Introduction
Bisphenol A (BPA) is a high-production-volume
chemical used in the manufacturing of polycarbonate and epoxy
resins, and is extensively used for the production of medical
devices and plastic food packaging (1). Trace amounts of unreacted or
additional BPA may be extracted from the materials, under
conditions of high temperature or extreme pH (2). Bio-monitoring of BPA in human blood
and urine indicates that human exposure to BPA is very universal
(3,4), primarily through oral exposure
(5).
Previous studies suggest that high doses of BPA
exposure may result in a variety of dysfunctions, including
abnormal reproduction (6),
metabolism (7) and mammary
carcinogenesis (8). Conflicting
data exists over the effects of low doses of BPA eon animal and
human reproduction (9,10). These contrasting data sets
regarding effects of BPA may result from experiments on differing
cell types and tissues, or BPA exposure routes and levels. However,
the effect on mouse spermatogonia and the associated underlying
mechanism of different doses of BPA exposure and subsequent
effects, remains to be elucidated. The present study investigated
whether the effects of high and low doses of BPA result in
differential responses in spermatogonial cells, using the mouse
cell line GC-1, which was immortalized by transformation with the
plasmid pSV3-neo in type B spermatogonia (11).
The results of the present study demonstrated that
high and low doses of BPA have differential effects on the levels
of global DNA methylation, in addition to H3K9Me3, H3K27Me3, and
mitogen activated protein kinase (MAPK) signaling, in mouse GC-1
spermatogonia cells.
Materials and methods
Cell culture
The mouse GC-1 spermatogonial cell line (American
Type Culture Collection, Manassas, VA, USA) was grown in Dulbecco's
modified Eagle's medium supplemented with 5% (v/v) fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), 2 mM L-glutamine, and antibiotics (100 mg streptomycin/ml,
100 IU penicillin/ml), in a humidified CO2 incubator at
37°C.
Cell proliferation assay
For cell proliferation experiments, cells were
seeded in 96-well cell culture plates at a density of 2000 cells
per well and grown in free phenol red (Gibco; Thermo Fisher
Scientific, Inc.) medium supplemented with 3% charcoal-stripped
FBS. BPA (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added
at the final concentrations of 0.001, 0.01, 0.1, 1 and 10 µg/ml
with five replicates and incubated for 5 days. Vehicle dimethyl
sulfoxide (Sigma-Aldrich; Merck KGaA) was used as a negative
control. A total of 10 µl of CCK-8 solution (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was added to each well of the
plate, then incubated for 2 h in the CO2 incubator, and
the absorbance was measured at a wavelength of 450 nm using
microplate reader.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The concentration and quality of total
RNA were determined by absorbance ratio 260:280 using a
spectrophotometer. cDNA was synthesized from 2 µg total RNA using
ReverTra Ace qPCR RT KIT (Toyobo Life Science, Osaka, Japan) for
reverse transcription of mRNA. The resulting cDNA was diluted
10-fold and used as template. qPCR was then performed using
SYBRGreen real-time PCR Mix (Toyobo Life Science) using a BioRad
MiniOpticon™ System under the following conditions: 95°C for 1 min,
and 40 cycles of 95°C for 15 sec, 56–58°C for 15 sec and 72°C for
45 sec. Immediately following the PCR, the melting curve was
generated by raising the temperature from 60°C to 95°C and reading
the fluorescence 1°C/2 sec. Each gene was analyzed in triplicate.
The expression of β-actin was used as an endogenous control and the
2−ΔΔCq method (12) was
used to determine the relative expression of the genes. The primer
sequences are listed in Table
I.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|
| DNMT1 |
ACTGCGTCTCGGTCATT |
GTCTGTGCCTCCCTCCAT |
| Ezh2 |
GTAGACACTCCTCCAAGAAAGAA |
GATGGTCACAGGGTTGATAGTT |
| Suv39h1 |
CTCTGCATCTTCCGCACTAAT |
AGTCGCTCATCAAGGTTGTCTAT |
| β-actin |
GTCCCTCACCCTCCCAAAAG |
GCTGCCTCAACACCTCAACCC |
Western blotting
Cell lysates were prepared using a cell lysis buffer
for western blotting and immunoprecipitation (cat. no. P0013J,
Beyotime Institute of Biotechnology), according to the
manufacturer's protocol. The protein concentration was determined
using a bicinchoninic acid assay (Beyotime Institute of
Biotechnology). Subsequently, protein samples added with loading
buffer were boiled for 5 min prior to sample electrophoresis. A
total of 20 µg proteins per lane was resolved by 10 or 15% SDS-PAGE
according to different target protein and blotted on a 0.22 µm
polyvinylidene fluoride membrane (EMD Millipore, Billerica, MS,
USA), which was blocked with 5% skim milk in PBS-T buffer for 1 h
at room temperature, followed by incubation with diluted primary
antibodies for 1 h at 37°C (for anti-β-actin) or overnight at 4°C
for the remaining antibodies. Following washing with PBS-T, the
membrane was incubated with horseradish peroxidase-labeled goat
anti-mouse immunoglobulin (Ig)G (1:3,000 in PBST; cat. no. A0216,
Beyotime Institute of Biotechnology) or horseradish
peroxidase-labeled goat anti-rabbit IgG (1:3,000 in PBST; cat. no.
A0208; Beyotime Institute of Biotechnology) at 37°C for 1 h. The
blot was detected using SuperSignal® West Pico
Chemiluminescent Substrate (Pierce; Thermo Fisher Scientific, Inc.)
on X-ray films. The band intensity was measured by Gel-Pro analyzer
(version 4.0; Media Cybernetics, Inc., Rockville, MD, USA) and
normalized to a reference loading control.
The primary antibodies included: 1:500 diluted mouse
anti-DNA methyltransferase (DNMT)1 monoclonal antibody (catalog no.
ab13537; Abcam, Cambridge, UK); 1:2,000 diluted mouse anti-β-actin
monoclonal antibody (catalog no. AA128; Beyotime Institute of
Biotechnology); rabbit anti-extracellular signal-regulated kinase
(ERK)1/2 polyclonal antibody (catalog no. AM076; Beyotime Institute
of Biotechnology); 1:1,000 diluted mouse anti-phosphor-ERK1/2
(Thr202/Tyr204) monoclonal antibody (catalog no. AM071; Beyotime
Institute of Biotechnology); 1:500 diluted mouse anti-p38 MAPK
monoclonal antibody (catalog no. AM065; Beyotime Institute of
Biotechnology); 1:500 diluted mouse anti-phospho-p38 MAPK
(Thr180/Tyr182) antibody (catalog no. AM063; Beyotime Institute of
Biotechnology); rabbit anti-H3K27Me3 polyclonal antibody (catalog
no. Dam1387952; Upstate Biotechnology, Inc., Lake Placid, NY, USA);
rabbit anti-H3K9Me3 polyclonal antibody (catalog no. Ab8898-100;
Abcam); rabbit anti-histone H3 polyclonal antibody (catalog no.
AH433; Beyotime Institute of Biotechnology).
Dot-blot assay
The dot blot assay was conducted as previously
described (13). Briefly, cells
were harvested for DNA isolation following BPA exposure for 5 days.
Genomic DNA was extracted by classical proteinase
K/phenol-chloroform methods, precipitated by mixing with 3 M sodium
acetate (pH 4.8) and cold ethanol. A total of 500 ng genomic DNA
was denatured in 0.1 N NaOH, 10 mM EDTA at 95°C for 5 min, and
spotted on the nitrocellulose membrane. The membranes were washed
with 2×SSC buffer, air-dried and baked at 80°C for 2 h. Following
blocking with 5% skim milk, the monoclonal 5-methylcytosine (mC)
antibody (diluted 1:1,000 in PBST with 5% skim milk; cat. no.
I-MECY-0100, Eurogentec, Ltd., Southampton, UK) was incubated
overnight at 4°C. Following washing with TBST, membranes were
treated with 1:2,000 dilution of horseradish peroxidase-conjugated
goat anti-mouse IgG (catalog no. A0216, Beyotime Institute of
Biotechnology) for 1 h at room temperature. The signals were
visualized using SuperSignal® West Pico Chemiluminescent
Substrate (Pierce; Thermo Fisher Scientific, Inc.). The membranes
were further stained with hematoxylin solution for 5 min at room
temperature for quantity loading control.
Statistical analysis
Data are presented as the mean ± and standard error
of the mean. Statistical analysis was performed using one-way
analysis of variance followed by the least significant difference
post hoc test. All analyses were conducted using SPSS software,
version 20.0 (IBM SPSS, Armonk, NY, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
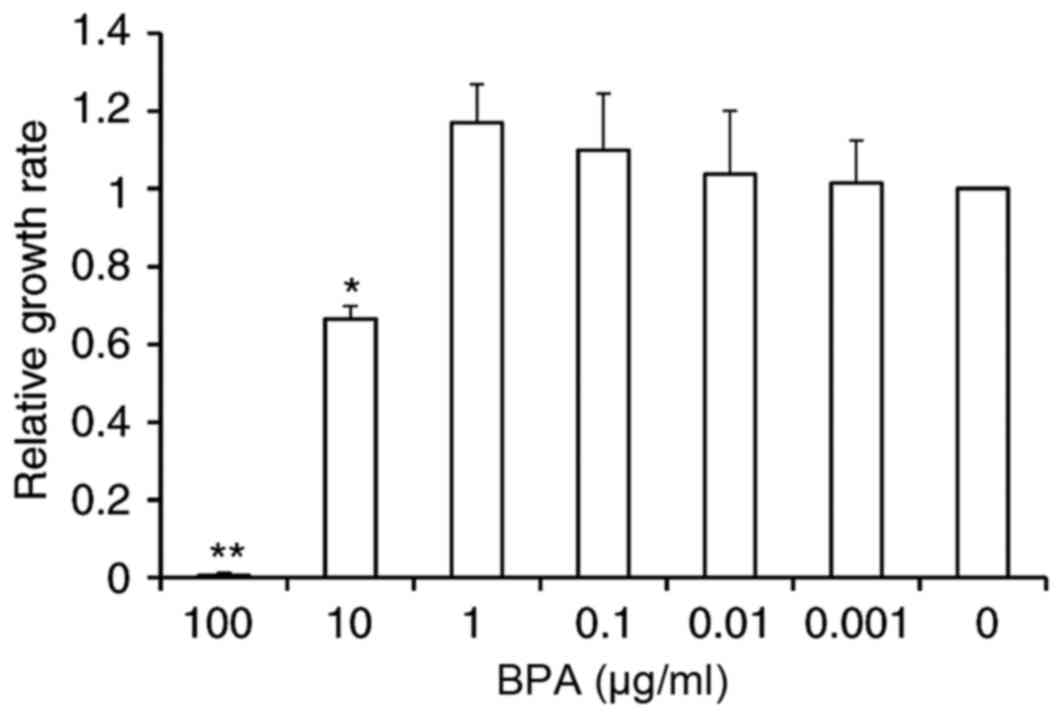
BPA exhibits varied effects on
proliferation of GC-1 spermatogonia cell line
Effects of BPA on GC-1 cell proliferation were
evaluated using a Cell Counting Kit-8 assay. Exposure to 100 µg/ml
of BPA resulted in cell toxicity, leading to cell death. Exposure
to 10 µg/ml BPA inhibited cell growth by 33.5% relative to
controls. However, there were no significant alterations in cell
proliferation at other doses of BPA, although a slight increase of
10 and 17.1% compared to control was observed at concentrations of
0.1 and 1 µg/ml BPA, respectively (Fig. 1).
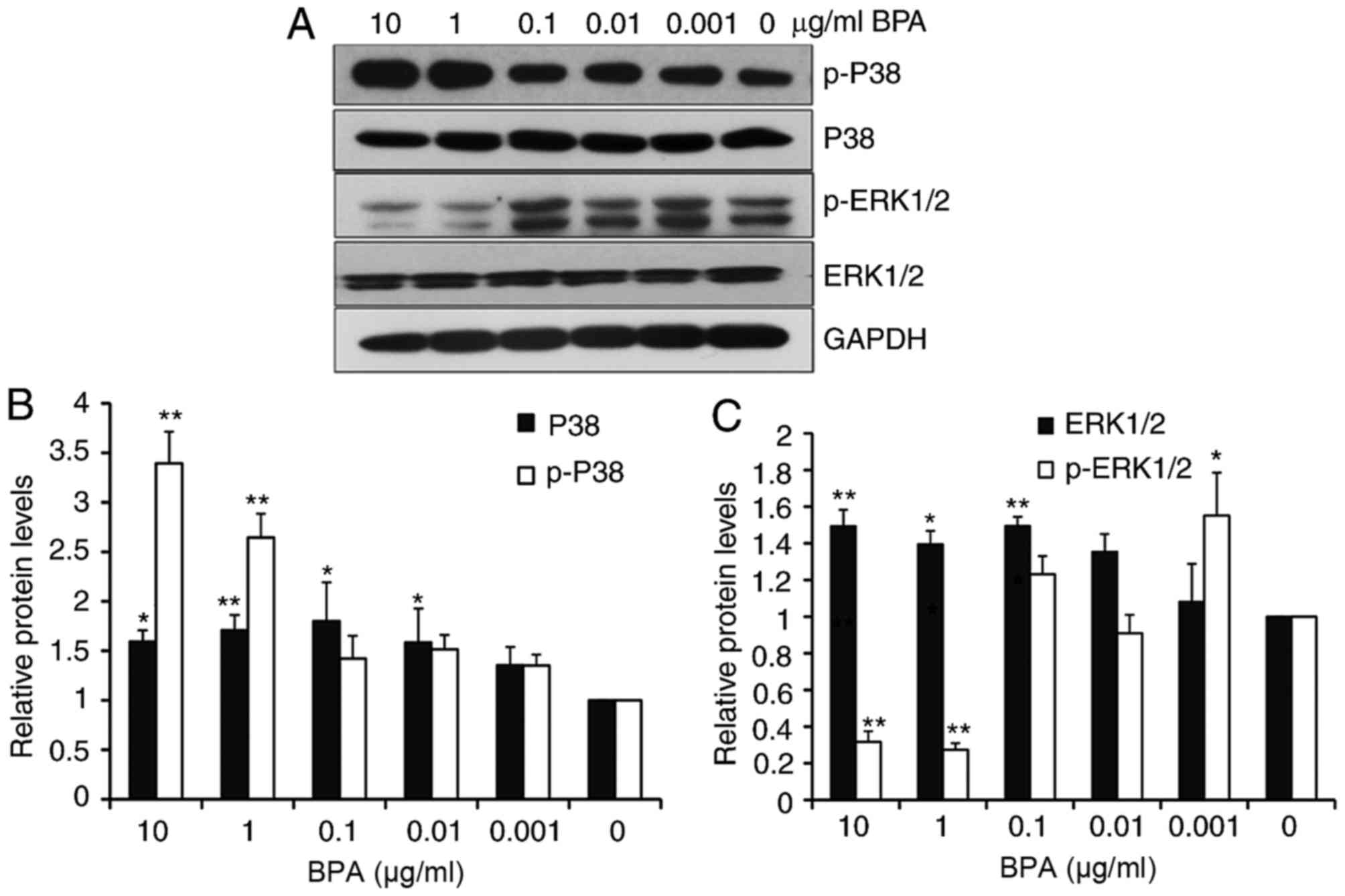
BPA exhibits varied effects on MAPK
signaling
The MAPK signaling pathways (p38, ERK1/2) exhibit
important roles in cell proliferation and development (14). The present study next evaluated
whether different doses of BPA had differential effects on the MAPK
pathways in GC-1 spermatogonia cells. Western blot analysis
revealed that BPA slightly increased the levels of p38 at all
tested concentrations, except for 0.001 µg/ml of BPA, and enhanced
the levels of phosphorylated p38 (p-p38) in a dose-dependent manner
(Fig. 2A and B). BPA additionally
increased the levels of extracellular ERK1/2 from 0.1 to 10 µg/ml
BPA exposure. However, ERK1/2 phosphorylation markedly decreased at
increased BPA concentrations (10 and 1 µg/ml; Fig. 2A and C).
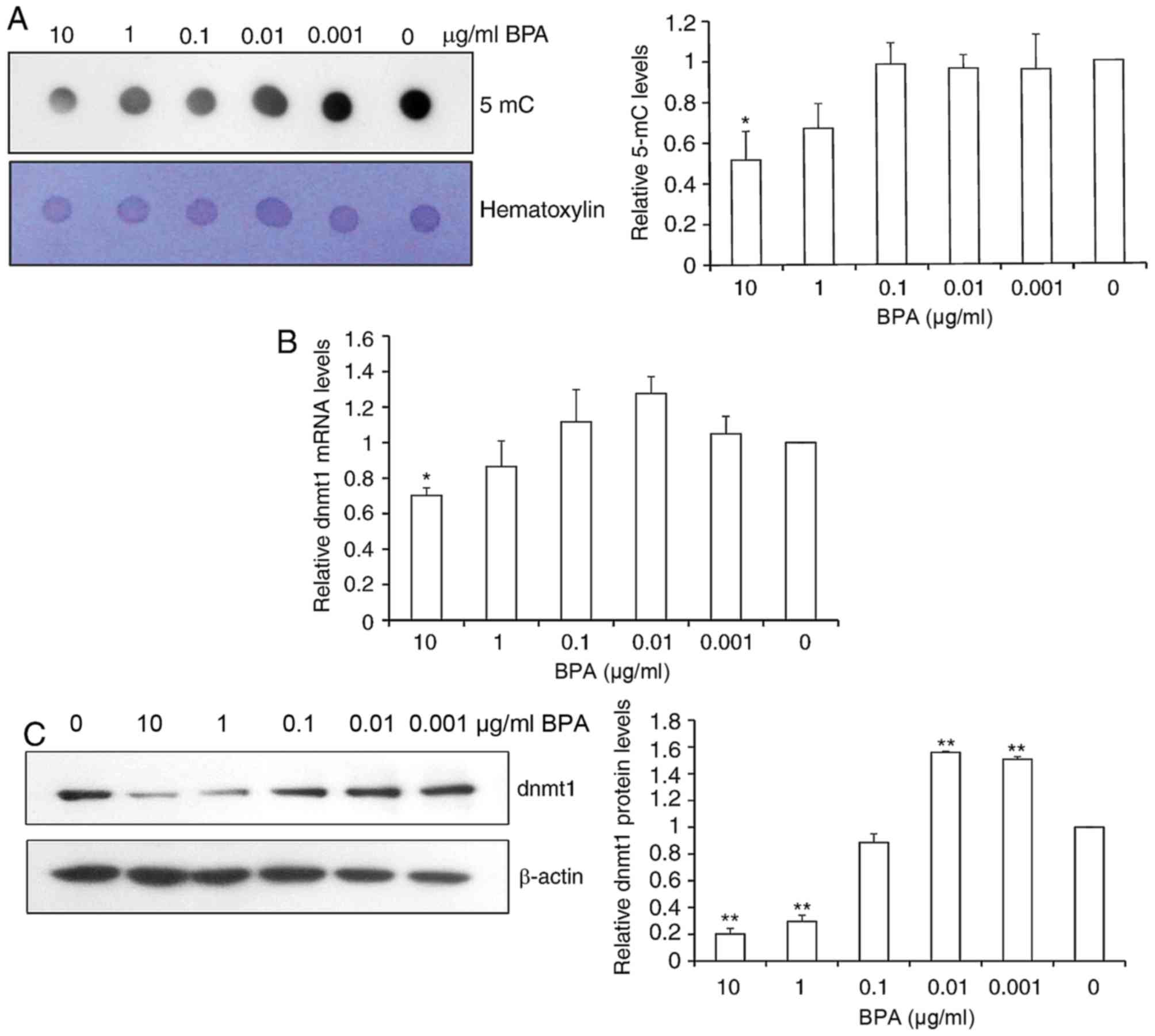
BPA decreases global DNA
methylation
In order to investigate the role of BPA in global
DNA methylation in mouse germ cells, dot-blot assays of DNA probed
with an antibody specific to 5-mC were performed. Quantification of
spot intensity revealed that the global 5-mC level was
significantly reduced 2-fold at the high dose of 10 µg/ml BPA and
slightly decreased by 21.2% at 1 µg/ml BPA (Fig. 3A).
Since global DNA methylation was altered following
BPA exposure and DNA DNMTs control DNA methylation, the present
study next determined the relative mRNA and protein expression of
DNMT1 via RT-qPCR and western blot analysis. BPA exposure resulted
in the significant reduction of DNMT1 mRNA expression at 10 µg/ml
of BPA, and a smaller decrease in DNMT1 mRNA levels was observed at
other concentrations of BPA (Fig.
3B). Consistent with the alterations in 5-mC levels, DNMT1
protein levels were significantly decreased by 85.6 and 77.2% at 10
and 1 µg/ml BPA, respectively. DNMT1 protein levels were increased
1.5-fold at 0.01 and 0.001 µg/ml BPA exposure (Fig. 3C).
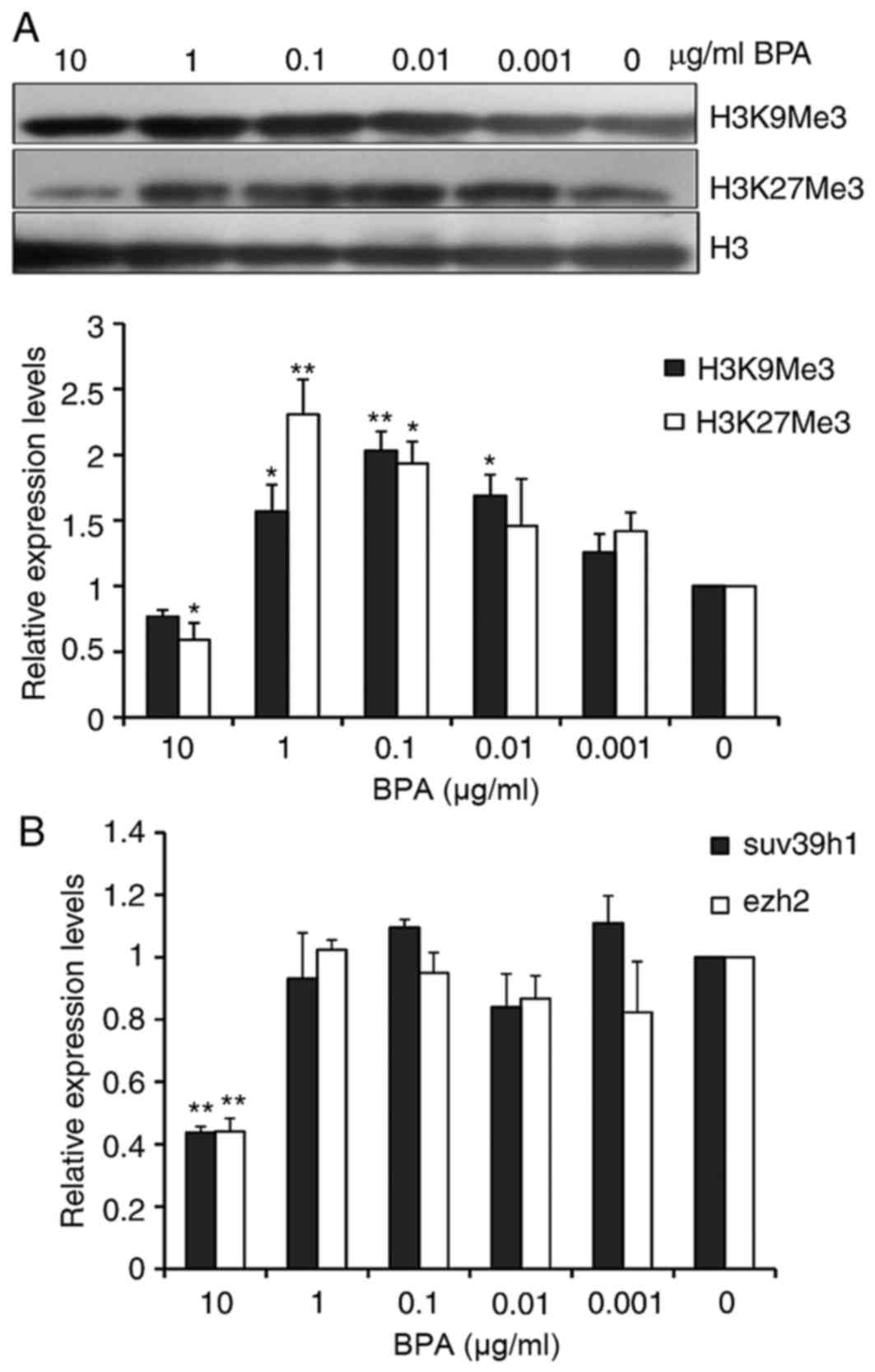
BPA exhibits varied effects on histone
methylation
Methylation of lysine on histone H3 is the primary
epigenetic modification of chromatin structure. Global levels of
H3K9Me3, which is trimethylated H3K9, and H3K27Me3, trimethylated
H3K27, in BPA-treated GC-1 cells were analyzed by western blotting.
As presented in Fig. 4A and B, the
global levels of H3K27me3 were decreased at the 10 µg/ml of BPA.
However, the global H3K9me3 and H3K27me3 levels were increased from
0.01 to 1 µg/ml of BPA. Suppressor of Variegation 3–9 Homolog 1
(Suv39h1) and Enhancer of Zeste Homolog 2 (Ezh2) have been reported
to respectively tri-methylate H3K9 and H3K27, therefore the present
study evaluated the mRNA levels of Suv39h1 and Ezh2 in BPA-treated
GC-1 cells using RT-qPCR. The analysis indicated that the mRNA
levels of Suv39h1 and ezh2 were markedly decreased 2.5-fold at the
high dose of 10 µg/ml of BPA; however, treatment with BPA at other
concentrations did not significantly alter Suv39h1 and Ezh2 mRNA
expression (Fig. 4C).
Discussion
The present study examined the effects of BPA on
cellular proliferation of GC-1 cells, an immortalized type B
spermatogonial cell line. Exposure to BPA for 5 days inhibited GC-1
cell proliferation at the high dose of 10 µg/ml of BPA, which
suggested that BPA results in abnormal sperm development. An
epidemiological study reported that urine BPA levels among BPA
occupationally exposed workers are significantly associated with
lower semen quality, including decreased sperm count and decreased
sperm motility (15), revealing
the deleterious effects of high doses of BPA exposure on male
reproduction. Low concentrations of BPA have been reported to
promote cell growth in the GC-1 and other cell lines. Stimulation
of human testicular seminoma JKT-1 cell proliferation was observed
at 24 h exposure to 10−5 M −10−12 M of BPA
(16). Sheng et al
(17) reported that
10−7 M-10−12 M BPA stimulates GC-1 cell
proliferation following exposure to BPA for 12 h. However, the
present study did not observe cell stimulation at low
concentrations of BPA exposure for 5 days. It was demonstrated that
lower concentrations of BPA increased the GC-1 cell growth,
although there was no statistical significance in the study, due to
variance between tests. These contrasting data sets may result from
different exposure patterns and duration. These data suggested the
underlying mechanisms may be different between high doses and low
doses of BPA exposure.
MAPK pathways are important in regulating germ cell
cycles and function in spermatogenesis. The present study observed
that the significantly increased phosphorylation of p38 was
paralleled by a decreased phosphorylation of ERK1/2 at high
concentrations of BPA exposure (10 and 1 µg/ml). Xia et al
(18) reported that p38 MAPK
activation and ERK inhibition concurrently are crucial for
apoptosis in rat PC-12 pheochromocytoma cells. Therefore, this
finding in the study suggested that increased doses of BPA promote
apoptosis of GC-1 cells and inhibit cell growth by activating p38
MAPK and suppressing ERK1/2 MAPK. Lower doses of BPA exposure
activated p38 and ERK1/2 MAPK. Considering cell proliferation or
apoptosis depends on the dynamic balance of the multiple signaling
pathways, at present, the specific association between p38, ERK1/2
MAPK alterations and cell growth at low doses of BPA remains to be
elucidated.
DNA methylation, an epigenetic modification in which
a methyl group is added to the cytosine in a CpG dinucleotide to
produce 5-methylcytosine (5-mC), is important in spermatogenesis,
development and germ cell functions. The epigenetic effect of BPA
was first demonstrated in the yellow agouti (Avy) mouse
model, in which BPA exposure altered the coat color towards yellow
by decreasing DNA methylation of Avy IAP sequence
(19). The present study
demonstrated that only high doses of BPA exposure (10 and 1 µg/ml
of BPA), rather than lower doses of BPA (0.001 to 0.1 µg/ml),
reduced the global DNA methylation levels in spermatogonial cells,
using a dot-blot assay. In addition, high doses of BPA induced the
reduction of DNMT1 protein and mRNA levels, which resulted in
global DNA hypo-methylation and further disrupted the normal
development of spermatogonia. A previous study reported that gene
expression of DNMT1 is significantly downregulated at 200 µM BPA
and not at lower concentrations. Expression of DNMT1 is less
susceptible to lower doses of BPA in developing hypothalamic cells
(20). These results suggested
that global DNA methylation alteration occurs in response to BPA
exposure.
Research regarding BPA effects on histone
methylation has only recently been introduced. However, H3K9Me3 and
H3K27Me3 have been demonstrated to exhibit repressive effects on
gene transcription. In the present study, the mRNA expression of
Suv39h1 and Ezh2, and the global levels of H3K27me3 were
significantly decreased at the high concentration of 10 µg/ml BPA.
Zhao et al (21) reported
gene silencing of Suv39h1 induces inhibition of cell proliferation
via downregulation of histone methylation of H3K9.
Levels of H3K9me3 and H3K27Me3 were upregulated with
exposure to lower concentrations of BPA (0.01 to 1 µg/ml); however,
there were no significant alterations in histone methyltransferase
Suv39h1 and Ezh2 mRNA expression. However, it remains unknown
whether protein levels or enzyme activities of Suv39h1 and Ezh2
alter. The authors previously demonstrated that H3K9me3 levels of
zebrafish mature sperm are significantly increased with exposure to
1.5 mg/l BPA in the present laboratory (data not published).
Doherty et al (22)
reported that the level of H3K27Me3 increases in breast cancer
MCF-7 cells exposed to BPA at 2.5×10−6 M, equivalent to
1 µg/ml of BPA in the present study, however the H3K27Me3 level and
Ezh2 expression at increased doses (10 µg/ml) of BPA were not
analyzed.
Epigenetic modifications, including DNA methylation,
histone post-translational modifications and ERK1/2 MAPK typically
have intimate interactions. DNA methylation promotes histone
methylation. Ezh2 has been reported to directly control DNA
methylation, thus linking histone methylation and DNA methylation
(23). It has previously been
demonstrated that DNMTs influence histone methylation (24). ERK signaling also has an important
role in activity-dependent modifications of histone proteins
(25). However, it is not
currently clear how this cross talk occurs to precisely regulate
the activities in response to diverse circumstances.
The present study demonstrated that inhibition of
spermatogonia cell growth occurred at the high doses of BPA
exposure (10 µg/ml). It was hypothesized that p38 MAPK activation
is triggered and concurrently inhibits the activity of ERK1/2 to
induce cell apoptosis, and also induce global DNA hypomethylation
and histone hypomethylation of H3K27Me3, which may result in genome
instability and finally, inhibition of cell proliferation.
Alteration in genomic and epigenetic responses at high and low
doses of BPA were observed, however there did not appear to be an
association with cell proliferation at the low doses of BPA.
Epigenetic alterations resulting from BPA may affect spermatogonia
development and complex activities associated with cell
proliferation. Therefore, BPA is associated with altered DNA
methylation, however the biological effects are still unclear.
In conclusion, the findings using the GC-1
spermatogonia cell model suggested that high and low doses of BPA
exposure demonstrated differential effects on cell growth, global
DNA methylation and histone H3K9Me3 and H3K27Me3 levels, in
addition to MAPK signaling pathways. The varied effects of BPA
depending on high or low doses in spermatogonia, implied that
complicated underlying mechanisms are involved and remain to be
investigated in future studies.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81270760
and 81571495), the National Basic Research Program of China (grant
no. 2014CB943104) and Shanghai Municipal Committee of Science and
Technology (grant nos. 10ZR1425500 and 15431902800).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
YHL performed western blotting, statistical analysis
and drafted the manuscript. FD performed the cell culture and cell
proliferation assay, XYZ performed the RT-qPCR assay, HJP performed
the dot-blot assay, RSL obtained the funding and designed the
study. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chapin RE, Adams J, Boekelheide K, Gray LE
Jr, Hayward SW, Lees PS, McIntyre BS, Portier KM, Schnorr TM,
Selevan SG, et al: NTP-CERHR expert panel report on the
reproductive and developmental toxicity of bisphenol A. Birth
Defects Res B Dev Reprod Toxicol. 83:157–395. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Geens T, Apelbaum TZ, Goeyens L, Neels H
and Covaci A: Intake of bisphenol A from canned beverages and foods
on the Belgian market. Food Addit Contam Part A Chem Anal Control
Expo Risk Assess. 27:1627–1637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lakind JS, Levesque J, Dumas P, Bryan S,
Clarke J and Naiman DQ: Comparing United States and Canadian
population exposures from National Biomonitoring Surveys: Bisphenol
A intake as a case study. J Expo Sci Environ Epidemiol. 22:219–226.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takeuchi T and Tsutsumi O: Serum bisphenol
a concentrations showed gender differences, possibly linked to
androgen levels. Biochem Biophys Res Commun. 291:76–78. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wilson NK, Chuang JC, Morgan MK, Lordo RA
and Sheldon LS: An observational study of the potential exposures
of preschool children to pentachlorophenol, bisphenol-A, and
nonylphenol at home and daycare. Environ Res. 103:9–20. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Susiarjo M, Hassold TJ, Freeman E and Hunt
PA: Bisphenol A exposure in utero disrupts early oogenesis in the
mouse. PLoS Genet. 3:e52007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Batista TM, Alonso-Magdalena P, Vieira E,
Amaral ME, Cederroth CR, Nef S, Quesada I, Carneiro EM and Nadal A:
Short-term treatment with bisphenol-A leads to metabolic
abnormalities in adult male mice. PLoS One. 7:e338142012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jenkins S, Wang J, Eltoum I, Desmond R and
Lamartiniere CA: Chronic oral exposure to bisphenol A results in a
nonmonotonic dose response in mammary carcinogenesis and metastasis
in MMTV-erbB2 mice. Environ Health Perspect. 119:1604–1609. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Motoharu S, Seiichiroh O, Ryuta I, Shuichi
K, Masamichi K, Yoshihiro H, Yasunobu A, Junzo Y and Chiharu T:
Bisphenol-A affects spermatogenesis in the adult rat even at a low
dose. J Occup Health. 43:185–190. 2001. View Article : Google Scholar
|
|
10
|
Nagao T, Saito Y, Usumi K, Yoshimura S and
Ono H: Low-dose bisphenol A does not affect reproductive organs in
estrogen-sensitive C57BL/6N mice exposed at the sexually mature,
juvenile, or embryonic stage. Reprod Toxicol. 16:123–130. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hofmann MC, Narisawa S, Hess RA and Millán
JL: Immortalization of germ cells and somatic testicular cells
using the SV40 large T antigen. Exp Cell Res. 201:417–435. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ito S, D'Alessio AC, Taranova OV, Hong K,
Sowers LC and Zhang Y: Role of Tet proteins in 5mC to 5hmC
conversion, ES-cell self-renewal and inner cell mass specification.
Nature. 466:1129–1133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim EK and Choi EJ: Pathological roles of
MAPK signaling pathways in human diseases. Biochim Biophys Acta.
1802:396–405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li DK, Zhou Z, Miao M, He Y, Wang J,
Ferber J, Herrinton LJ, Gao E and Yuan W: Urine bisphenol-A (BPA)
level in relation to semen quality. Fertil Steril. 95(625–630):
e1–e4. 2011.PubMed/NCBI
|
|
16
|
Bouskine A, Nebout M, Brücker-Davis F,
Benahmed M and Fenichel P: Low doses of bisphenol A promote human
seminoma cell proliferation by activating PKA and PKG via a
membrane G-protein-coupled estrogen receptor. Environ Health
Perspect. 117:1053–1058. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sheng ZG and Zhu BZ: Low concentrations of
bisphenol A induce mouse spermatogonial cell proliferation by G
protein-coupled receptor 30 and estrogen receptor-α. Environ Health
Perspect. 119:1775–1780. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xia Z, Dickens M, Raingeaud J, Davis RJ
and Greenberg ME: Opposing effects of ERK and JNK-p38 MAP kinases
on apoptosis. Science. 270:1326–1331. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dolinoy DC, Huang D and Jirtle RL:
Maternal nutrient supplementation counteracts bisphenol A-induced
DNA hypomethylation in early development. Proc Natl Acad Sci USA.
104:13056–13061. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Warita K, Mitsuhashi T, Ohta K, Suzuki S,
Hoshi N, Miki T and Takeuchi Y: Gene expression of epigenetic
regulatory factors related to primary silencing mechanism is less
susceptible to lower doses of bisphenol A in embryonic hypothalamic
cells. J Toxicol Sci. 38:285–289. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao T, Ma XD and Huang YQ: Experimental
study of SUV39H1 gene specific siRNA in human leukemia cell line.
Zhonghua Xue Ye Xue Za Zhi. 34:49–54. 2013.(In Chinese). PubMed/NCBI
|
|
22
|
Doherty LF, Bromer JG, Zhou Y, Aldad TS
and Taylor HS: In utero exposure to diethylstilbestrol (DES) or
bisphenol-A (BPA) increases EZH2 expression in the mammary gland:
An epigenetic mechanism linking endocrine disruptors to breast
cancer. Horm Cancer. 1:146–155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Viré E, Brenner C, Deplus R, Blanchon L,
Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden
JM, et al: The polycomb group protein EZH2 directly controls DNA
methylation. Nature. 439:871–874. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fuks F, Hurd PJ, Deplus R and Kouzarides
T: The DNA methyltransferases associate with HP1 and the SUV39H1
histone methyltransferase. Nucleic Acids Res. 31:2305–2312. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ciccarelli A and Giustetto M: Role of ERK
signaling in activity-dependent modifications of histone proteins.
Neuropharmacology. 80:34–44. 2014. View Article : Google Scholar : PubMed/NCBI
|