Introduction
There is an increasing body of evidence
demonstrating that oxidative stress may be associated with cancer.
Oxidative stress triggers the modification of DNA bases, DNA
fragmentation and strand breaks, which contribute to the
development of numerous cancer types as well as various other
diseases (1). Numerous studies
concerning the association between oxidative stress and cancer have
been previously conducted (2–4).
When confronted with external oxidative damage, the body will have
evolved numerous defense systems to remove the stimulus. The
kelch-like ECH-associated protein 1 (KEAP1)/nuclear factor
erythroid 2-related factor 2 (NRF2) system is one of the most
important cell defense systems and survival pathways in vivo
(5). NRF2 serves a core role in
this pathway. NRF2 is anchored in the cytoplasm by KEAP1 in the
resting state and translocates into the nucleus to activate the
antioxidant response element (ARE) under oxidative stress
conditions, which may lead to an increase in the expression of
downstream antioxidative proteins, including NAD(P)H quinone
oxidoreductase 1 (NQO1) and heme oxygenase 1 (HO1) (6). NQO1 and HO1 are regarded as inducible
phaseIIdetoxifying enzymes. NQO1 is a flavoprotein that protects
the body from oxidative damage via stabilization of the p53 tumor
suppressor (7). HO1 catalyzes the
initial and rate-limiting steps in heme catabolism and exhibits a
protective effect by decreasing the intracellular pro-oxidant
levels (8). However, it has been
reported that as well as protecting normal cells from oxidative
damage, NRF2 also protects tumor cells. This finding has been
confirmed within numerous cell lines and tissues, including
non-small cell lung carcinoma, pancreatic cancer and ovarian cancer
(7,9–11).
Selective knockdown of KEAP1 with small interfering (si)RNA was
reported to promote the nuclear migration and expression of NRF2
and its downstream genes in human umbilical vein endothelial cells
(12). Furthermore, research by
Wakabayashi et al reported that KEAP1−/− mice are
more likely to die postnatally due to malnutrition resulting from
hyperkeratosis in the esophagus and forestomach; however,
simultaneous ablation of NRF2 may reverse KEAP1
deficiency-associated phenotypes (13).
To the best of our knowledge, no previous studies
concerning an association between the Hep2 cell line and the
KEAP1/NRF2 signaling pathway have been reported. Therefore, in the
present study, the effects of KEAP1 knockdown on NRF2 and its
downstream elements were investigated using RNA interference (RNAi)
to reveal the integrity of the KEAP1/NRF2 system and the effect on
oxidative stress in the Hep2 cell line following the addition of
hydrogen peroxide (H2O2).
Materials and methods
Cell lines and cell culture
The Animal Ethics Committee of the Eye, Ear, Nose
and Throat Hospital of Fudan University (Shanghai, China) reviewed
and approved the study protocol. The Hep2 cell line employed in the
present study was from our own laboratory (Laboratory Center, Eye,
Ear, Nose and Throat Hospital of Fudan University, Shanghai,
China). Cells were maintained in RPMI-1640 (Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and penicillin
(50 U/ml)-streptomycin (50 µg/ml) solution (Gibco; Thermo Fisher
Scientific, Inc.). The cell line was incubated at 37°C in a
humidified atmosphere of 95% air and 5% CO2. The mixed
cancer Hep2 cell line, which was originally considered to be of the
laryngeal carcinoma type but was later reported to be contaminated
with cervical carcinoma HeLa cells, was used as a cancer cell model
in the current study (14–16).
Construction of lentivirus
vectors
According to the human KEAP1 transcript in GenBank
(https://www.ncbi.nlm.nih.gov/nuccore/; NM_203500),
three target RNA interference sequences that silence the KEAP1 gene
were identified. Lentiviral vectors expressing RNAi specific for
the KEAP1 gene and a scrambled sequence encoding a green
fluorescent protein (GFP) sequence were designed and constructed by
Obio Technology Co., Ltd. (Shanghai, China). The following
sequences were used: 5′-GCAAGGACTACCTGGTCAAGA-3′ (shKEAP1),
5′-CGGGAGTACATCTACATGCAT-3′ (shKEAP1-1),
5′-GTGGCGAATGATCACAGCAAT-3′ (shKEAP1-2) and
5′-TTCTCCGAACGTGTCACGT-3′ (scRNA). Pairs of complementary
oligonucleotides with these sequences were synthesized, annealed
and cloned into the lentiviral plasmid vector
[pLKD-CMV-G&PR-U6-short hairpin (sh)RNA] (Obio Technology,
Ltd., Shanghai, China) using the AgeI and EcoRI
enzymes (Takara Bio, Inc., Otsu, Japan). The recombinant plasmid
vectors (32 µg) containing shRNA were co-transfected into 293T
cells along with the helper plasmids psPAX2 and pHCMV-VSV-G (Takara
Bio, Inc.) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Subsequently, the supernatants were
collected and purified after 48 h of transfection and then
titrated.
The constructed lentiviral vectors contained a green
fluorescence gene that emits green fluorescence in response to
excitation with blue light. This characteristic may be utilized to
optimize the transfection conditions. Following the screening of a
series of preliminary experiments using 2 µg/ml puromycin, the
optimal transfection efficiency was determined. Briefly, for cell
transfection, Hep2 cells in the exponential growth phase were
collected, counted and seeded in 24-well tissue culture plates
(Corning Incorporated, Corning, NY, USA) at a density of
2×104 cells/well to attain 30% confluence on the day of
transfection. The lentivirus containing specific KEAP1 shRNA and
the negative control lentivirus were applied to Hep2 cells with the
multiplicity of infection of 10 or 20. In addition, polybrene was
added into each well with a final concentration of 5 µg/ml. At
12–20 h post-transfection, the medium was completely replaced and
the cells were incubated at 37°C in a 5% CO2 incubator
for an additional 72 h. The transfection rate was evaluated by
counting GFP-positive cells under an inverted fluorescence
microscope; silencing efficacy was verified using real-reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
western blotting and an immunofluorescence technique. Hep2 cells
that exhibited the highest degree of KEAP1 silencing following
transfection with lentivirus containing KEAP1-shRNA were selected
for the establishment of a stable cell line in the presence of
puromycin at a final concentration of 2 µg/ml. The media containing
puromycin were changed every 2–3 days, for 2 weeks. Parent Hep2 and
scHep2 cells served as the controls.
Detection of apoptosis by flow
cytometry
Cells were seeded in 6-well plates (Corning
Incorporated) at a density of 4×105 cells/well and
cultured for 24 h. Subsequently, cells were treated with two
different concentrations of H2O2 (0.1 and
0.25 mmol/l) for 24 h at 37°C. The floating original medium was
collected and the cells were digested with 0.25% trypsin without
EDTA (Hyclone; GE Healthcare Life Sciences) at 37°C for 1 min. Cell
suspensions were centrifuged for 5 min at 1,000 × g at room
temperature. Cells were resuspended and gently washed twice with
ice-cold phosphate-buffered saline (PBS; pH 7.2–7.4; Hyclone; GE
Healthcare Life Sciences). Finally, the cells were collected in 1.5
ml Eppendorf tube. Cell density was adjusted to 1×106
cells/tube. According to the operating instructions of an Annexin
V-allophycocyanin (APC)/7-aminoactinomycin D (7-AAD) double
staining apoptosis detection kit (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China), 500 µl binding buffer was added to suspend cells.
A total of 5 µl Annexin V-APC and 7-AAD was subsequently added
respectively for flow cytometry within 1 h using a Beckman MoFlo
XDP flow cytometer with Summit 5.2 (Beckman Coulter, Inc., Brea, CA
USA). Cells not treated with any H2O2 served
as the controls.
Cell viability assay
shKEAP1 Hep2 and scHep2 cell lines in the
logarithmic growth phase were trypsinized, counted, resuspended and
plated at a density of 2×103 cells/well in a volume of
200 µl RPMI-1640 medium per well in a 96-well plate in triplicate.
After 6, 24, 48, 72 and 96 h incubation individually (data not
shown), cell proliferation was measured using a Cell Counting Kit-8
(CCK-8; Dojindo Moleular Technologies Inc., Kumamoto, Japan)
according to the manufacturer's protocols. The absorbance value was
recorded at 450 nm using a microplate spectrophotometer. To
quantify the ability of H2O2 to inhibit the
shKEAP1 Hep2 cell line, a cell growth assay was also performed
using CCK-8. Briefly, cells were seeded in a 96-well tissue culture
plate (2×104/well), incubated overnight at 37°C and
subsequently treated with 0, 0.25, 0.5, 0.75 and 1.0 mmol/l of
H2O2 for 24 h. CCK-8 was subsequently added
to each well to detect the inhibitory effect of
H2O2 on the cells using a microplate
spectrophotometer at a wavelength of 450 nm.
Immunofluorescence staining
Cells in the exponential growth phase were digested
with trypsin, resuspended and the cell density was adjusted to
1×106 cells/ml with culture media. Prior to cell density
adjustment, one glass coverslip (22×22 mm) was placed into each
well of a 6-well dish. A total of 5 µl cell suspensions was seeded
onto the glass coverslip in order to grow up to 30–50% confluence
24 h later. H2O2 was added into the wells at
final concentrations of 0.25 and 0.5 mmol/l for 6 and 9 h. Equal
volumes of PBS were added into the wells that were not treated with
H2O2 for the same aforementioned durations.
Following incubation for the specified durations, the media were
removed and cells were washed three times with ice-cold PBS. The
slides of cells were fixed in fresh 4% formaldehyde for 20 min at
room temperature. Subsequently, the cells were washed three times
with PBS and incubated in PBS containing 1% (v/v) Triton X-100 and
1% bovine serum albumin (Beyotime Institute of Biotechnology) for 1
h on ice to permeabilize the cells and block non-specific
protein-protein interactions. The glass coverslips were removed
from the 6-well plates and subsequently incubated with 30 µl NRF2
primary antibody (cat. no. ab31163; Abcam, Cambridge, UK) diluted
at 1:100 and placed in a humidified chamber overnight at 4°C. The
secondary antibody used was cyanine 3-conjugated goat anti-rabbit
IgG (H+L; cat. no. A01516; Beyotime Institute of Biotechnology)
used at a 1:1,000 dilution for 1 h. DAPI was used to stain the cell
nuclei for 5 min at room temperature (cat. no. C1002; Beyotime
Institute of Biotechnology). Finally, the coverslips were sealed
with antifade mounting medium (Beyotime Institute of Biotechnology)
and observed under fluorescence microscopy (magnification, ×40).
The immunofluorescence detection of NRF2 without
H2O2 and the staining of KEAP1 were performed
as described above but using a KEAP1-specific antibody (cat. no.
ab139729; Abcam) also diluted at 1:100 and the same secondary
antibody (cyanine 3-conjugated goat anti-rabbit IgG; H+L; cat. no.
A01516).
RT-qPCR
Cells were seeded in 6-well plates and treated with
0, 0.5 and 1.0 mmol/l H2O2 Total RNA was
extracted from cells using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). RNA quality and quantity was measured
using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc.). A total of 1 µg RNA was reverse
transcribed into cDNA using the PrimeScript RT reagent kit (Perfect
Real Time; Takara Bio, Inc.) in a final 20 µl volume reaction,
according to the manufacturer's protocol. A volume of 1 µl RT
reaction mixture and 9 µl qPCR mixture were mixed and SYBR Green
I-based RT-qPCR analyses of human KEAP1, NRF2, NQO1 and HO1 were
performed by using the SYBR Premix Ex Taq system (Tli RNaseH Plus;
Takara Bio, Inc.), according to the manufacturer's protocol in
triplicate on an ABI 7500 system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Negative RT controls were included in each
assay. All primers were designed and synthesized by Sangon Biotech
Co., Ltd. (Shanghai, China), as presented in Table I. GAPDH was used as a reference
gene. Relative quantitative levels of samples were determined by
the 2−∆∆Cq method (17).
 | Table I.Sequences of reverse
transcription-quantitative polymerase chain reaction
oligonucleotide primers. |
Table I.
Sequences of reverse
transcription-quantitative polymerase chain reaction
oligonucleotide primers.
| Gene | Forward
sequence | Reverse
sequence |
|---|
| NRF2 |
CGTCCCAGCAGGACATGGAT |
AGCTCATACTCTTTCCGTCGC |
| KEAP1 |
TCCCCTACAGCCAAGGTCC |
TCAGTGGAGGCGTACATCAC |
| NQO1 |
CCTTCCGGAGTAAGAAGGCAG |
TCCAGGCGTTTCTTCCATCC |
| HO1 |
ATGACACCAAGGACCAGAGC |
GCATAAAGCCCTACAGCAACT |
| GAPDH |
CAGGAGGCATTGCTGATGAT |
GAAGGCTGGGGCTCATTT |
Western blot analysis
Total protein was acquired by lysing cells in a
radioimmunoprecicpitation assay buffer containing
phenylmethylsulfonyl fluoride with a final concentration of 1
mmol/l (Beijing Cowin Biotech Co., Ltd., Beijing, China). The
cytosolic and nuclear fractions of NRF2 were prepared using a
NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Pierce;
Thermo Fisher Scientific, Inc.). Protein extracts were obtained via
centrifugation at 4°C and 10,000 × g for 10 min; proteins were
quantified using a Bicinchoninic Acid Protein assay kit (Beyotime
Institute of Biotechnology). A total of 40 µg total protein was
electrophoretically separated via 10% SDS-PAGE and transferred to
polyvinylidene fluoride membranes. To block the membranes, 5%
non-fat milk in Tris-buffered saline containing 0.1% Tween-20
(TBST; pH 7.4–7.5) was applied at room temperature for 1 h.
Membranes were subsequently incubated overnight with anti-NRF2
(cat. no. ab31163; Abcam), anti-KEAP1 (cat. no. ab139729; Abcam),
anti-NQO1 (cat. no. A180; Cell Signaling Technology, Inc., Danvers,
MA, USA), anti-HO1 (cat. no. ab68477; Abcam), and anti-β-actin
(cat. no. GB13001-3; Servicebio, Inc.) or lamin B primary
antibodies (GB11408; Servicebio, Inc.) in blocking buffer at 4°C at
1:1,000 dilution. Subsequently, membranes were washed three times
in TBST followed by incubation for 1 h with HRP-labeled goat
anti-rabbit/mouse IgG (H+L) (GB23303/GB23301; Servicebio, Inc.) at
a 1:2,500 dilution and washed in TBST again. Bands were visualized
using hypersensitive enhanced chemiluminescence kit (cat. no.
G2020; Servicebio, Inc. Wuhan China). β-actin was used for
cytoplasmic extracts and lamin B used for nuclear extracts.
Statistical analysis
The significance of intergroup differences among the
multiple groups was determined using one-way analysis of variance.
The Least Significant Difference test was used for the post hoc
test. P<0.05 was considered to indicate a statistically
significant difference. The comparisons of the cell viability among
the groups were performed by Bonferroni following one-way analysis
of variance. All statistical analyses were conducted using the SPSS
17.0 statistical package (SPSS, Inc., Chicago, IL, USA). The
experiments were repeated at least three times and data are
presented as the mean ± standard deviation.
Results
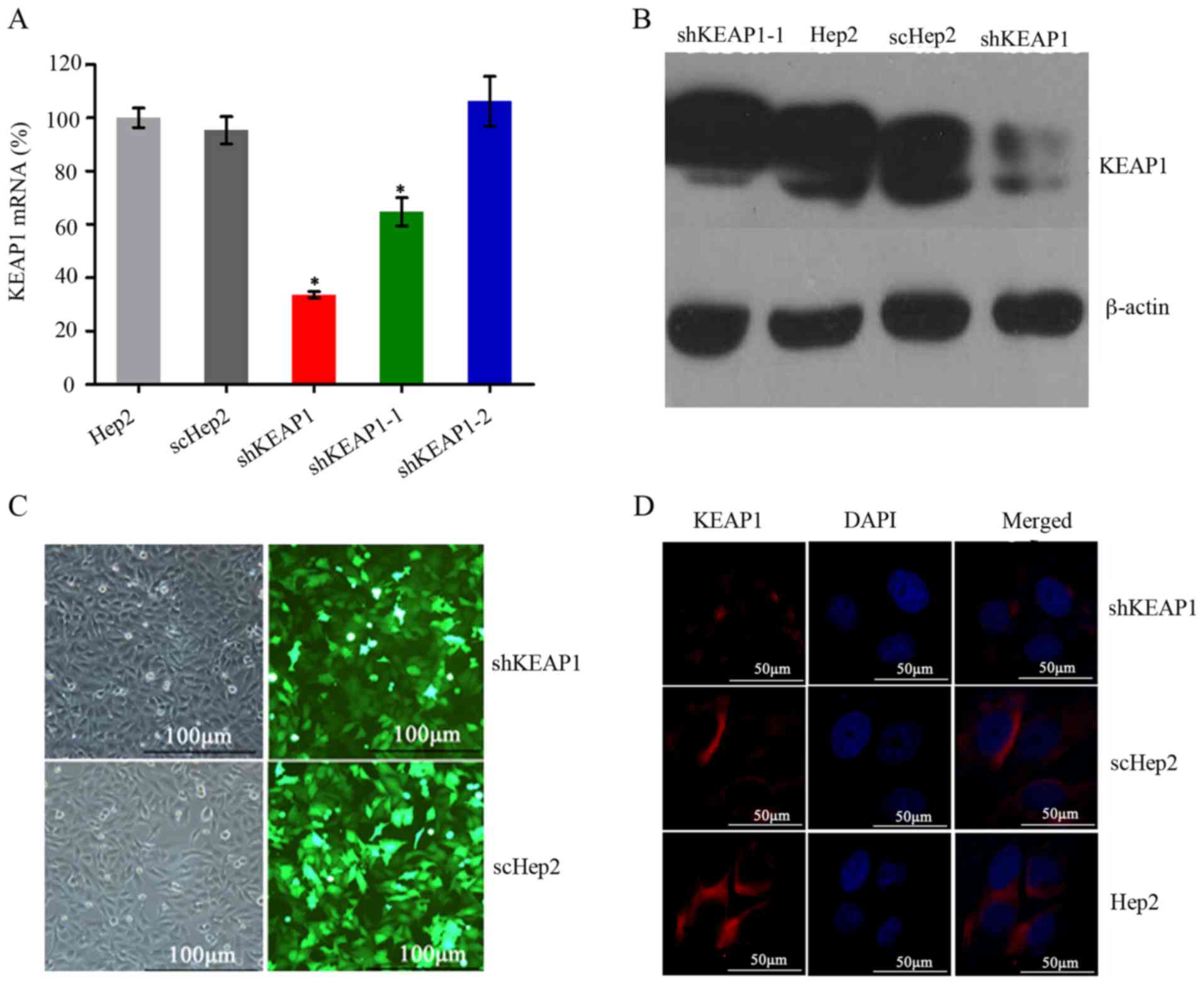
Confirmation of efficient KEAP1
knockdown
In order to establish a stable shKEAP1 Hep2 cell
line with effective knockdown of the KEAP1 gene, Hep2 cells were
transfected with three KEAP1 shRNAs and one scrambled shRNA
expression lentiviral plasmids bearing GFP, which was used to
assess the efficacy of transfection. RT-qPCR was performed on
exponentially growing cells to determine the expression levels of
KEAP1 following 72 h transduction. Of the three shKEAP1 sequences,
one exhibited the highest knockdown efficiency, which reduced KEAP1
mRNA expression levels by up to 67±1% compared with the Hep2 cells
transfected with the other shRNAs (P<0.05; Fig. 1A). Based on this, this sequence was
selected to establish a stable shKEAP1 Hep2 cell line with
puromycin for 2 weeks. Subsequently, the protein expression levels
of KEAP1 in shKEAP1 Hep2 and scHep2 cell lines were measured by
western blotting. KEAP1 protein expression levels were markedly
reduced following KEAP1 knockdown using the shKEAP1 construct
(Fig. 1B). Furthermore, the
transfection efficiency of scHep2 cells was demonstrated to be
equivalent to that of shHep2, which was ~80% (Fig. 1C). Immunofluorescence staining
demonstrated that KEAP1 protein was primarily expressed in the
cytoplasm. Weak fluorescence and bands were detected within the
shKeap1 group, which were markedly lower compared with in scHep2
and Hep2 cells; however, within the control groups, expression
levels were similar (Fig. 1D),
indicating that a stable and effective shKEAP1 Hep2 cell line was
established.
KEAP1 knockdown increases the
expression of NRF2, NQO1 and HO1 in Hep2 cells
The effects of KEAP1 knockdown on the mRNA and
protein expression of NRF2 and NRF2-associated genes were
investigated. The results demonstrated that shRNA-mediated
depletion of KEAP1 was associated with significant increases in the
mRNA expression levels of NRF2, NQO1 and HO1 by 1.42±0.05-,
1.75±0.10- and 1.59±0.07-fold, respectively, compared with the
scHep2 group (Fig. 2A). Notably,
immunofluorescence staining demonstrated that the expression levels
of nuclear NRF2 were low, with NRF2 primarily located in the
cytoplasm within parent Hep2 cells; however, NRF2-reactive
fluorescent signals were observed within the nucleus of shKEAP1
cells, indicating that KEAP1 knockdown may cause NRF2 migration
into the nuclei (Fig. 2B).
Furthermore, western blot analysis demonstrated that delivery of
specific shKEAP1 by lentiviral transduction reduced the cytosolic
levels of NRF2, which was observed alongside a marked increase in
nuclear NRF2 levels (Fig. 2C).
Total protein levels of NQO1 and HO1 were markedly elevated in
shKEAP1 Hep2 cells compared with the scHep2 cell line and the blank
control Hep2 group (Fig. 2D).
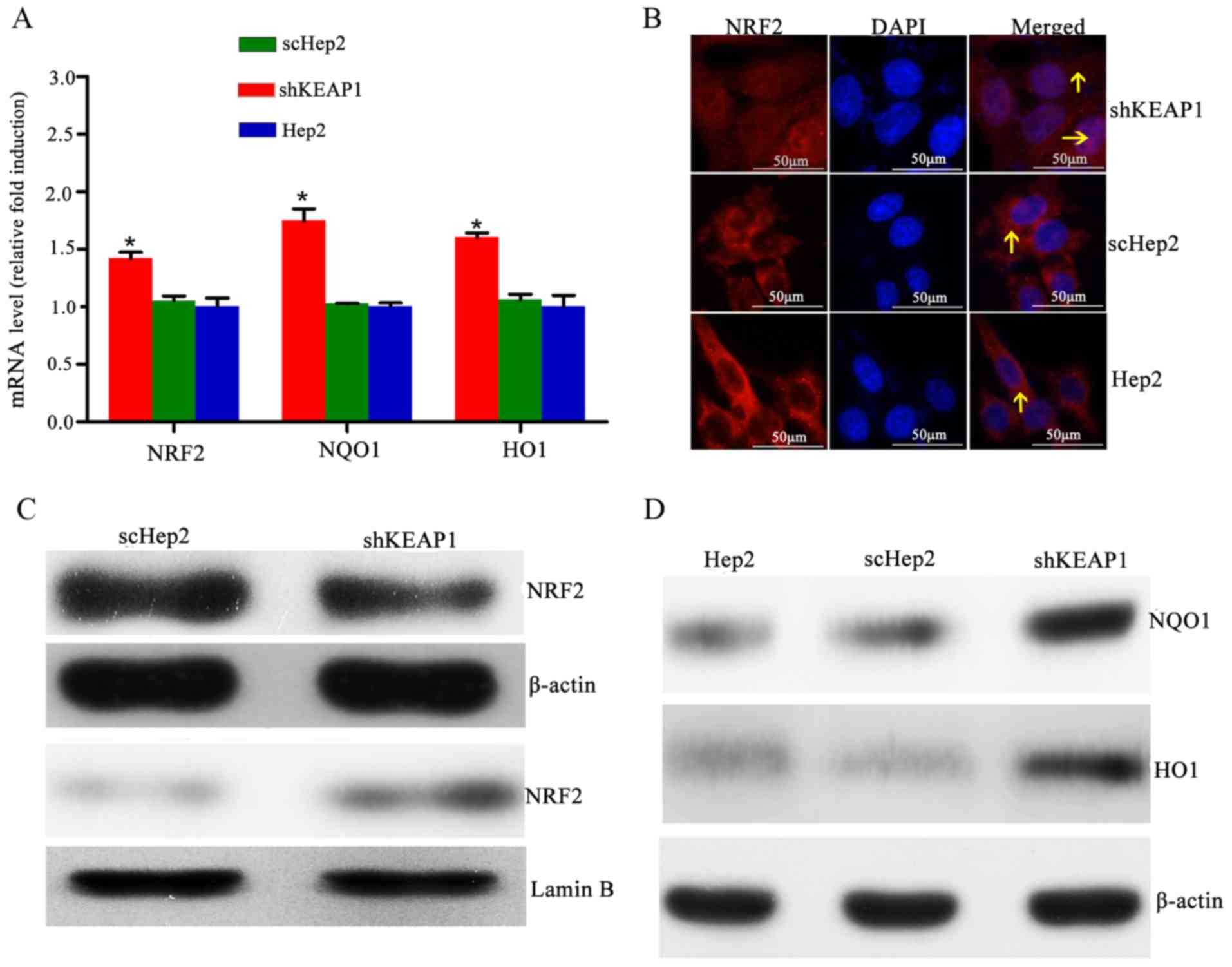 | Figure 2.Effect of KEAP1 knockdown on the
expression of NRF2 and downstream targets. (A) mRNA expression
levels of NRF2, NQO1 and HO1 in Hep2, scHep2 and shKEAP1 Hep2
cells. Expression levels of NRF2, NQO1 and HO1 were increased
following the knockdown of KEAP1 in Hep2 cells. (B) Representative
NRF2 immunofluorescence staining images indicate that NRF2
translocated into the nuclei from the cytoplasm following knockdown
of KEAP1 in Hep2 cells (magnification, ×40). (C) Western blotting
demonstrated that nuclear NRF2 protein expression levels were
elevated, while cytoplasmic NRF2 protein expression levels were
reduced, following the knockdown of KEAP1 in Hep2 cells. (D)
Western blotting demonstrated that total NQO1 and HO1 protein
expression levels were increased within shKEAP1 Hep2 cells,
compared with the scHep2 group. *P<0.05 vs. scHep2 group. The
arrows indicate NRF2. KEAP1, kelch-like ECH-associated protein 1;
NRF2, nuclear factor erythroid 2-related factor 2; NQO1, NAD(P)H
quinone oxidoreductase 1; HO1, heme oxygenase 1; scHep2, scrambled
control-transfected Hep2 cells; sh, short hairpin RNA. |
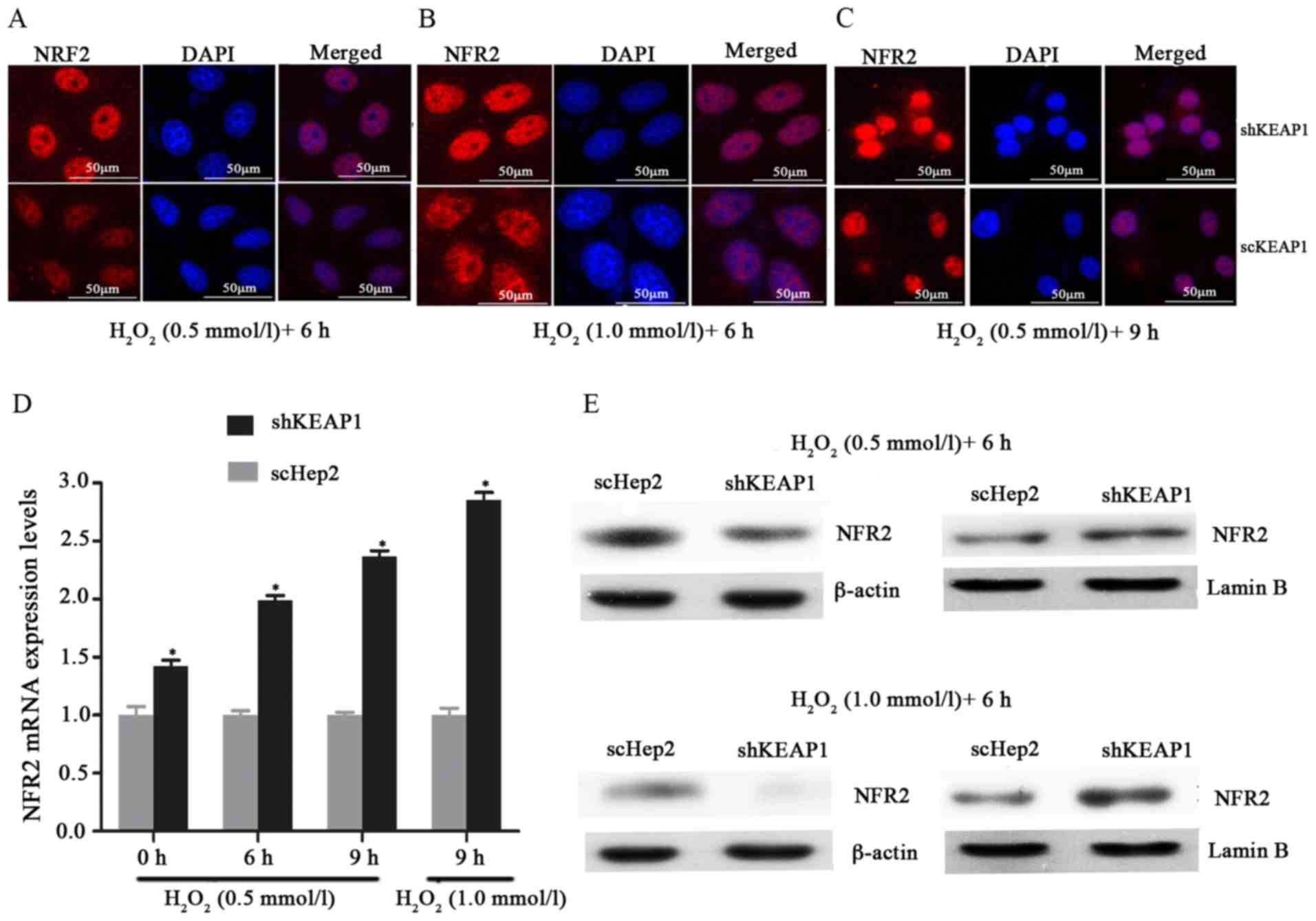
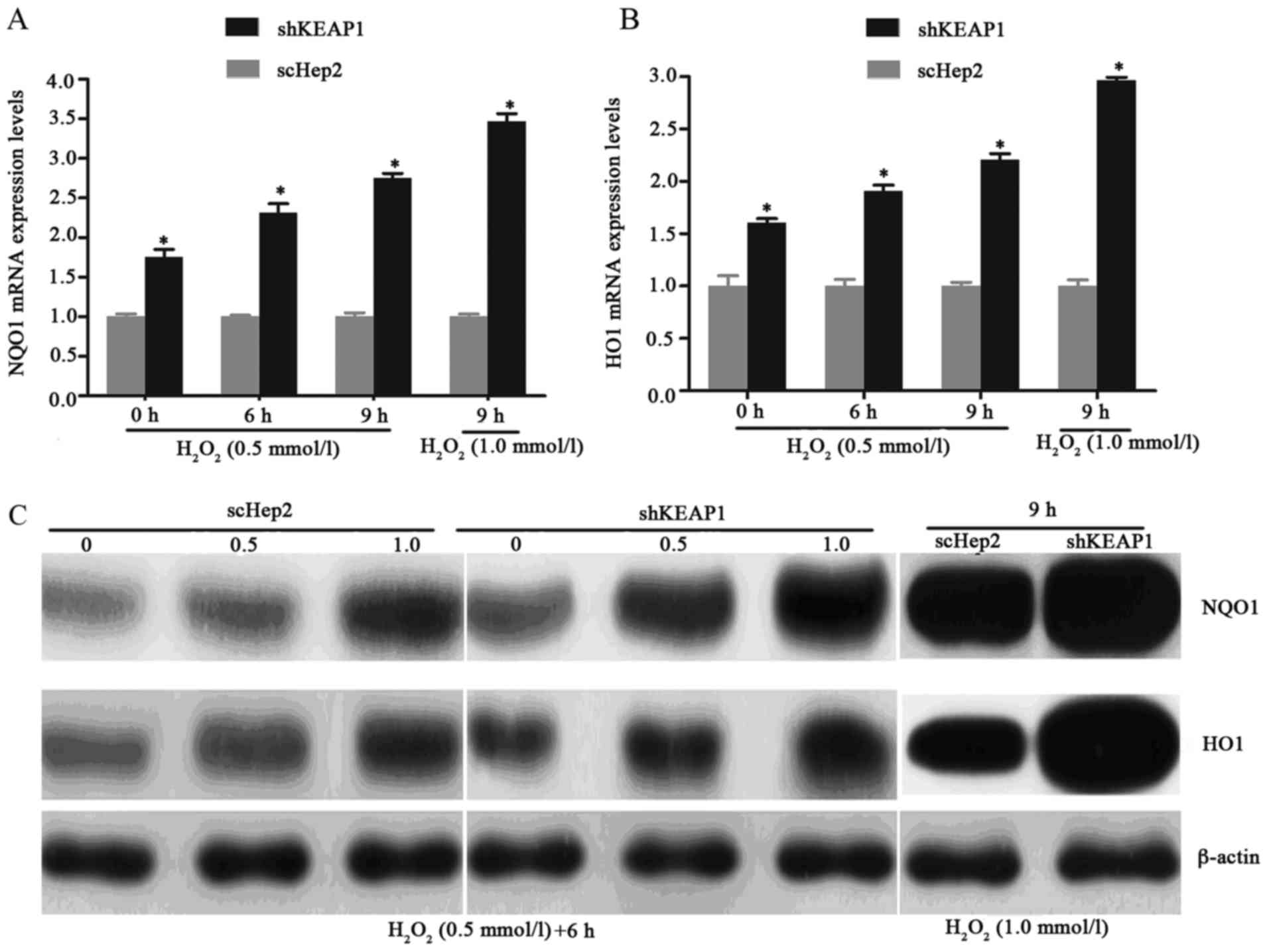
Effect of KEAP1 knockdown on oxidative
damage in Hep2 cells
To investigate the role of KEAP1 knockdown against
oxidative damage in the Hep2 cell line, the expression of this
system and the resistance of the cells to oxidative stresses
induced by H2O2 were analyzed.
Immunofluorescence analysis demonstrated that when cultured shKEAP1
Hep2 cells were treated with 0.5 and 1 mmol/l
H2O2 for 6 h, NRF2 markedly migrated into the
cell nucleus compared with in scHep2 cells. In addition, the
expression of NRF2 appeared to be markedly higher in response to
the concentration of 1 mM/l l H2O2 for 6 h
compared with 0.5 mmol/l (Fig.
3A-C). As presented in Figs. 3D
and E, and 4, the results of
RT-qPCR analysis and western blotting demonstrated consistencies
with immunofluorescence analysis; the increases in the NRF2, NQO1
and HO1 transcript and protein levels demonstrated time- and
dose-dependency within the knockdown cell line. Knockdown of KEAP1
via shRNA in Hep2 cells increased the NRF2, NQO1 and HO1 mRNA
expression levels by 1.99, 2.32 and 1.91-fold respectively,
compared with scHep2 cells when treated with 0.5 mM/l
H2O2 for 6 h (Figs. 3D, 4A
and B). At the concentration of 1 mM/l
H2O2 for 9 h, NRF2, NQO1 and HO1 mRNA levels
were 2.85, 3.47 and 2.96-fold higher in the KEAP1 knockdown group,
respectively, compared with in the scHep2 cells. mRNA expression
levels were analyzed upon exposure to 0.5 mmol/l
H2O2 for 6 and 9 h. NRF2, NQO1 and HO1 were
all expressed highly within shKEAP1 cells compared with in scHep2
cells at both concentrations. Western blot analyses revealed that
the protein expression levels of nuclear NRF2, and total NQO1 and
HO1, were markedly upregulated in shKEAP1 Hep2 cells compared with
in scHep2 control cells at the concentration of 0.5 and 1 mmol/l
H2O2 for 6 h (Figs. 3E and 4C). Supporting the results of
immunofluorescence, KEAP1 knockdown led to a reduction in the
protein expression levels of NRF2 within the cytoplasm and an
increase within the nuclei at 0.5 and 1 mM/l
H2O2 concentrations, compared with scHep2
cells (Fig. 3E). The expression
patterns of NQO1 and HO1 were similar to the nuclear NRF2 in
shKEAP1 Hep2 cells upon exposure to H2O2
(Fig. 4C).
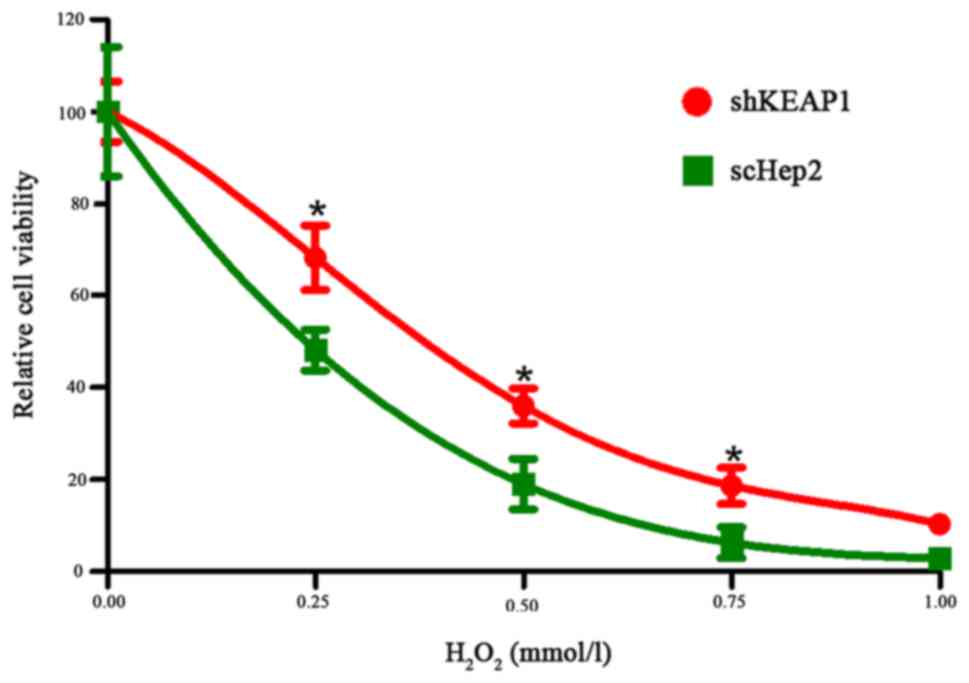
Effects of KEAP1 knockdown on Hep2
cell function
As the expression levels of NRF2, NQO1 and HO1 were
altered within NRF2-activated Hep2 cells, the effects of KEAP1
knockdown on cell function were subsequently investigated. Cell
viability decreased in a dose-dependent manner within the shKEAP1
and scHep2 cell lines with increasing doses of
H2O2 with the range of final concentrations
from 0–1 mM/l for 24 h. At concentrations of 0.25, 0.5 and 0.75
mM/l, the mean relative viabilities of shKEAP1 and scHep2 cells
were 75 and 51, 33 and 12, and 18 and 6%, respectively. It was
observed that transfected shKEAP1 Hep2 cells demonstrated
significantly higher cell viabilities following
H2O2 treatment compared with in the scHep2
group, as demonstrated in Fig. 5.
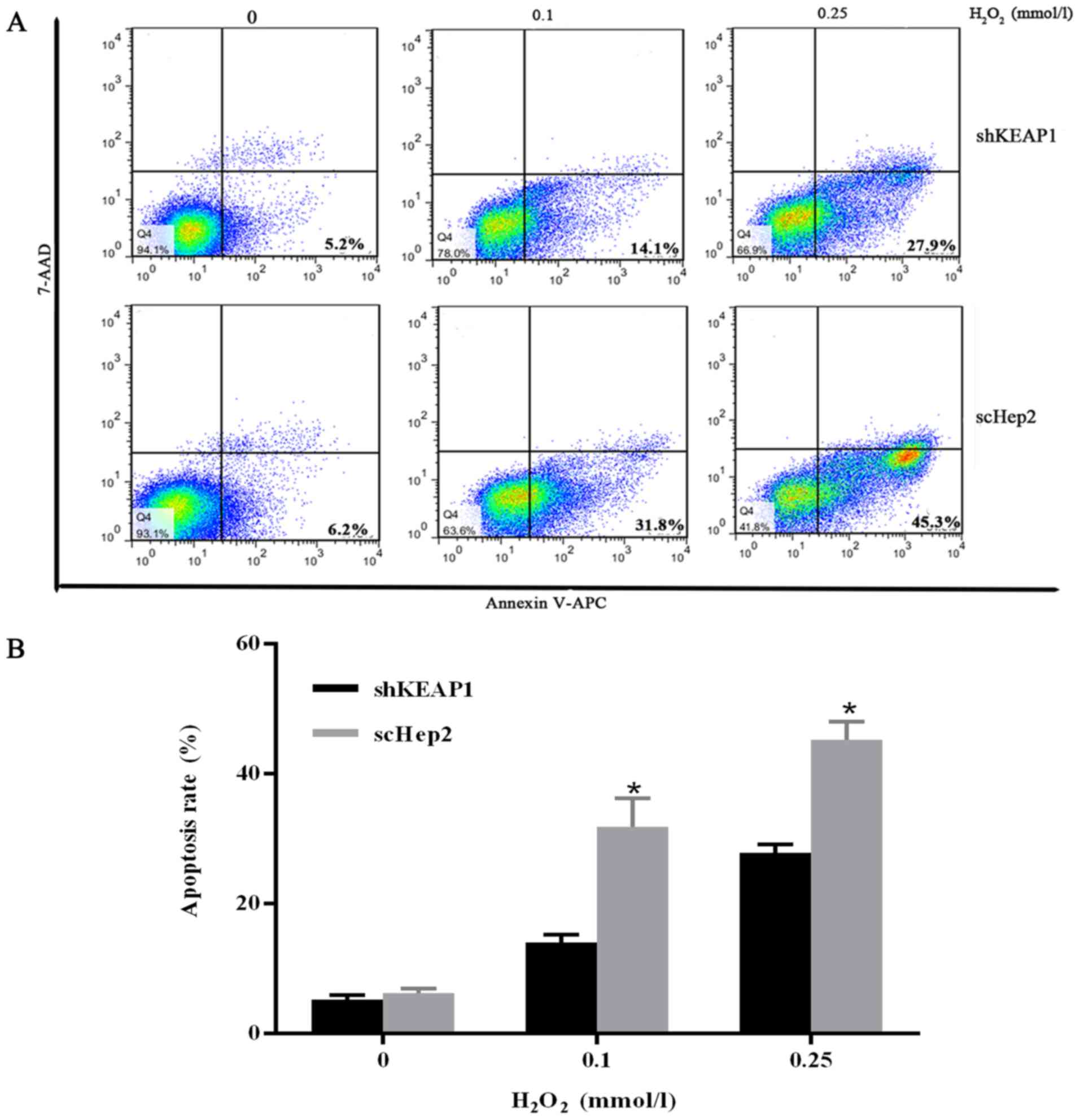
To validate the results obtained from the CCK-8 assay and to
investigate whether apoptosis served a role against oxidative
stress injury, the percentage of the apoptotic cells was analyzed
by flow cytometry. Cells were cultured with varying concentrations
of H2O2 (0, 0.1 and 0.25 mmol/l) for 24 h.
Marked increases were observed in the apoptosis rates of scHep2
cells at 0.1 and 0.25 mmol/l H2O2 from 31.8
and 45.3%, while 14.1 and 27.9% in shKEAP1 cells (Fig. 6).
Discussion
The association between oxidative damage and cancer
has been increasingly emphasized. High antioxidant levels have been
reported within the serum and tissue samples of numerous patients
with cancer. Suzuki et al reported that increasing the
intake of antioxidants, such as renieratene, vitamin C and vitamin
E, may not only reduce the risk of developing head and neck cancer
of the general population, but also those who smoke and consume
alcohol (18). One study
demonstrated that cells exhibit elevated sensitivity to the
external harmful materials, when the activities of antioxidants and
enzymes are inhibited (19). These
findings have demonstrated that oxidative damage may exert notable
effects in the process of the cancer occurrence, progression and
treatment.
When confronted with oxidative stress, antioxidative
systems within the body are activated to remove oxidative
stress-inducing substances. Knockout of the NRF2 gene in mice
revealed a reduction of antioxidant enzymes, including glutathione
S-transferase and HO1, and an increased sensitivity of mice to
carcinogens as knockout mice exhibited higher incidences of cancer
compared with wild-type mice (20). Research has also indicated that
interfering with KEAP1 gene expression may upregulate the
expression of NRF2; Singh et al reported that within lung
cancer cells with low expression levels of KEAP1, the nuclear NRF2
expression levels were significantly higher compared with lung
cancer cells with high KEAP1 expression. NQO1 expression was
increased in lung cancer cells with lower KEAP1 expression,
compared with cells with high KEAP1 expression. In addition, NRF2
and its downstream genes were demonstrated to be downregulated via
transfection of NRF2-siRNA into cells or through increasing KEAP1
expression (10). These findings
have also been observed within pancreatic cancer cell lines, where
KEAP1 was also inversely associated with NRF2 and its downstream
genes, including NQO1 and HO1. Therefore, high expression levels of
KEAP1 may lead to low expression levels of NRF2 and antioxidants.
Elevated levels of KEAP1 expression may cause the expansion ability
of cells to decrease (21). In
addition, these findings were also verified within ovarian
carcinoma cells. Following the introduction of KEAP1-siRNA into
ovarian carcinoma cells, the expression levels of KEAP1 mRNA
decreased by 60% and the target protein NQO1 was markedly lowered,
which accelerated the cell growth (11). Jung and Kwak identified the same
phenomenon within colorectal cells. Specific KEAP1-shRNA was
transfected into colorectal cells to reduce the expression levels
of KEAP1 mRNA by 50%, resulting in the marked upregulation of
nuclear translocation of NRF2 and the expression of its target
proteins, including NQO1 and HO1 (22).
In the present study, the experimental results
regarding the role of the KEAP1-NRF2-ARE signaling pathway within
Hep2 cells were consistent with the above motioned studies. A
highly effective shKEAP1 stable Hep2 cell line was established in
the present study. Low expression levels of NQO1 and HO1 were
detected within normal Hep2 cells, while shKEAP1 cells exhibited
increased expression levels. Importantly, the findings of the
present study confirmed that the downregulation of KEAP1 was
associated with the migration of NRF2 from the cytoplasm to the
nuclei. This demonstrated that the activation of NRF2 was
associated with high expression levels of NQO1 and HO1. In
addition, the effects of KEAP1 knockdown on the proliferation of
Hep2 cells were analyzed. The results demonstrated that the cells
of the shKEAP1 group exhibited a marked increase in their
proliferation compared with the scHep2 control group. Notably, the
cell viability of shKEAP1 Hep2 cells was greater compared with in
scHep2 cells from 48 to 96 h following the transfection of
KEAP1-shRNA (data not shown). This indicated that the effects of
KEAP1 knockdown via lentiviral transfection may be long lasting. To
investigate the mechanisms involved in this process, the apoptotic
rate was measured within the scHep2 and shKEAP1 Hep2 cell lines.
Marked alterations 24 h post-transfection between the shKEAP1 and
control groups were not observed at 0 mmol/l
H2O2; however, at 0.1 and 0.25 mmol/l
H2O2 the apoptosis rate was higher within
scHep2 cells compared with in shKEAP1 Hep2 cells. These results
demonstrated that the inhibition of KEAP1 expression within Hep2
cells may reduce cell apoptosis to improve survivability. In
addition, similar findings have been observed in other cell types.
Li et al reported that NRF2 was primarily located within
nuclei and that overexpression of NRF2 may promote proliferation in
endometrial cancer cells (23). Ma
et al revealed that the downregulation of NRF2 reduced the
expression levels of NQO1 and HO1 within cervical cancer cells.
Furthermore, the volume of shNRF2-transfected cervical cancer cell
xenograft tumors was lower compared with wild-type NRF2 expression
cell xenograft tumors (24).
Numerous studies have demonstrated that there are
close associations between H2O2 and the
KEAP1-NRF2-ARE signaling pathway. Following the addition of
H2O2 to astrocyte cells, the KEAP1/NRF2
system was activated and NRF2 migrated into the nuclei from the
cytoplasm. The expression levels of NRF2 and its target protein,
NQO1, were notably upregulated, which was positively associated
with increasing H2O2 concentrations and the
duration of treatment (25,26).
Pi et al (27) also
demonstrated that H2O2 markedly induced
nuclear NRF2 expression and reduced cytoplasmic NRF2 expression.
The longer the duration of treatment and the higher the
concentration was, the higher expression levels of NQO1 and nuclear
NRF2 (27). Additionally, an in
vivo study revealed that nuclear NRF2 accumulated within living
tissues induced by H2O2 in a dose-dependent
manner. Collectively, these previous studies revealed that
H2O2 may activate the KEAP1-NRF2-ARE
signaling pathway and affect NRF2 nuclear transfer in a time- and
dose-dependent manner. In the present study, research into NRF2
nuclear migration was performed using varying
H2O2 concentrations and durations of
treatment. As expected, marked nuclear transfer of NRF2 was
observed in response to H2O2, and this
phenomenon was more profound within shKEAP1 Hep2 cells compared
with scHep2 cells. The upregulation of nuclear NRF2, NQO1 and HO1
may have reinforced the antioxidant capacity in Hep2 cells. Habib
et al reported that following the addition of varying
concentrations of H2O2 to a breast carcinoma
cell line, NQO1 and HO1 expression, and nuclear NRF2 expression,
were notably increased in a time- and dose-dependent manner
(28).
The present study identified the typical
KEAP1-NRF2-ARE signaling pathway within Hep2 cells. Suppression of
KEAP1 may lead to a decline in cytoplasmic NRF2 and migration of
NRF2 to the nuclei, which is a core factor promoting the expression
of antioxidants, including NQO1 and HO1. It may be advantageous to
inhibit the antioxidant activity of the Hep2 cells via the
inactivation of the KEAP1-NRF2-ARE signal transduction pathway
(29). Once the signaling pathway
was inactivated, the cytoprotective proteins, including NQO1 and
HO1 are downregulated and survival of cells decreased. This may be
useful for inhibiting the growth of cancer. The results of the
present study may contribute to the prevention, diagnosis and
treatment of cancer.
Acknowledgements
The present study was supported by Science and
Technology Commission of Shanghai Municipality (grant no.
15401971600).
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LC, CL, HW and PH conceived and designed the study.
CL, YZ, JC, YY and MC performed the experiments, and completed the
acquisition, analysis and interpretation of data for the work. LC
and CL wrote the paper. HW and PH reviewed and edited the
manuscript. All authors read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jałoszyński P, Jaruga P, Oliński R,
Biczysko W, Szyfter W, Nagy E, Möller L and Szyfter K: Oxidative
DNA base modifications and polycyclic aromatic hydrocarbon DNA
adducts in squamous cell carcinoma of larynx. Free Radic Res.
37:231–240. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seven A, Civelek S, Inci E, Inci F, Korkut
N and Burçak G: Evaluation of oxidative stress parameters in blood
of patients with laryngeal carcinoma. Clin Biochem. 32:369–373.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dwivedi R, Raturi D, Kandpal N, Dwivedi R,
Singh R and Puri V: Oxidative stress in patients with laryngeal
carcinoma. Indian J Cancer. 45:97–99. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Inci E, Civelek S, Seven A, Inci F, Korkut
N and Burçax G: Laryngeal cancer: In relation to oxidative stress.
Tohoku J Exp Med. 200:17–23. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Motohashi H and Yamamoto M: Nrf2-Keap1
defines a physiologically important stress response mechanism.
Trends Mol Med. 10:549–557. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uruno A and Motohashi H: The Keap1-Nrf2
system as an in vivo sensor for electrophiles. Nitric Oxide.
25:153–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Awadallah NS, Dehn D, Shah RJ, Nash
Russell S, Chen YK, Ross D, Bentz JS and Shroyer KR: NQO1
expression in pancreatic cancer and its potential use as a
biomarker. Appl Immunohistochem Mol Morphol. 16:24–31.
2008.PubMed/NCBI
|
|
8
|
Dunn L, Midwinter RG, Ni J, Hamid HA,
Parish CR and Stoker R: New insights into intracellular locations
and functions of heme oxygenase-1. Antioxid Redox Signal.
20:1723–1742. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kensler TW and Wakabayashi N: Nrf2: Friend
or foe for chemoprevention? Carcinogenesis. 31:90–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Singh A, Misra V, Thimmulappa RK, Lee H,
Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E,
et al: Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung
cancer. PLoS Med. 3:e4202006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Konstantinopoulos PA, Spentzos D,
Fountzilas E, Francoeur N, Sanisetty S, Grammatikos AP, Hecht JL
and Cannistra SA: Keap1 mutations and Nrf2 pathway activation in
epithelial ovarian cancer. Cancer Res. 71:5081–5089. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim JH, Choi YK, Lee KS, Cho DH, Baek YY,
Lee DK, Ha KS, Choe J, Won MH, Jeoung D, et al: Functional
dissection of Nrf2-dependent phase II genes in vascular
inflammation and endotoxic injury using Keap1 siRNA. Free Radic
Biol Med. 53:629–640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wakabayashi N, Itoh K, Wakabayashi J,
Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F,
Roop DR, et al: Keap1-null mutation leads to postnatal lethality
due to constitutive Nrf2 activation. Nat Genet. 35:238–245. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Spanou C, Stagos D, Aligiannis N and
Kouretas D: Influence of potent antioxidant leguminosae family
plant extracts on growth and antioxidant defense system of Hep2
cancer cell line. J Med Food. 13:149–55. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Spanou C, Stagos D, Aligiannis N and
Kouretas D: Influence of potent antioxidant leguminosae family
plant extracts on growth and antioxidant defense system of Hep2
cancer cell line. J Med Food. 13:149–155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jewett A, Wang MY, Teruel A, Poupak Z,
Bostanian Z and Park NH: Cytokine dependent inverse regulation of
CD54 (ICAM1) and major histocompatibility complex class I antigens
by nuclear factor kappaB in HEp2 tumor cel: Effect on the function
of natural killer cells. Hum Immunol. 64:505–520. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suzuki T, Wakai K, Matsuo K, Hirose K, Ito
H, Kuriki K, Sato S, Ueda R, Hasegawa Y and Tajima K: Effect of
dietary antioxidants and risk of oral, pharyngeal and laryngeal
squamous cell carcinoma according to smoking and drinking habits.
Cancer Sci. 97:760–767. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khor TO, Huang MT, Prawan A, Liu Y, Hao X,
Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS and Kong AN: Increased
susceptibility of Nrf2 knockout mice to colitis-associated
colorectal cancer. Cancer Prev Res (Phila). 1:187–191. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iida K, Itoh K, Kumagai Y, Oyasu R,
Hattori K, Kawai K, Shimazui T, Akaza H and Yamamoto M: Nrf2 is
essential for the chemopreventive efficacy of oltipraz against
urinary bladder carcinogenesis. Cancer Res. 64:6424–6431. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lister A, Nedjadi T, Kitteringham NR,
Campbell F, Costello E, Lloyd B, Copple IM, Williams S, Owen A,
Neoptolemos JP, et al: Nrf2 is overexpressed in pancreatic cancer:
Implications for cell proliferation and therapy. Mol Cancer.
10:372011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jung KA and Kwak MK: Enhanced
4-hydroxynonenal resistance in KEAP1 silenced human colon cancer
cells. Oxid Med Cell Longev. 2013:4239652013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li K, Zhong C, Wang B, He J and Bi J: Nrf2
expression participates in growth and differentiation of
endometrial carcinoma cells in vitro and in vivo. J Mol Histol.
45:161–167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma X, Zhang J, Liu S, Huang Y, Chen B and
Wang D: Nrf2 knockdown by shRNA inhibits tumor growth and increases
efficacy of chemotherapy in cervical cancer. Cancer Chemother
Pharmacol. 69:485–494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dell'Orco M, Milani P, Arrigoni L,
Pansarasa O, Sardone V, Maffioli E, Polveraccio F, Bordoni M,
Diamanti L, Ceroni M, et al: Hydrogen peroxide-mediated induction
of SOD1 gene transcription is independent from Nrf2 in a cellular
model of neurodegeneration. Biochim Biophys Acta. 1859:315–323.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Erlank H, Elmann A, Kohen R and Kanner J:
Polyphenols activate Nrf2 in astrocytes via H2O2, semiquinones, and
quinones. Free Radic Biol Med. 51:2319–2327. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pi J, Qu W, Reece JM, Kumagai Y and
Waalkes MP: Transcription factor Nrf2 activation by inorganic
arsenic in cultured keratinocytes: Involvement of hydrogen
peroxide. Exp Cell Res. 290:234–245. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Habib E, Linher-Melville K, Lin HX and
Singh G: Expression of xCT and activity of system xc(−) are
regulated by NRF2 in human breast cancer cells in response to
oxidative stress. Redox Biol. 5:33–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen J, Yu Y, Ji T, Ma R, Chen M, Li G, Li
F, Ding Q, Kang Q, Huang D, et al: Clinical implication of Keap1
and phosphorylated Nrf2 expression in hepatocellular carcinoma.
Cancer Med. 5:2678–2687. 2016. View Article : Google Scholar : PubMed/NCBI
|