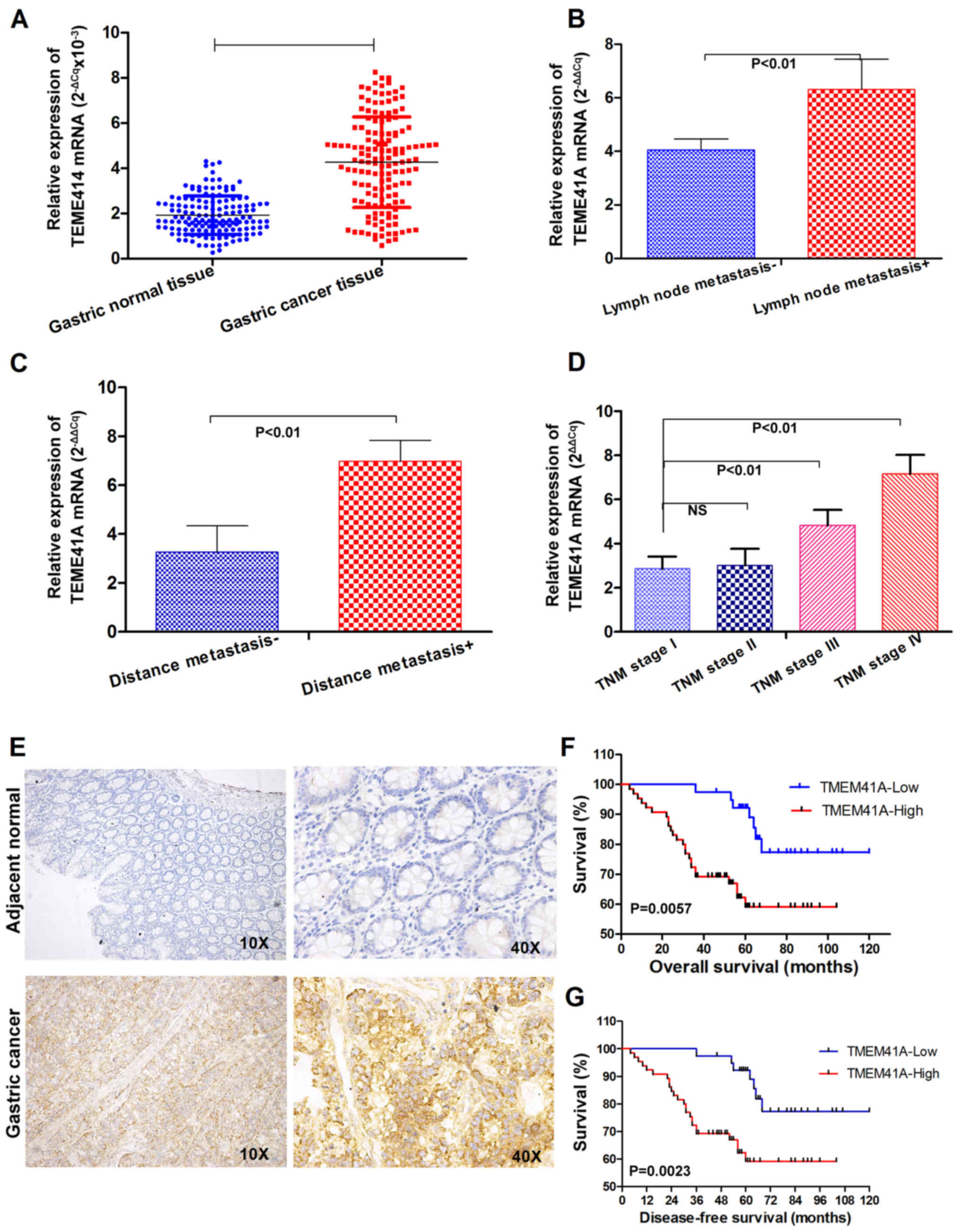
To investigate the potential role of TMEM41A in
GC-associated metastasis, RT-qPCR was used to assess TMEM41A
expression in 147 cases of GC and paired adjacent normal tissues,
which revealed the overexpression of TMEM41A mRNA in the GC tissue
exhibited by 123/147 (83.67%) of the pairs, with a mean fold change
of 2.22 for the cancer tissues; expression levels were
significantly higher within the tumor tissues compared with in that
of the normal tissues (Fig. 2A).
In addition, the relative mean expression levels of TMEM41A were
significantly higher in subjects with lymph node involvement
compared with those without (P<0.01; Fig. 2B). Consistently, TMEM41A expression
was significantly higher in subjects with distant metastasis
compared to those without (P<0.01; Fig. 2C). The associations of TMEM41A
expression with the clinicopathological characteristics of patients
with GC are summarized in Table I.
TMEM41A expression was observed to be significantly associated with
advanced TNM stages (III and IV), lymph node involvement and
distant metastasis (P<0.01; Fig.
2B-D); however, TMEM41A expression was not significantly
associated with other clinicopathological characteristics,
including patient age, sex, tumor location, differentiation or size
(all P>0.05).
TMEM41A overexpression is correlated
with lymph node metastasis, distant metastasis and advanced TNM
stage in human GC tissues
Subsequently, the present study determined the
expression levels of TMEM41A protein in the same pathological
specimens. TMEM41A protein was suggested to be localized on the
cell plasma membranes of GC samples (Fig. 2E). As presented in Table I, of the 147 patients with GC, 104
(selected based on follow-up, staining, and other factors,
including variations in therapy) were divided into two subgroups
based on the expression levels of TMEM41A. The high-expression
group included 65 patients with a higher expression levels of
TMEM41A in the carcinoma tissue compared with in adjacent normal
gastric tissues, whereas in the low-expression group (n=39), the
mean TMEM41A staining intensity obtained during semi-quantitative
analysis of carcinoma tissue samples decreased compared with the
adjacent normal gastric tissues (data not shown).
Survival analyses were performed on patients with
GC. Patients in the TMEM41A high-expression group exhibited poorer
overall and disease-free survival compared with the low-expression
group (Fig. 2F and G).
There were no significant differences in the 1-year
overall survival rate between the two groups; however, the 3- and
5-year overall survival rates were poorer in the TMEM41A
high-expression group. The 5-year overall survival rates for the
high- and low-expression groups were 75.6 and 45.8%, respectively
(P=0.012; Table II). This finding
suggested that high expression levels of TMEM41A in GC may be
considered as a predictor of poor prognoses of GC.
 | Table II.Association between TMEM41A
expression and survival rate. |
Table II.
Association between TMEM41A
expression and survival rate.
|
| TMEM41A
density |
|---|
|
|
|
|---|
| Survival
measurement (%) | Low-TMEM41A | High-TMEM41A | P-value |
|---|
| 1-year overall
survival | 95.6±4.0 | 89.5±4.4 |
|
| 3-year overall
survival | 90.4±8.8 | 75.5±8.7 |
|
| 5-year overall
survival | 75.6±6.0 | 45.8±6.8 | 0.012a |
TMEM41A does not promote the
proliferation of GC cells
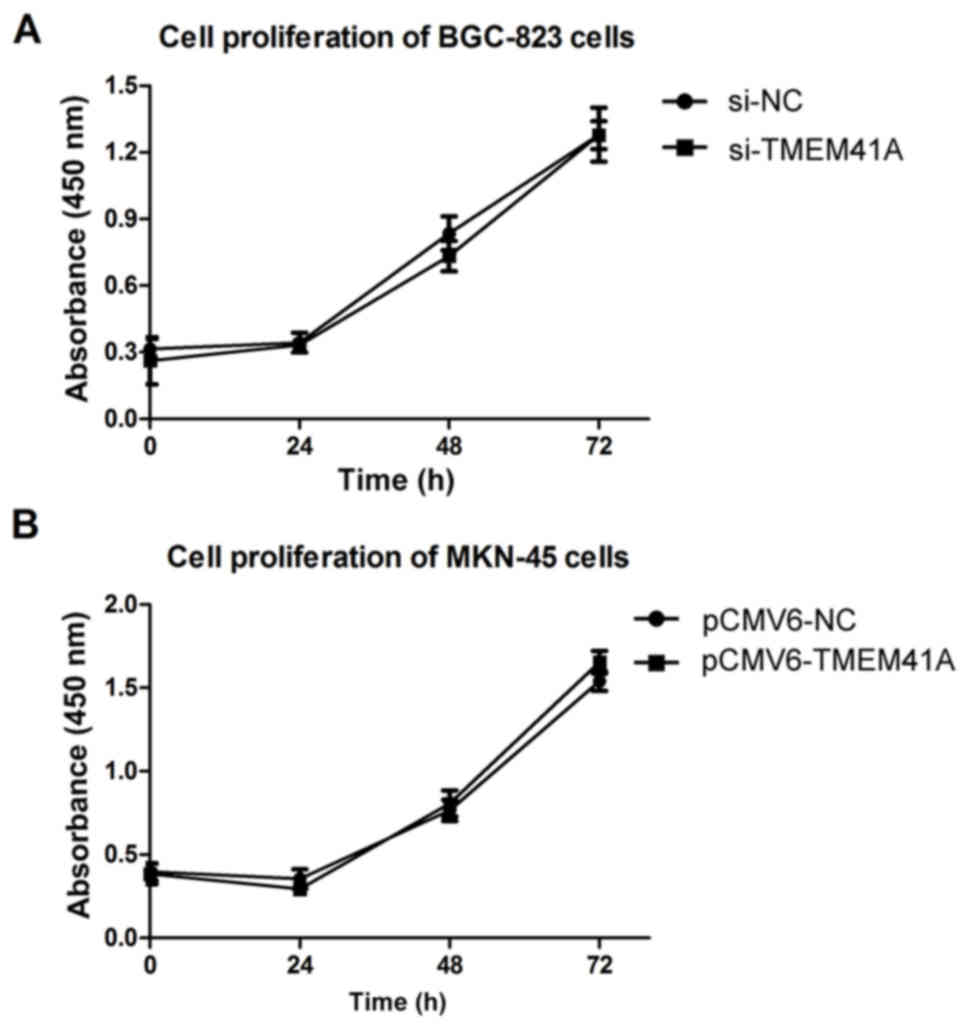
The present study performed Cell Counting kit-8
assays to investigate the effects of TMEM41A on GC cell
proliferation. The Cell Counting kit-8 assays demonstrated that the
stable knockdown of TMEM41A mediated by siRNA was not associated
with variations in BGC-823 cell proliferation (P>0.0; Fig. 3A). Similar results were observed in
TMEM41A-overexpressing MKN-45 cells (P>0.05; Fig. 3B).
Association between TMEM41A expression
and the migration of human GC cells
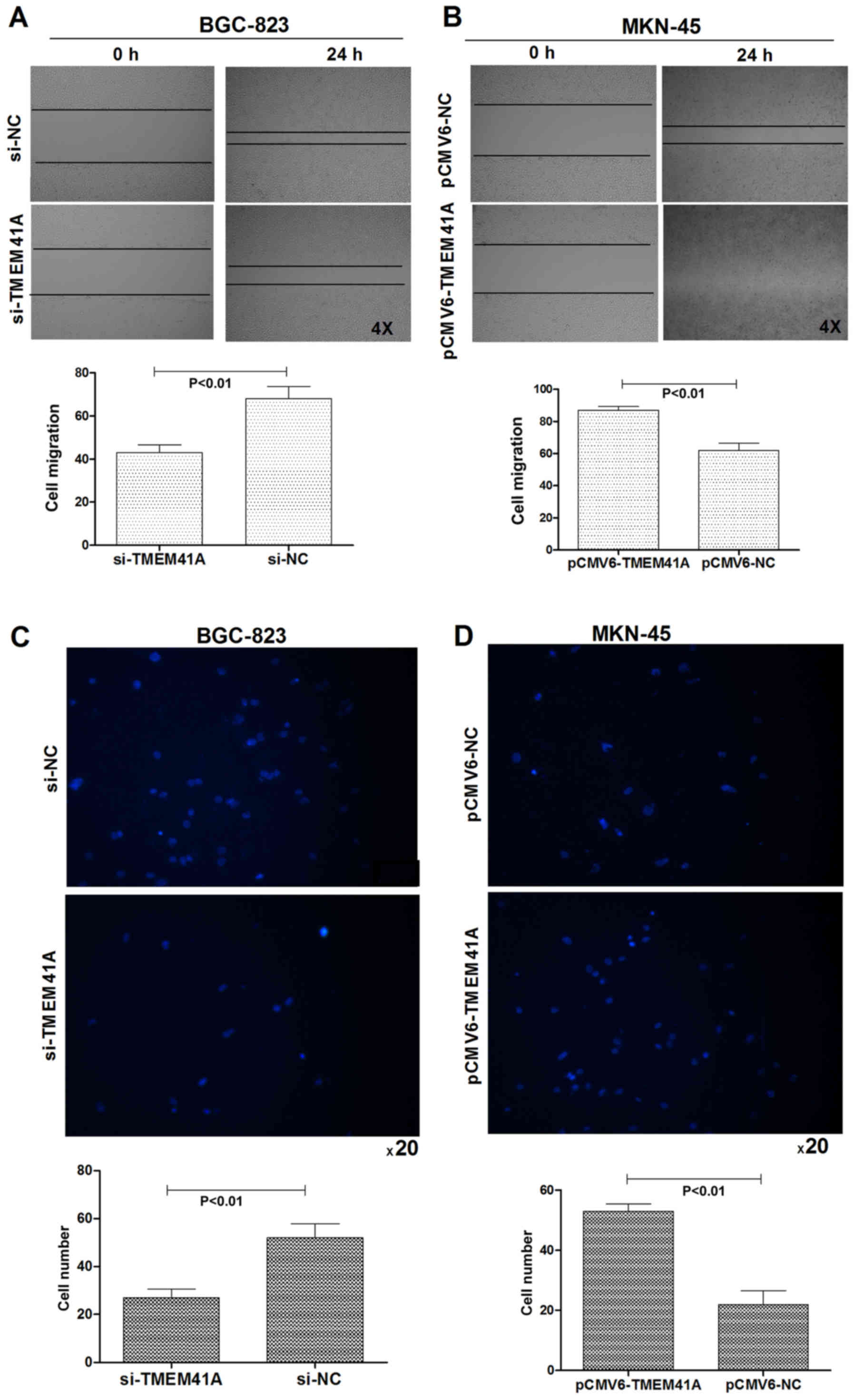
In order to investigate the role of TMEM41A in cell
migration, wound-healing and Transwell assays were performed. The
results indicated that knockdown of TMEM41A significantly decreased
the migration ability of BGC-823 cells (Fig. 4A; P<0.01). Conversely, a
significant increase in the rate of migration was observed when
TMEM41A was overexpressed in MKN-45 cells compared with in the
control (Fig. 4B; P<0.01).
Transwell assays indicated that the migration ability of BGC-823
cells was significantly decreased following the silencing of
endogenous TMEM41A compared with in the control (Fig. 4C; P<0.01). Conversely, the
overexpression of TMEM41A significantly increased the migration
ability of MKN-45 cells compared with in the control (Fig. 4D; P<0.01).
Different expression of TMEM41A leads
to cytoskeletal rearrangement and inhibition of cell adhesion and
spreading
To determine the role of TMEM41A in the regulation
of cytoskeletal dynamics, rhodamine-conjugated phalloidin staining
was applied (Fig. 5A). Inside the
cells, actin filaments were distributed in a disorderly manner, and
no evident stress fiber formation was observed in TMEM41A knockdown
cells, indicating that the knockdown of TMEM41A may inhibit stress
fiber formation. On the cell membranes, membrane ruffling and the
formation of pseudopodia, reflective of cell migration, were
markedly observed in control and siTMEM41A transfected cells; in
the overexpression group, opposing findings were observed. These
results indicate that high expression levels of TMEM41A may promote
metastatic properties of GC cells, consistent with the
wound-healing assay as aforementioned.
TMEM41A may induce autophagy within GC
cells
To further verify the hypothesis of the present
study, RFP-LC3B plasmids were transiently transfected into the
cells. Punctate fluorescence demonstrated the induction of
autophagy. As presented in Fig.
5B, no notable punctate fluorescence was observed in
si-TMEM41A-treated BGC-823 cells, but not in control
vector-transfected or TMEM41A-overexpressing cells. These results
suggested that TMEM41A may be associated with autophagy.
Effect of TMEM41A downregulation on
E-cadherin expression and autophagy in GC cells
EMT is crucial for the invasion and migration of
tumor cells, and the development of metastasis. To investigate the
mechanisms underlying the effects of TMEM41A on cell migration, the
effects of TMEM41A knockdown or overexpression on EMT were examined
by analyzing the expression levels of EMT-associated factors
(Fig. 6A). The knockdown or
overexpression of TMEM41A affected the expression levels of
E-cadherin and N-cadherin, and the autophagy-associated factors p62
and LC3-1/2, indicating that the knockdown or overexpression of
TMEM41A may affect EMT and the autophagy process (Fig. 6A).
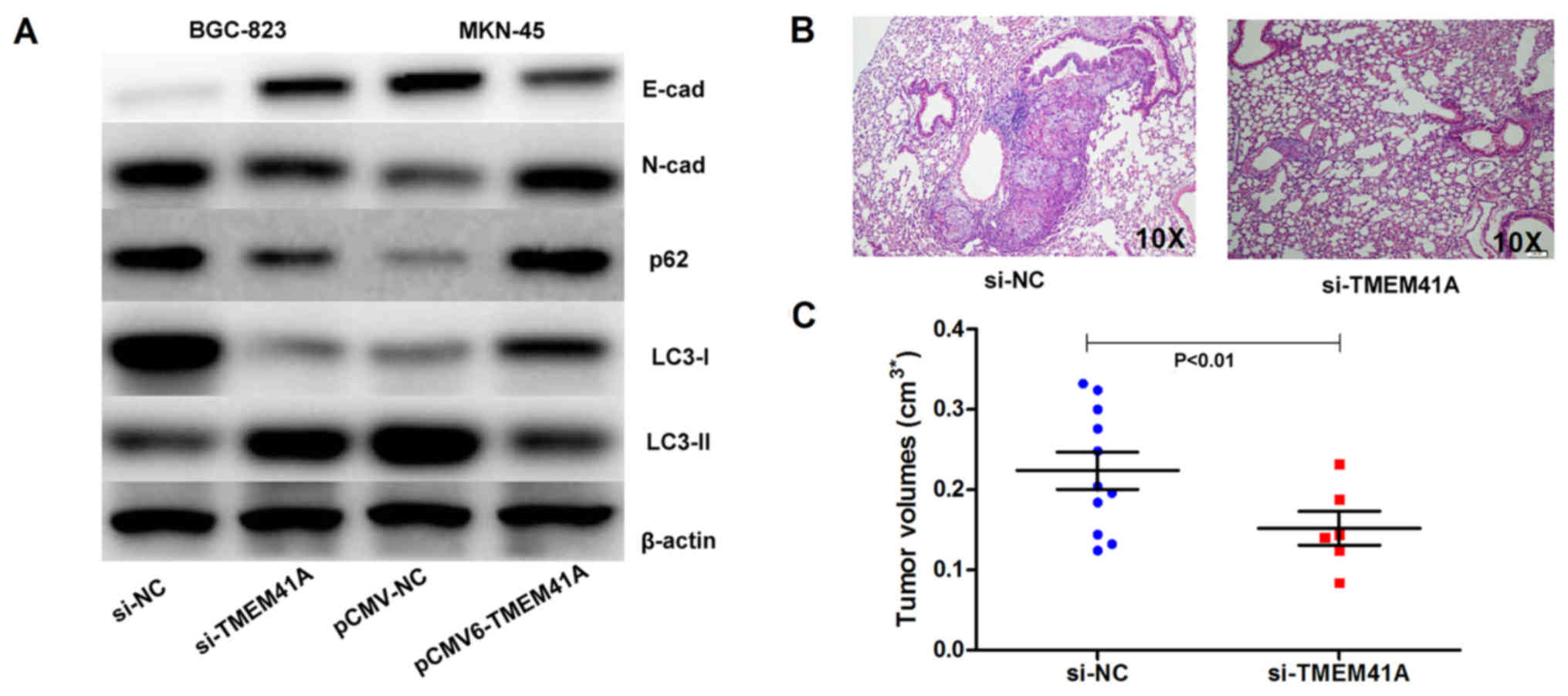 | Figure 6.Knockdown of TMEM41A decreases the
migration ability associated with EMT and autophagy in GC cells.
(A) Western blot analysis demonstrated the effects of TMEM41A
expression on EMT factors, including E-cad and N-cad expression and
autophagy via the dysregulation of p62, and the conversion of LC3-I
into LC3-II. (B) Knockdown of TMEM41A in BGC-823 cells impaired the
formation of tumor metastases in the lungs of nude mice, as
demonstrated by hematoxylin and eosin staining. (C) Knockdown of
TMEM41A in the BGC-823 cells was associated with smaller tumor
volume. Cad, cadherin; EMT, epithelial-mesenchymal transition; GC,
gastric cancer; LC3, light chain 3; NC, negative control; si, small
interfering RNA; TMEM41A, transmembrane protein 41A; E-cad,
E-cadherin; N-cad, N-cadherin. |
Knockdown of TMEM41A affects tumor
metastasis in nude mice
To further determine the effect of TMEM41A on GC
tumor metastasis in vivo, the present study employed an
siRNA-mediated stable knockdown cell model. H&E staining
indicated that tumors were formed in vivo at a lower rate in
the si-TMEM41A group compared with in the si-NC group (Fig. 6B). Both groups exhibited successful
tumor formation in the lung, and tumor size of the si-TMEM41A group
was significantly smaller compared with in the si-NC group
(Fig. 6C; P<0.01). These
results indicated alterations in the expression of TMEM41A and
EMT-associated markers may be associated with metastatic behavior
in vivo.
Discussion
To retain normal organ size and specific organ
morphology, cell numbers in tissues of high proliferative potential
are strictly controlled by the balance between cell proliferation
and apoptosis during organ development (17). This suggests that the dysregulation
of genes associated with these processes may be detected in human
malignancy.
Metastasis is one of the key processes affecting the
prognosis of GC, but it is a complex process which requires further
investigation (3). A total of ~70%
of patients with GC develop metastasis (18). Advances in treatments for GC have
been made; however, patients with advanced or metastatic GC exhibit
poor prognosis (19). Therefore,
the identification of novel diagnostic or prognostic biomarkers for
GC is one of the major goals in this field.
The TMEM protein superfamily comprises >310
different members, including membrane proteins, which are
extensively expressed on cell and lysosomal membranes, and the
endoplasmic reticulum, are strongly associated with membrane
integrity, transport and signaling pathways (20). An increasing number of studies have
investigated the TMEM superfamily and tumor-associated genes,
particularly the oncogenes, TMEM16a (7,8),
TMEM17a and TMEM17b (9) and the
tumor suppressor gene TMEM100 (21,22).
Certain TMEM genes have been reported to be involved in the
development and metastasis of GC, such as TMEM16a (20).
TMEM41A has been considered to be a potential
cancer-associated gene, as it is located on the long arm of
chromosome 3 (10,11). Various human cancer-associated gene
loci are present on chromosome 3. One of the most common
abnormalities in human cancers is the loss of the short arm of
chromosome 3 (23). Therefore, the
tumor suppressor gene(s) located on 3p may serve a key role in the
development of numerous types of cancer (13,14).
In addition, the amplification of chromosome 3q is associated with
cancer; the amplification of the distal portion of chromosome 3q
has been reported to be an important signature associated with lung
squamous cell carcinoma (24). The
role of TMEM41A in cancer has not been previously investigated. The
present study revealed that the TMEM41A expression levels were high
in patients with GC. Increased TMEM41A expression levels were
positively associated with distant metastasis, advanced TNM stage
and lymph node involvement, as observed in the present study.
Patients with low TMEM41A expression levels exhibited improved
overall survival compared with those with high TMEM41A expression
levels. To the best of the authors' knowledge, the present study is
the first to investigate the prognostic potential of TMEM41A in
patients with GC.
An increasing number of studies (25–28)
have investigated the mechanisms underlying the aberrant expression
of TMEM41A; however, the regulation of TMEM41A protein expression
remains unclear. It has previously been demonstrated that its
expression may be regulated by numerous mechanisms, including
chromosomal abnormalities (25),
polymorphisms (26), epigenetic
mechanisms (27) and histone
modifications (28). Further
investigation in identifying the underlying mechanisms of irregular
TMEM41A expression in GC is required. In particular, whether the
upregulation of TMEM41A may be due to chromosomal abnormalities
requires further investigation.
EMT is a series of events including the alteration
of cell-cell and cell-extracellular matrix interactions, and cancer
cell transformation from an epithelial to a mesenchymal phenotype
(29). These alterations are often
accompanied with increased cell motility. Therefore, EMT is a
hallmark of cancer, and is associated with a poor clinical outcome
and metastasis (30,31). During tumor progression and
metastasis, EMT may be induced by autonomous oncogenic activation
or inactivation of signaling molecules, in the presence or absence
of external stimuli (32). In this
process, markers of epithelial differentiation are downregulated,
including E-cadherin; however, N-cadherin, Slug, vimentin,
Twist-related protein-1 and Snail are upregulated (33). EMT is a key process in the
metastasis of GC (34). The
findings of present study suggested that TMEM41A may facilitate the
metastasis of GC cells via EMT. Furthermore, TMEM41A may
participate in the regulation of E-cadherin and N-cadherin
expression; however, further experiments may be conducted in the
future to elucidate the association between TMEM41A and the EMT
signaling pathway.
It was previously suggested that autophagy may
contribute towards the regulation of the migration and invasive
abilities of cancer cells, and the activation of autophagy has been
associated with the inhibition of EMT (35–37).
It has been reported that autophagy accelerates the invasion of
glioblastoma stem cells by regulating DNA damage-regulated
autophagy modulator 1 and p62 (38). The present study demonstrated
similar results, as the upregulation of E-cadherin was accompanied
with that of p62 and the altered conversion of LC3-I to LC3-II. In
addition, immunofluorescence analysis in the present study was not
conducted with a control and the pCMV-TMEM41A group. Therefore,
further analysis in the future is required to verify the findings
of the present study. However, whether TMEM41A regulates EMT via
autophagy requires further investigation.
Additionally, the data presented in The Cancer
Genome Atlas dataset, revealed that TMEM41A was highly expressed in
normal gastric tissues; however, immunohistochemistry analysis did
not demonstrate such expression in tumor samples in the present
study. Furthermore, analysis with Protein Atlas (www.proteinatlas.org) also revealed that the mRNA and
protein expression levels of TMEM41A may be inconsistent.
As the expression levels of TMEM41A at the gene and
protein levels have not been fully investigated, contradictory
findings may be reported. The present study reported that the mRNA
expression levels of TMEM41A were similar at the protein level;
however, the protein expression levels revealed by
immunohistochemistry were contradictory. Therefore, the present
study suggested that the dilution of antibody (1:50 vs. 1:400),
post-translational modulation, or other unknown components may be
associated with these differing results (data not shown).
Conversely, the aforementioned databases have demonstrated a high
correlation between TMEM41A mRNA expression and poor prognoses in
endometrial, liver and pancreatic cancers, but not gastric cancer.
Therefore, further investigation is required to understand these
differing expression profiles.
The present study demonstrated a reduction in
TMEM41A expression levels in a pair of GC cell lines, and revealed
that high TMEM41A expression levels may promote GC-associated
metastasis, which may be mediated by the downregulation of
E-cadherin expression. The present study aimed to highlight the
potential of TMEM41A as a novel target against GC-associated
metastasis and suggest that TMEM41A may be considered to be an
important oncogene in GC. Further studies examining the mechanisms
underlying the regulation of TMEM41A expression are required to
investigate the possible association between TMEM41A with
E-cadherin expression in GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
BL and YX made substantial contributions to the
design of the present study and wrote the manuscript. CQ made
substantial contributions to the design of the present study and
performed the animal experiment, XC conducted software analysis and
revised the manuscript and WM directed and managed the design of
the present study. All authors read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Qingdao University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GC
|
gastric cancer
|
|
TMEM41A
|
transmembrane protein 41A
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
PBS
|
phosphate-buffered saline
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yuan DD, Zhu ZX, Zhang X and Liu J:
Targeted therapy for gastric cancer: Current status and future
directions (Review). Oncol Rep. 35:1245–1254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jin X, Zhu Z and Shi Y: Metastasis
mechanism and gene/protein expression in gastric cancer with
distant organs metastasis. Bull Cancer. 101:E1–E12. 2014.PubMed/NCBI
|
|
4
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bates RC and Mercurio AM: The
epithelial-mesenchymal transition (EMT) and colorectal cancer
progression. Cancer Biol Ther. 4:365–370. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferrera L, Caputo A and Galietta LJ:
TMEM16A protein: A new identity for Ca(2+)-dependent Cl- channels.
Physiology (Bethesda). 25:357–363. 2010.PubMed/NCBI
|
|
7
|
Zeng X, Huang P, Chen M, Liu S, Wu N, Wang
F and Zhang J: TMEM16A regulates portal vein smooth muscle cell
proliferation in portal hypertension. Exp Ther Med. 15:1062–1068.
2018.PubMed/NCBI
|
|
8
|
Zhang X, Zhang Y, Miao Y, Zhou H, Jiang G
and Wang E: TMEM17 depresses invasion and metastasis in lung cancer
cells via ERK signaling pathway. Oncotarget. 8:70685–70694.
2017.PubMed/NCBI
|
|
9
|
Huber MA, Kraut N and Beug H: Molecular
requirements for epithelial-mesenchymal transition during tumor
progression. Curr Opin Cell Biol. 17:548–558. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Strausberg RL, Feingold EA, Grouse LH,
Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler
GD, Altschul SF, et al: Generation and initial analysis of more
than 15,000 full-length human and mouse cDNA sequences. Proc Natl
Acad Sci USA. 99:16899–16903. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maestro R, Gasparotto D, Vukosavljevic T,
Barzan L, Sulfaro S and Boiocchi M: Three discrete regions of
deletion at 3p in head and neck cancers. Cancer Res. 53:5775–5779.
1993.PubMed/NCBI
|
|
13
|
Ricciardi R, Burks E, Schoetz DJ, Verma Y,
Kershnar E, Kilpatrick MW, Tsipouras P and Walat RJ: Is there a
gain in chromosome 3q in the pathway to anal cancer? Dis Colon
Rectum. 57:1183–1187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu CL, Sloan P, Read AP, Harris R and
Thakker N: Deletion mapping on the short arm of chromosome 3 in
squamous cell carcinoma of the oral cavity. Cancer Res.
54:6484–6488. 1994.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng X, Zhu K, Liu J, Chen J, Tang J,
Liang Y, Jin R, Liang X and Cai X: The evaluative value of Sema3C
and MFN2 co-expression detected by immunohistochemistry for
prognosis in hepatocellular carcinoma patients after hepatectomy.
Onco Targets Ther. 9:3213–3221. 2016.PubMed/NCBI
|
|
17
|
Kumar R, Vadlamudi RK and Adam L:
Apoptosis in mammary gland and cancer. Endocr Relat Cancer.
7:257–269. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qinghai Z, Yanying W, Yunfang C, Xukui Z
and Xiaoqiao Z: Effect of interleukin-17A and interleukin-17F gene
polymorphisms on the risk of gastric cancer in a Chinese
population. Gene. 537:328–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park S, Ha S, Kwon HW, Kim WH, Kim TY, Oh
DY, Cheon GJ and Bang YJ: Prospective evaluation of changes in
tumor size and tumor metabolism in patients with advanced gastric
cancer undergoing chemotherapy: Association and clinical
implication. J Nucl Med. 58:899–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zeng X, Huang P, Chen M, Liu S, Wu N, Wang
F and Zhang J: TMEM16A regulates portal vein smooth muscle cell
proliferation in portal hypertension. Exp Ther Med. 15:1062–1068.
2018.PubMed/NCBI
|
|
21
|
Han Z, Wang T, Han S, Chen Y, Chen T, Jia
Q, Li B, Li B, Wang J, Chen G, et al: Low-expression of TMEM100 is
associated with poor prognosis in non-small-cell lung cancer. Am J
Transl Res. 9:2567–2578. 2017.PubMed/NCBI
|
|
22
|
Ou D, Yang H, Hua D, Xiao S and Yang L:
Novel roles of TMEM100: Inhibition metastasis and proliferation of
hepatocellular carcinoma. Oncotarget. 6:17379–17390. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Al-Hussain T, Ali A and Akhtar M: Wilms
tumor: An update. Adv Anat Pathol. 21:166–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haga Y, Hiroshima K, Iyoda A, Kohno H,
Shibuya K, Iizasa T, Fujisawa T and Ohwada H: Frequency of loss of
heterozygosity at 3 p, 9 p, 13 q and 17 p is related to
proliferative activity in smokers with stage I non-small cell lung
cancer. Thorac Cardiovasc Surg. 53:114–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calin GA and Croce CM: MicroRNAs and
chromosomal abnormalities in cancer cells. Oncogene. 25:6202–6210.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Calvo SE, Pagliarini DJ and Mootha VK:
Upstream open reading frames cause widespread reduction of protein
expression and are polymorphic among humans. Proc Natl Acad Sci
USA. 106:7507–7512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: how the genome integrates intrinsic
and environmental signals. Nat Genet. 33 Suppl:S245–S254. 2003.
View Article : Google Scholar
|
|
28
|
Fahrner JA, Eguchi S, Herman JG and Baylin
SB: Dependence of histone modifications and gene expression on DNA
hypermethylation in cancer. Cancer Res. 62:7213–7282.
2002.PubMed/NCBI
|
|
29
|
Prieto-García E, Díaz-García CV,
García-Ruiz I and Agulló-Ortuño MT: Epithelial-to-mesenchymal
transition in tumor progression. Med Oncol. 34:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lefevre M, Rousseau A, Rayon T, Dalstein
V, Clavel C, Beby-Defaux A, Pretet JL, Soussan P, Polette M, Saint
Lacau Guily J and Birembaut P: Papillophar Study Group: Epithelial
to mesenchymal transition and HPV infection in squamous cell
oropharyngeal carcinomas: The papillophar study. Br J Cancer.
116:362–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y and Zhou BP: Epithelial-mesenchymal
transition-A hallmark of breast cancer metastasis. Cancer Hallm.
1:38–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scheel C, Eaton EN, Li SH, Chaffer CL,
Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL and
Weinberg RA: Paracrine and autocrine signals induce and maintain
mesenchymal and stem cell states in the breast. Cell. 145:926–940.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao Y, Guo Q, Chen J, Hu J, Wang S and
Sun Y: Role of long non-coding RNA HULC in cell proliferation,
apoptosis and tumor metastasis of gastric cancer: A clinical and
in vitro investigation. Oncol Rep. 31:358–364. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu M, Huang Z, Zang X, Pan L, Liang W,
Chen J, Qian H, Xu W, Jiang P and Zhang X: Long noncoding RNA
LINC00978 promotes cancer growth and acts as a diagnostic biomarker
in gastric cancer. Cell Prolif. 2018. View Article : Google Scholar
|
|
35
|
Gugnoni M, Sancisi V, Manzotti G, Gandolfi
G and Ciarrocchi A: Autophagy and epithelial-mesenchymal
transition: An intricate interplay in cancer. Cell Death Dis.
7:e25202016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo
Y, Song Z, Zheng Q and Xiong J: Autophagy promotes hepatocellular
carcinoma cell invasion through activation of
epithelial-mesenchymal transition. Carcinogenesis. 34:1343–1351.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H,
Yan J, Lv X, Chen X and Hu ZW: DEDD interacts with PI3KC3 to
activate autophagy and attenuate epithelial-mesenchymal transition
in human breast cancer. Cancer Res. 72:3238–3250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Catalano M, D'Alessandro G, Lepore F,
Corazzari M, Caldarola S, Valacca C, Faienza F, Esposito V,
Limatola C, Cecconi F and Di Bartolomeo S: Autophagy induction
impairs migration and invasion by reversing EMT in glioblastoma
cells. Mol Oncol. 9:1612–1625. 2015. View Article : Google Scholar : PubMed/NCBI
|