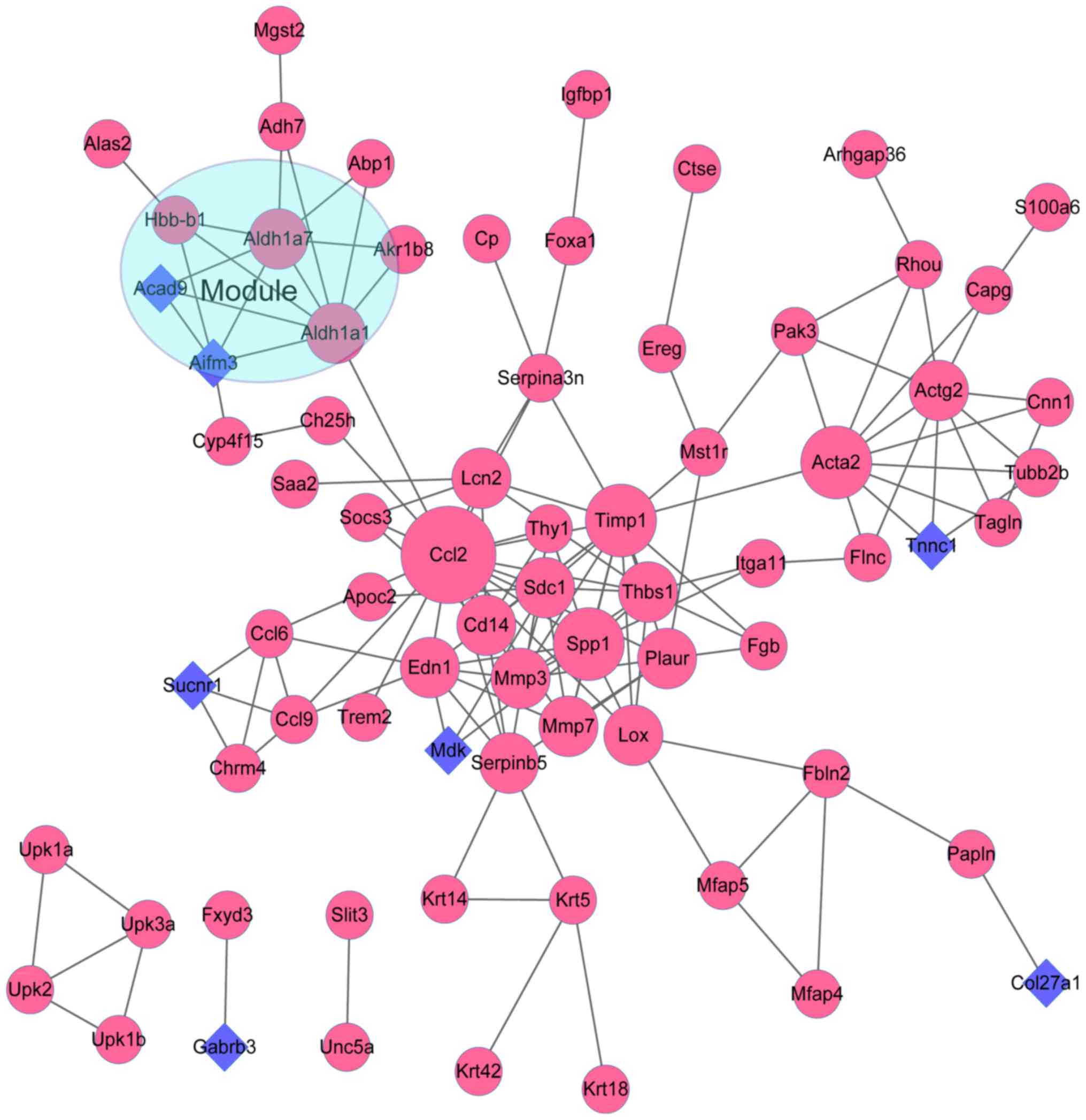
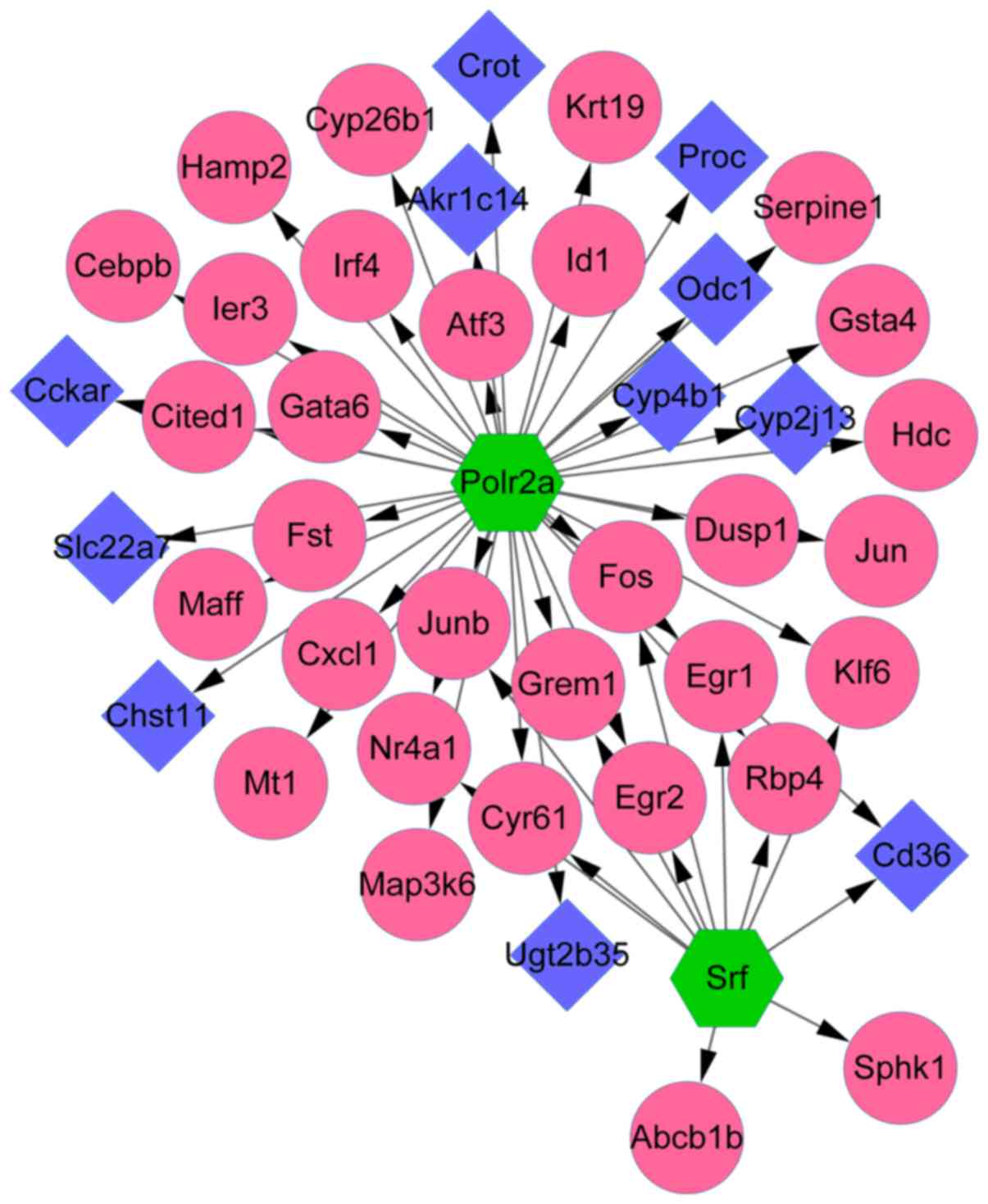
|
1
|
Piepsz A, Ham H and Josephson S: RE:
Biomarkers of congenital obstructive nephropathy: Past, present and
future. J Urol. 173:2207–2208. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ingraham SE and Mchugh KM: Current
perspectives on congenital obstructive nephropathy. Pediatr
Nephrol. 26:1453–1461. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
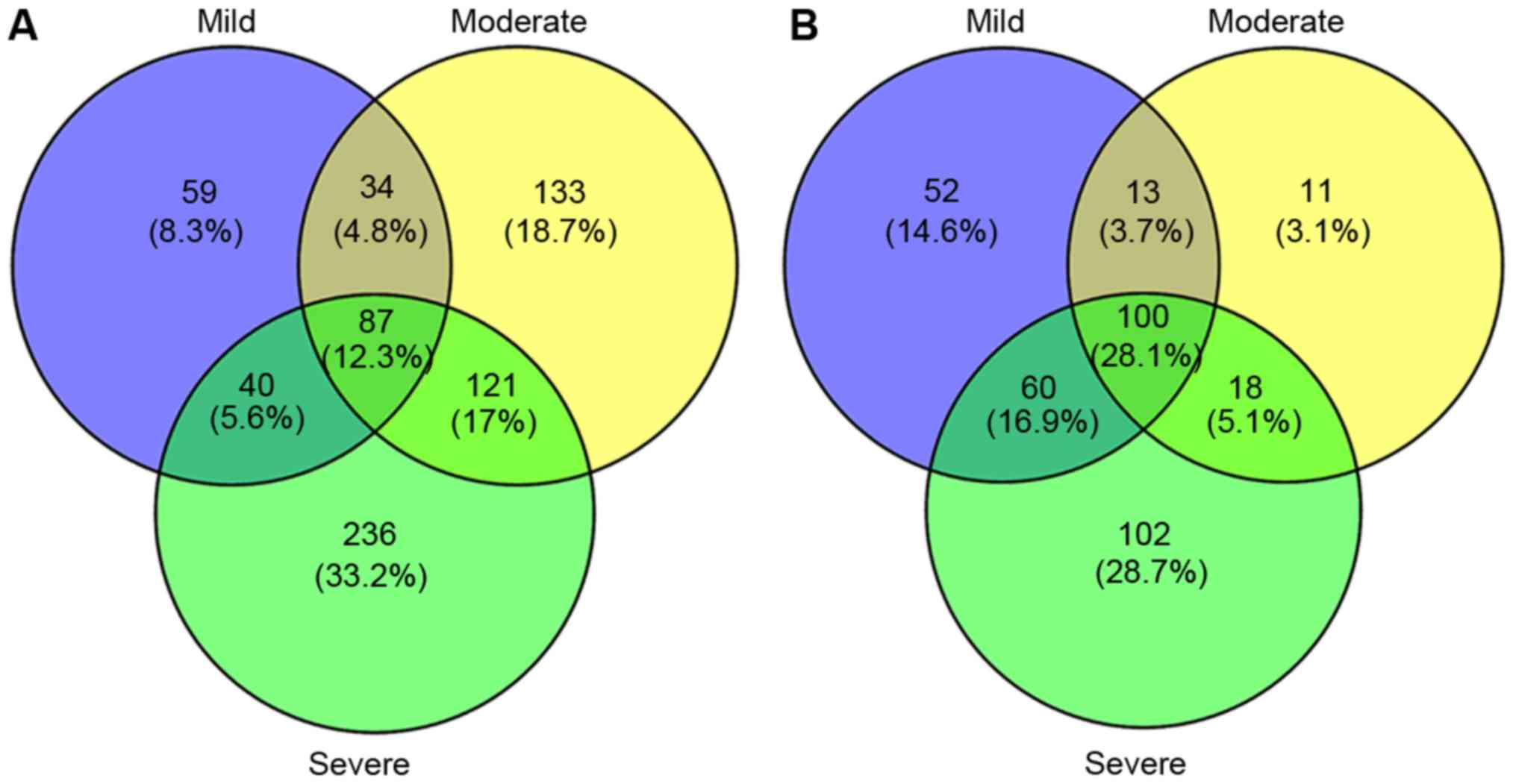
Chevalier RL, Thornhill BA, Forbes MS and
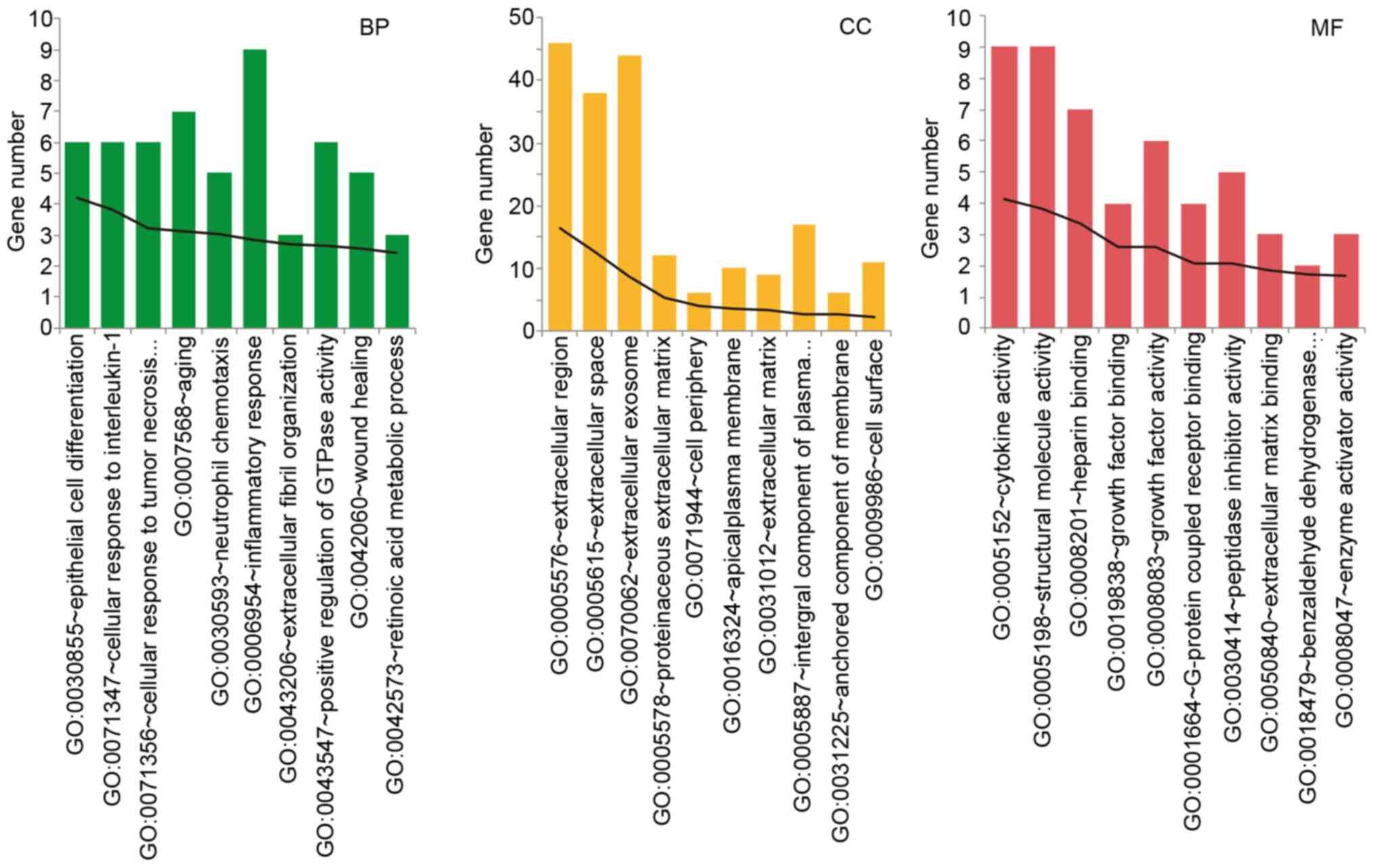
Kiley SC: Mechanisms of renal injury and progression of renal
disease in congenital obstructive nephropathy. Pediatr Nephrol.
25:687–697. 2010. View Article : Google Scholar : PubMed/NCBI
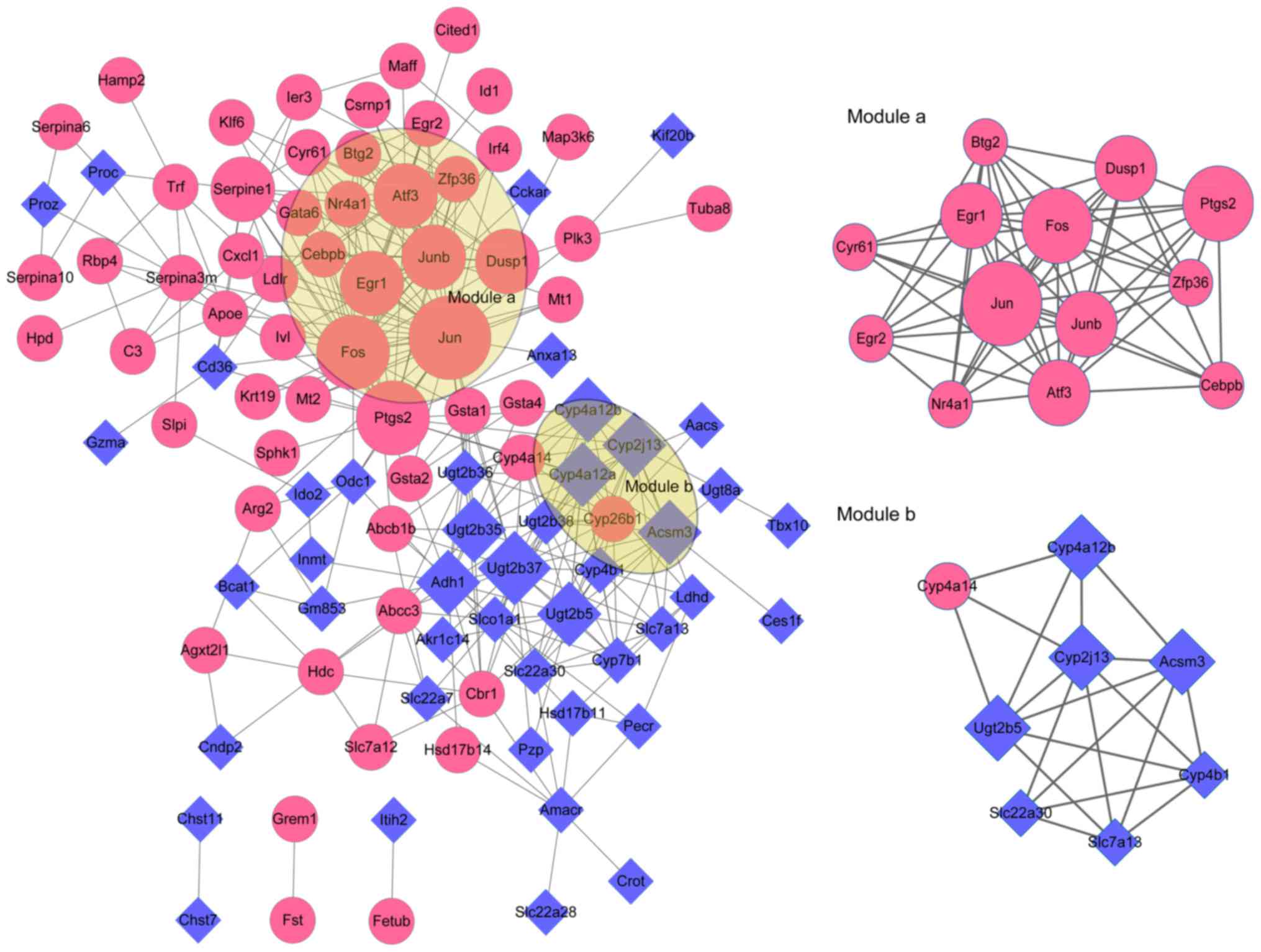
|
|
4
|
Liapis H: Biology of congenital
obstructive nephropathy. Nephron Exp Nephrol. 93:e87–e91. 2003.
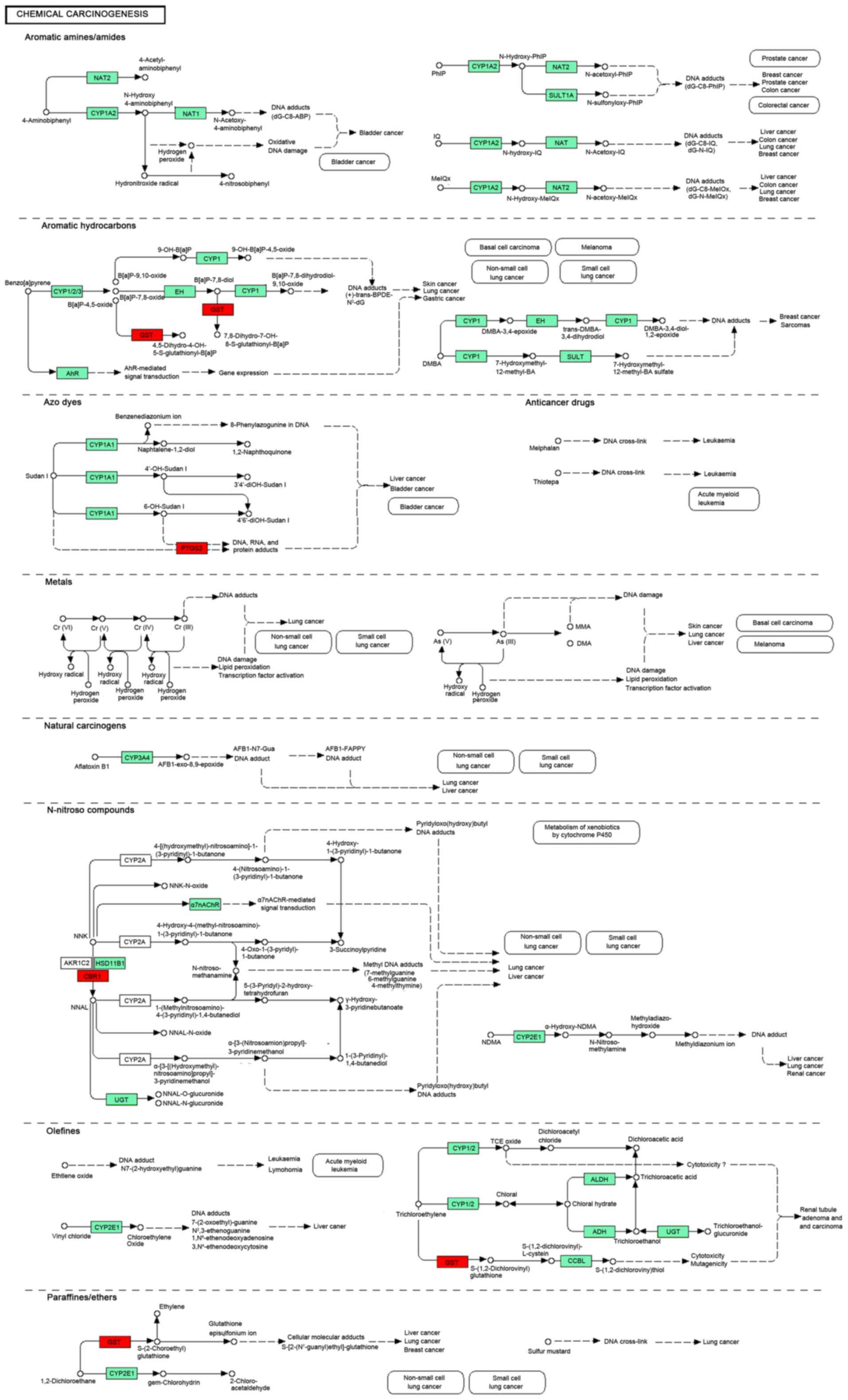
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chevalier RL: Prognostic factors and
biomarkers of congenital obstructive nephropathy. Pediatr Nephrol.
31:1411–1420. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hahn H, Ku SE, Kim KS, Park YS, Yoon CH
and Cheong HI: Implication of genetic variations in congenital
obstructive nephropathy. Pediatr Nephrol. 20:1541–1544. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grandaliano G, Gesualdo L, Bartoli F,
Ranieri E, Monno R, Leggio A, Paradies G, Caldarulo E, Infante B
and Schena FP: MCP-1 and EGF renal expression and urine excretion
in human congenital obstructive nephropathy. Kidney Int.
58:182–192. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inazaki K, Kanamaru Y, Kojima Y, Sueyoshi
N, Okumura K, Kaneko K, Yamashiro Y, Ogawa H and Nakao A: Smad3
deficiency attenuates renal fibrosis, inflammation, and apoptosis
after unilateral ureteral obstruction. Kidney Int. 66:597–604.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hermens JS, Thelen P, Ringert RH and
Seseke F: Alterations of selected genes of the Wnt signal chain in
rat kidneys with spontaneous congenital obstructive uropathy. J
Pediatr Urol. 3:86–95. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chevalier RL: Promise for gene therapy in
obstructive nephropathy. Kidney Int. 66:1709–1710. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chevalier RL: Obstructive nephropathy:
Towards biomarker discovery and gene therapy. Nat Clin Pract
Nephrol. 2:157–168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Becknell B, Carpenter AR, Allen JL,
Wilhide ME, Ingraham SE, Hains DS and McHugh KM: Molecular basis of
renal adaptation in a murine model of congenital obstructive
nephropathy. PLoS One. 8:e727622013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carpenter AR, Becknell B, Ingraham SE and
McHugh KM: Ultrasound imaging of the murine kidney. Methods Mol
Biol. 886:403–410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Limma: Linear models for
microarray data. In: Bioinformatics and Computational Biology
Solutions Using R and BioconductorStatistics for Biology and
Health. Gentleman R, Carey VJ, Huber W, Irizarry RA and Dudoit S:
Springer; New York, NY: pp. 397–420. 2005, View Article : Google Scholar
|
|
17
|
Oliveros JC: Venny. An interactive tool
for comparing lists with Venn Diagrams. BioinfoGP of CNB-CSIC.
http://bioinfogp.cnnb.csic.es/tools/venny/index.htJuly
1–2017
|
|
18
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gene Ontology Consortium: The gene
ontology in 2010: Extensions and refinements. Nucleic Acids Res.
38:(Database Issue). D331–D335. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast Delayed Enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Janky R, Verfaillie A, Imrichová H, Van de
Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K,
Sanchez Naval M, Potier D, et al: iRegulon: From a gene list to a
gene regulatory network using large motif and track collections.
PLoS Comput Biol. 10:e10037312014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davis AP, Grondin CJ, Lennon-Hopkins K,
Saraceni-Richards C, Sciaky D, King BL, Wiegers TC and Mattingly
CJ: The comparative toxicogenomics database's 10th year
anniversary: Update 2015. Nucleic Acids Res. 43:(Database Issue).
D914–D920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arlt VM, Zuo J, Trenz K, Roufosse CA, Lord
GM, Nortier JL, Schmeiser HH, Hollstein M and Phillips DH: Gene
expression changes induced by the human carcinogen aristolochic
acid I in renal and hepatic tissue of mice. Int J Cancer.
128:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin K, Su KK, Li T, Zhu XQ, Wang Q, Ge RS,
Pan ZF, Wu BW, Ge LJ, Zhang YH, et al: Hepatic premalignant
alterations triggered by human nephrotoxin aristolochic acid I in
canines. Cancer Prev Res (Phila). 9:324–334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ismail E, Al-Mulla F, Tsuchida S, Suto K,
Motley P, Harrison PR and Birnie GD: Carbonyl reductase: A novel
metastasis-modulating function. Cancer Res. 60:1173–1176.
2000.PubMed/NCBI
|
|
30
|
Gao S, Tang K, Zhang J, Yang Z, Cui H, Li
P, Tang H and Zhou M: Department of Orthopedics, Orthopedic Center
of Chinese PLA, Southwest Hospital, Third Military Medical
University; Department of Neurobiology, Third Military Medical
University: Effect of cyclic stretch on expression of c-fos gene in
rat Achilles-derived tendon stem cells. Chin J Rep Reconstr Surg.
2017.
|
|
31
|
Ge R, Wang Z, Zeng Q, Xu X and Olumi AF:
F-box protein 10, an NF-κB-dependent anti-apoptotic protein,
regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP
pathway. Cell Death Differ. 18:1184–1195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Otsuka M, Hatakenaka M, Ishigami K and
Masuda K: Expression of the c-myc and c-fos genes as a potential
indicator of late radiation damage to the kidney. Int J Radiat
Oncol Biol Phys. 49:169–173. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou F, Zhang L, Su Y, Zhang J and An Z:
Inhibitory effect of artemisinin on the upregulation of c-fos and
c-jun gene expression in kidney tissue of diabetic rats. Modern
Journal of Integrated Traditional Chinese & Western Medicine.
23:2294–2295. 2014.
|
|
34
|
Yazdi AS and Ghoreschi K: The
interleukin-1 family. Adv Exp Med Biol. 941:21–29. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rusai K, Huang H, Sayed N, Strobl M, Roos
M, Schmaderer C, Heemann U and Lutz J: Administration of
interleukin-1 receptor antagonist ameliorates renal
ischemia-reperfusion injury. Transpl Int. 21:572–580. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pindjakova J, Hanley SA, Duffy MM, Sutton
CE, Weidhofer GA, Miller MN, Nath KA, Mills KH, Ceredig R and
Griffin MD: Interleukin-1 accounts for intrarenal Th17 cell
activation during ureteral obstruction. Kidney Int. 81:379–390.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wajant H, Pfizenmaier K and Scheurich P:
Tumor necrosis factor signaling. Cell Death Differ. 10:45–65. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meldrum K, Misseri R, Rink R and Meldrum
D: Tumor necrosis factor alpha (TNF) mediates renal tubular cell
apoptosis during obstructive uropathy. J Am Coll Surg.
197:S912003.
|
|
39
|
Awad AS, You H, Gao T, Cooper TK,
Nedospasov SA, Vacher J, Wilkinson PF, Farrell FX and Reeves Brian
W: Macrophage-derived tumor necrosis factor-α mediates diabetic
renal injury. Kidney Int. 88:722–733. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shu KH, Lee SH, Cheng CH, Wu MJ and Lian
JD: Impact of interleukin-1 receptor antagonist and tumor necrosis
factor-alpha gene polymorphism on IgA nephropathy. Kidney Int.
58:783–789. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
World Health Organization, . Stuart MC,
Kouimtzi M and Hill S: WHO model formulary 2008. World Health
Organization; Geneva: 2009
|
|
42
|
Sattarinezhad E, Panjehshahin MR,
Torabinezhad S, Kamali-Sarvestani E, Farjadian S, Pirsalami F and
Moezi L: Protective effect of edaravone against
cyclosporine-induced chronic nephropathy through antioxidant and
nitric oxide modulating pathways in rats. Iran J Med Sci.
42:170–178. 2017.PubMed/NCBI
|