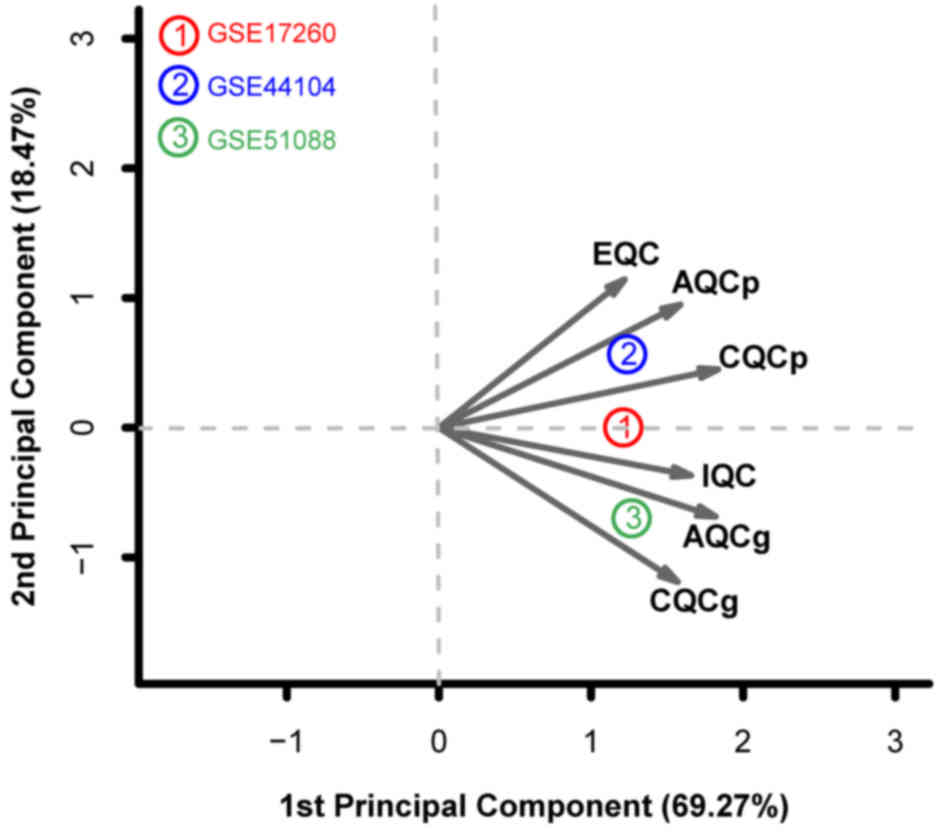
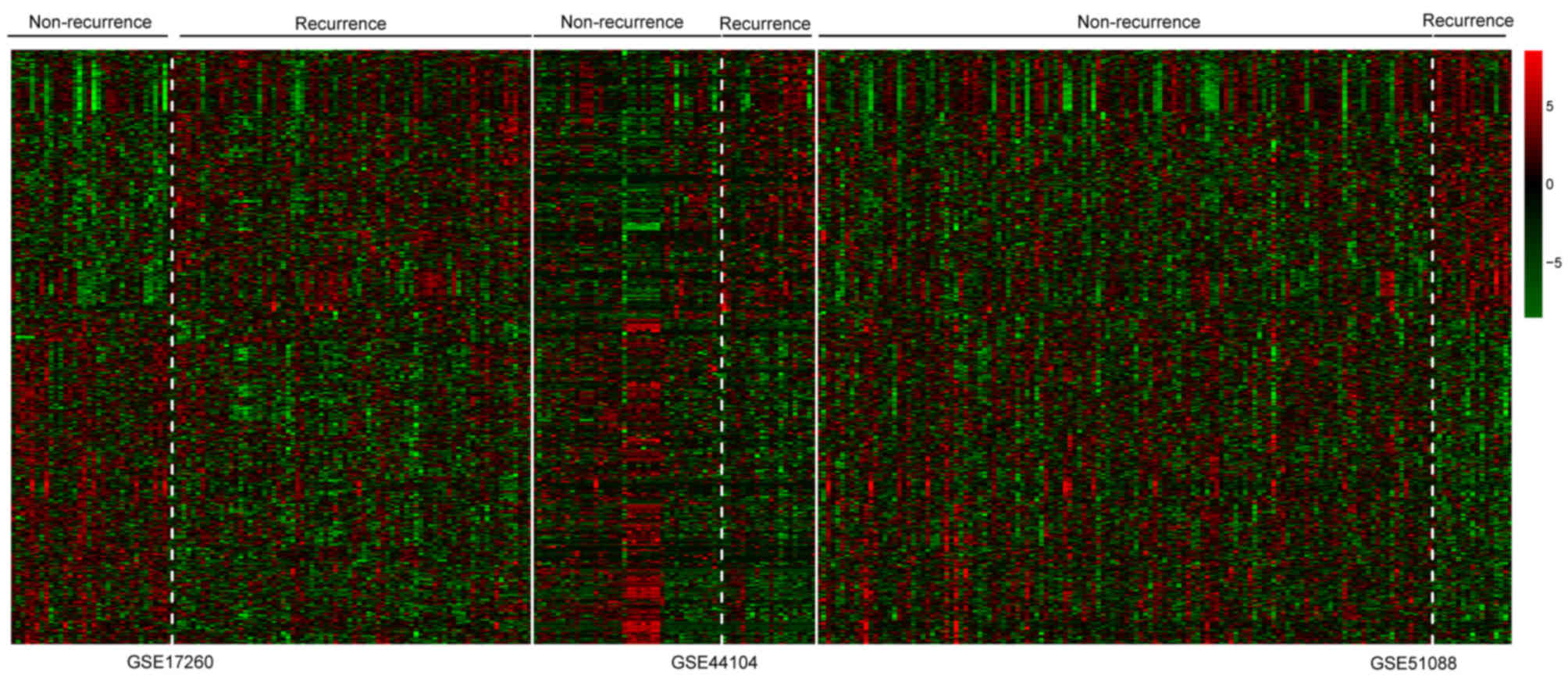
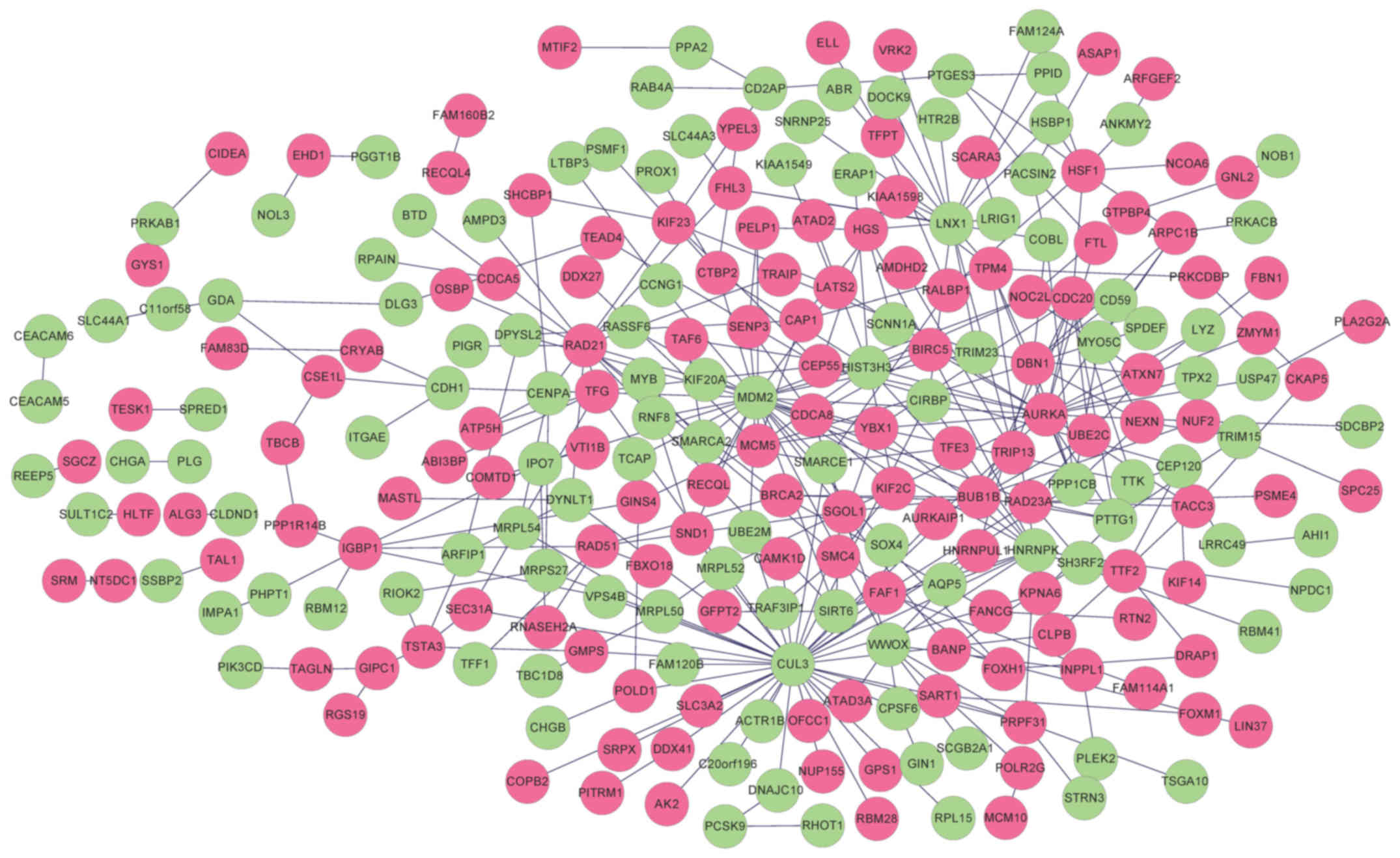
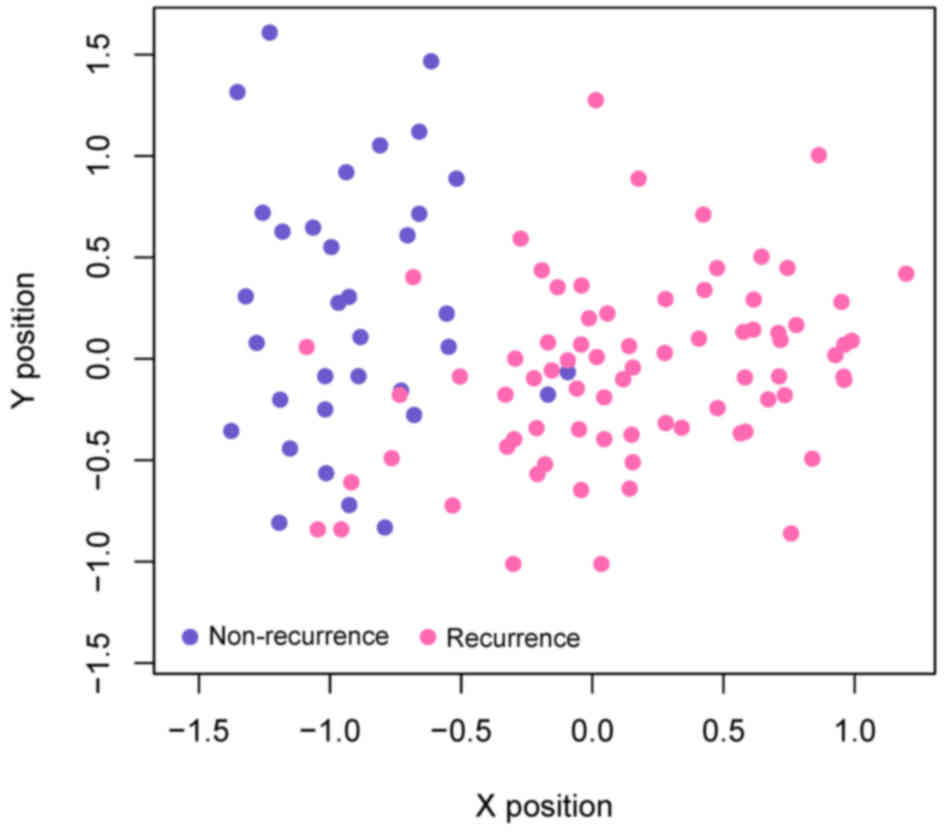
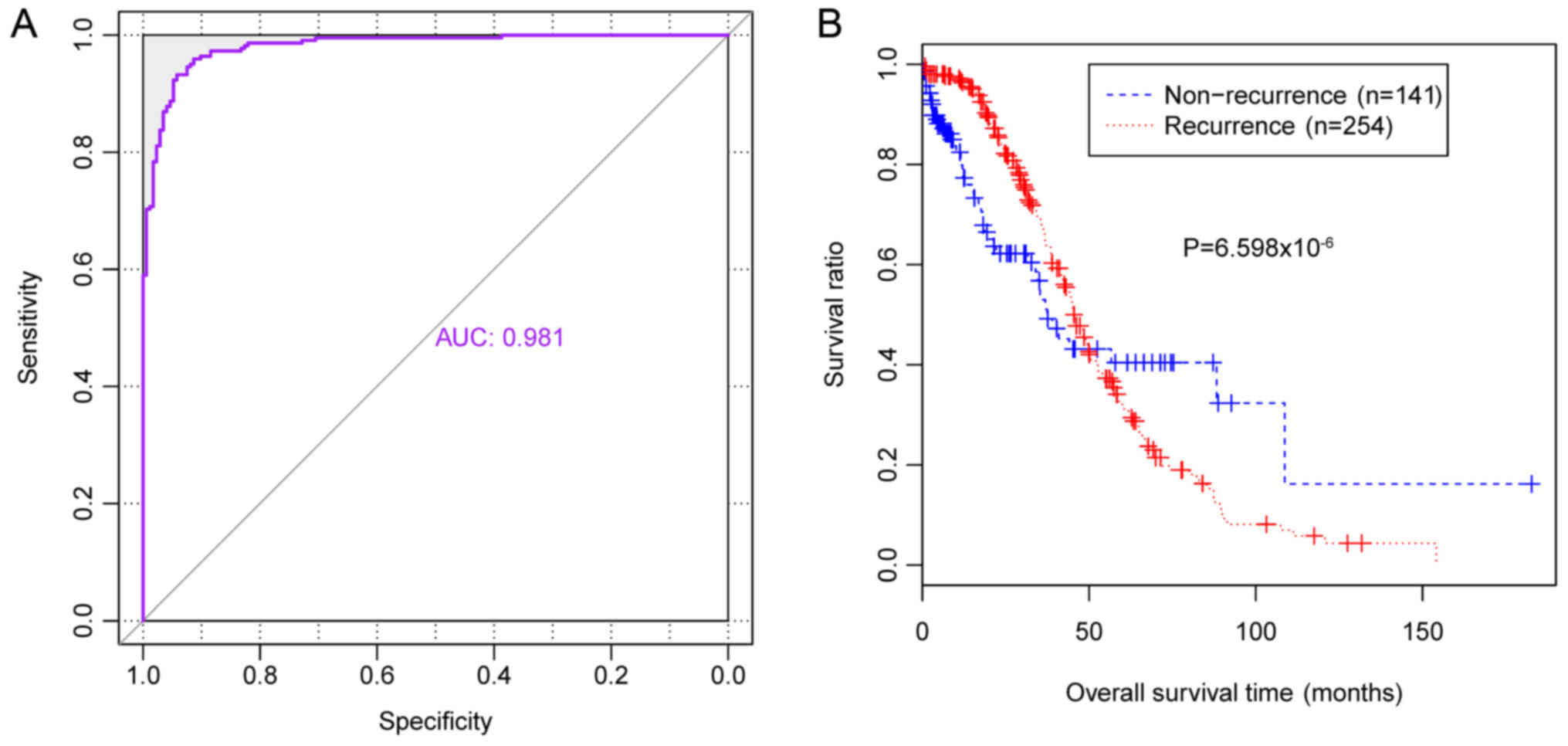
|
1
|
Howlader N, Noone AM, Krapcho M, Garshell
J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z,
et al: SEER Cancer Statistics Review 1975–2012. National Cancer
Institute; Bethesda, MD: 2015, https://seer.cancer.gov/archive/csr/1975_2012/November
18–2015
|
|
2
|
Davidson B and Tropé CG: Ovarian cancer:
Diagnostic, biological and prognostic aspects. Womens Health
(Lond). 10:519–533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gloss BS and Samimi G: Epigenetic
biomarkers in epithelial ovarian cancer. Cancer Lett. 342:257–263.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leung F, Diamandis EP and Kulasingam V:
Ovarian cancer biomarkers: Current state and future implications
from high-throughput technologies. Adv Clin Chem. 66:25–77. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Au KK, Josahkian JA, Francis JA, Squire JA
and Koti M: Current state of biomarkers in ovarian cancer
prognosis. Future Oncol. 11:3187–3195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huo J, Hu J, Liu G, Cui Y and Ju Y:
Elevated serum interleukin-37 level is a predictive biomarker of
poor prognosis in epithelial ovarian cancer patients. Arch Gynecol
Obstet. 295:459–465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Masoumi-Moghaddam S, Amini A, Wei AQ,
Robertson G and Morris DL: Sprouty 2 protein, but not Sprouty 4, is
an independent prognostic biomarker for human epithelial ovarian
cancer. Int J Cancer. 137:560–570. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee JM, Trepel JB, Choyke P, Cao L,
Sissung T, Houston N, Yu M, Figg WD, Turkbey IB, Steinberg SM, et
al: CECs and IL-8 have prognostic and predictive utility in
patients with recurrent platinum-sensitive ovarian cancer:
Biomarker correlates from the randomized phase-2 trial of olaparib
and cediranib compared with olaparib in recurrent
platinum-sensitive ovarian cancer. Front Oncol. 5:1232015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng Y, He F, Wu H, Huang H, Zhang L, Han
X and Liu J: GOLPH3L is a novel prognostic biomarker for epithelial
ovarian cancer. J Cancer. 6:893–900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roque DM, Buza N, Glasgow M, Bellone S,
Bortolomai I, Gasparrini S, Cocco E, Ratner E, Silasi DA, Azodi M,
et al: Class III β-tubulin overexpression within the tumor
microenvironment is a prognostic biomarker for poor overall
survival in ovarian cancer patients treated with neoadjuvant
carboplatin/paclitaxel. Clin Exp Metastasis. 31:101–110. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Penzvalto Z, Lanczky A, Lenart J,
Meggyesházi N, Krenács T, Szoboszlai N, Denkert C, Pete I and
Győrffy B: MEK1 is associated with carboplatin resistance and is a
prognostic biomarker in epithelial ovarian cancer. BMC Cancer.
14:8372014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abdel-Fatah TM, Russell R, Albarakati N,
Maloney DJ, Dorjsuren D, Rueda OM, Moseley P, Mohan V, Sun H,
Abbotts R, et al: Genomic and protein expression analysis reveals
flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian
cancer. Mol Oncol. 8:1326–1338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei W, Mok SC, Oliva E, Kim SH, Mohapatra
G and Birrer MJ: FGF18 as a prognostic and therapeutic biomarker in
ovarian cancer. J Clin Invest. 123:4435–4448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu T, Gao H, Chen X, Lou G, Gu L, Yang M,
Xia B and Yin H: TNFAIP8 as a predictor of metastasis and a novel
prognostic biomarker in patients with epithelial ovarian cancer. Br
J Cancer. 109:1685–1692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoshihara K, Tajima A, Yahata T, Kodama S,
Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H, et
al: Gene expression profile for predicting survival in
advanced-stage serous ovarian cancer across two independent
datasets. PLoS One. 5:e96152010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu YH, Chang TH, Huang YF, Huang HD and
Chou CY: COL11A1 promotes tumor progression and predicts poor
clinical outcome in ovarian cancer. Oncogene. 33:3432–3440. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karlan BY, Dering J, Walsh C, Orsulic S,
Lester J, Anderson LA, Ginther CL, Fejzo M and Slamon D:
POSTN/TGFBI-associated stromal signature predicts poor prognosis in
serous epithelial ovarian cancer. Gynecol Oncol. 132:334–342. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu WM, Mei R, Di X, Ryder TB, Hubbell E,
Dee S, Webster TA, Harrington CA, Ho MH, Baid J and Smeekens SP:
Analysis of high density expression microarrays with signed-rank
call algorithms. Bioinformatics. 18:1593–1599. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
R Development Core Team, . R: a language
and environment for statistical computing. the R Foundation for
Statistical Computing; Vienna: 2016
|
|
20
|
de Souto MC, Jaskowiak PA and Costa IG:
Impact of missing data imputation methods on gene expression
clustering and classification. BMC Bioinformatics. 16:642015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Affymetrix® Microarray Suite.
User's Guide. Version 5.0. Affymetrix, Inc.; Santa Clara: 2001
|
|
22
|
Kang DD, Sibille E, Kaminski N and Tseng
GC: MetaQC: Objective quality control and inclusion/exclusion
criteria for genomic meta-analysis. Nucleic Acids Res. 40:e152012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qi C, Hong L, Cheng Z and Yin Q:
Identification of metastasis-associated genes in colorectal cancer
using metaDE and survival analysis. Oncol Lett. 11:568–574. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon PI, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Goh KI, Oh E, Jeong H, Kahng B and Kim D:
Classification of scale-free networks. Proc Natl Acad Sci USA.
99:pp. 12583–12588. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qureshi MN, Min B, Jo HJ and Lee B:
Multiclass classification for the differential diagnosis on the
ADHD subtypes using recursive feature elimination and hierarchical
extreme learning machine: Structural MRI study. PLoS One.
11:e01606972016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
28
|
Sporn MB and Liby KT: NRF2 and cancer: The
good, the bad and the importance of context. Nat Rev Cancer.
12:564–571. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liao H, Zhou Q, Zhang Z, Wang Q, Sun Y, Yi
X and Feng Y: NRF2 is overexpressed in ovarian epithelial carcinoma
and is regulated by gonadotrophin and sex-steroid hormones. Oncol
Rep. 27:1918–1924. 2012.PubMed/NCBI
|
|
30
|
Martinez VD, Vucic EA, Thu KL, Pikor LA,
Hubaux R and Lam WL: Unique pattern of component gene disruption in
the NRF2 inhibitor KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex in
serous ovarian cancer. Biomed Res Int. 2014:1594592014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ginath S, Menczer J, Friedmann Y, Aingorn
H, Aviv A, Tajima K, Dantes A, Glezerman M, Vlodavsky I and
Amsterdam A: Expression of heparanase, Mdm2, and erbB2 in ovarian
cancer. Int J Oncol. 18:1133–1144. 2001.PubMed/NCBI
|
|
32
|
Mi RR and Ni H: MDM2 sensitizes a human
ovarian cancer cell line. Gynecol Oncol. 90:238–244. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mir R, Tortosa A, Martinez-soler F, Vidal
A, Condom E, Pérez-Perarnau A, Ruiz-Larroya T, Gil J and
Giménez-Bonafé P: Mdm2 antagonists induce apoptosis and synergize
with cisplatin overcoming chemoresistance in TP53 wild-type ovarian
cancer cells. Int J Cancer. 132:1525–1536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang F, Guo X, Yang G, Rosen DG and Liu J:
AURKA and BRCA2 expression highly correlate with prognosis of
endometriofid ovarian carcinoma. Mod Pathol. 24:836–845. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Do TV, Xiao F, Bickel LE, Klein-Szanto AJ,
Pathak HB, Hua X, Howe C, O'Brien SW, Maglaty M, Ecsedy JA, et al:
Aurora kinase A mediates epithelial ovarian cancer cell migration
and adhesion. Oncogene. 33:539–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gourley C, Paige AJW, Taylor KJ, Scott D,
Francis NJ, Rush R, Aldaz CM, Smyth JF and Gabra H: WWOX mRNA
expression profile in epithelial ovarian cancer supports the role
of WWOX variant 1 as a tumour suppressor, although the role of
variant 4 remains unclear. Int J Oncol. 26:1681–1689.
2005.PubMed/NCBI
|
|
37
|
Gourley C, Paige AJ, Taylor KJ, Ward C,
Kuske B, Zhang J, Sun M, Janczar S, Harrison DJ, Muir M, et al:
WWOX gene expression abolishes ovarian cancer tumorigenicity in
vivo and decreases attachment to fibronectin via integrin alpha3.
Cancer Res. 69:4835–4842. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yan H, Tong J, Lin X, Han Q and Huang H:
Effect of the WWOX gene on the regulation of the cell cycle and
apoptosis in human ovarian cancer stem cells. Mol Med Rep.
12:1783–1788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feng S, Pan W, Jin Y and Zheng J: MiR-25
promotes ovarian cancer proliferation and motility by targeting
LATS2. Tumour Biol. 35:12339–12344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xia Y and Gao Y: MicroRNA-181b promotes
ovarian cancer cell growth and invasion by targeting LATS2. Biochem
Biophys Res Commun. 447:446–451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang J, Yin XJ, Xu CJ, Ning YX, Chen M,
Zhang H, Chen SF and Yao LQ: The histone deacetylase SIRT6 inhibits
ovarian cancer cell proliferation via down-regulation of Notch 3
expression. Eur Rev Med Pharmacol Sci. 19:818–824. 2015.PubMed/NCBI
|
|
42
|
Zhang G, Liu Z, Qin S and Li K: Decreased
expression of SIRT6 promotes tumor cell growth correlates closely
with poor prognosis of ovarian cancer. Eur J Gynaecol Oncol.
36:629–632. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lindquist D, Kvarnbrink S, Henriksson R
and Hedman H: LRIG and cancer prognosis. Acta Oncol. 53:1135–1142.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang H, Yao J, Yin J and Wei X: Decreased
LRIG1 in human ovarian cancer cell SKOV3 upregulates MRP-1 and
contributes to the chemoresistance of VP16. Cancer Biother
Radiopharm. 31:125–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lim SK and Gopalan G: Aurora-A kinase
interacting protein 1 (AURKAIP1) promotes Aurora-A degradation
through an alternative ubiquitin-independent pathway. Biochem J.
403:119–127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu L, Liu X, Cui K, Di Y, Xin L, Sun X,
Zhang W, Yang X, Wei M, Yao Z and Yang J: SND1 acts downstream of
TGFβ1 and upstream of Smurf1 to promote breast cancer metastasis.
Cancer Res. 75:1275–1286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang N, Du X, Zang L, Song N, Yang T, Dong
R, Wu T, He X and Lu J: Prognostic impact of metadherin–SND1
interaction in colon cancer. Mol Biol Rep. 39:10497–10504. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cappellari M, Bielli P, Paronetto MP,
Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O and Sette C:
The transcriptional co-activator SND1 is a novel regulator of
alternative splicing in prostate cancer cells. Oncogene.
33:3794–3802. 2014. View Article : Google Scholar : PubMed/NCBI
|