Introduction
Cardiovascular disease (CVD) is a leading cause of
mortality worldwide, and the rates will continue to increase in the
coming decades (1). One typical
CVD, myocardial infarction (MI; also known as heart attack), causes
heart failure or cardiac arrest (2), and leads to millions of mortalities
every year in developing countries. Epidemiological studies have
shown that high blood pressure, smoking and obesity are leading
factors in MI development (3,4).
However, the molecular mechanisms of MI, especially its recurrence,
remain unclear. Therefore, elucidation of the molecular mechanisms
underlying MI is crucial for reducing the risk of recurrence.
Advances in biotechnology have allowed for the
successful identification of the genes associated with biomarkers
and clinical outcomes regarding high-risk MI. For example,
mutations in the myocardial infarction-associated transcript have
been reported to cause susceptibility to MI (5). In addition, it was demonstrated that
mutations in the oxidized low-density lipoprotein receptor 1 gene
may significantly increase the risk of MI (6). Although some MI-related genes have
been detected, many were identified independently and functional
associations among the genes have rarely been explored. Therefore,
it is necessary to investigate MI from a systematic perspective, as
the complex disease was reported to occur due to the dysregulation
of functional gene sets (7). A
previous study reported that the examination of protein complexes
may provide a better understanding not only of cellular functions,
but also human diseases (8). For
instance, the BRAFT protein complex was reported to be involved in
Fanconi anemia and Bloom Syndrome (9). The mammalian target of rapamycin
complex 1 serves a crucial role in hematopoiesis, hematopoietic
differentiation and leukemogenesis (10). In addition, one previous study
revealed that the proteins in complexes may be responsible for
diseases (11). Therefore,
identification of the dysfunctional protein complexes may aid our
understanding of the molecular mechanisms of MI. However, the
protein complexes associated with MI have not been fully
investigated.
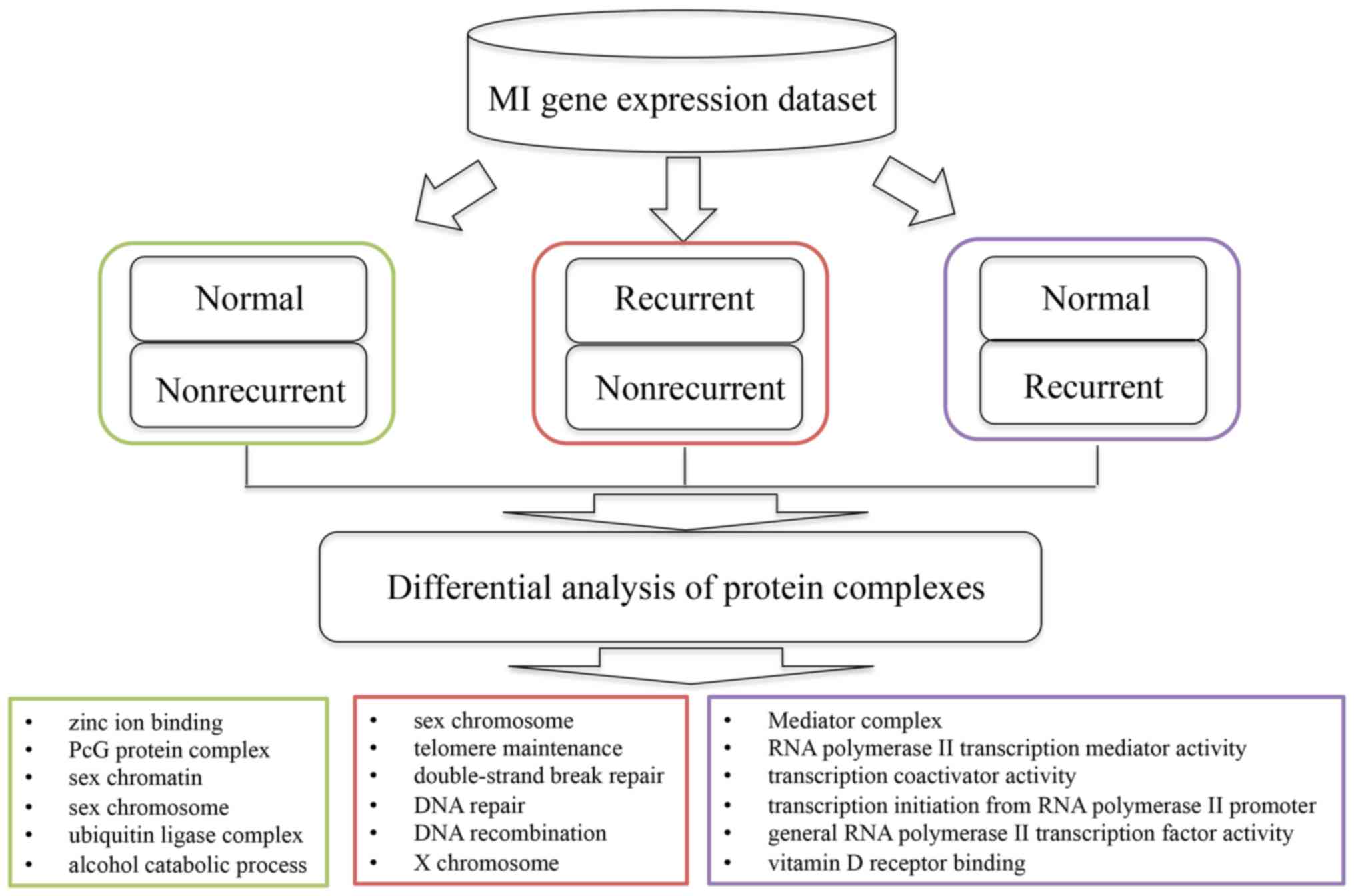
The present study proposed a bioinformatics approach
to identify protein complexes associated with MI development and
recurrence (Fig. 1). Based on the
gene expression profiles associated with MI, dysfunctional
complexes that may be involved in MI were identified, followed by
functional enrichment analysis on the protein complexes detected.
Combined with previous data, the present study revealed that some
proteins from the complexes were related to MI, which suggested an
important role for these protein complexes in the molecular
mechanism of MI.
Materials and methods
Data set
MI gene expression data set (GSE48060) was obtained
from the Gene Expression Omnibus depository (12). The data set contains 52 samples,
comprising 21 normal, 26 nonrecurrent and 5 recurrent samples. The
normal samples had no previous history of cardiac diseases or other
comorbidities, the nonrecurrent samples are the first-time patients
with MI, whereas recurrence referred to those patients with any
recurrent events within 18-months following the initial treatment.
All expression values were pre-processed with Robust Multi-array
Average (13). The expression
value of a gene associated with multiple probes was calculated as
the average expression value of all related probes. Gene expression
profiles were normalized with mean 0 and standard deviation 1.
The protein complexes and protein-protein
interactions were retrieved from the Human Protein Reference
Database (HPRD) (14). Functional
enrichment analysis of genes in each protein complex was performed
by DAVID (15), which is an online
tool for understanding biological functions behind a list of
genes.
Identification of differentially
expressed genes (DEGs)
Genes that are differentially expressed between two
conditions may be related to the condition and therefore may help
explain how the differences occurred. In the present study, the
data set was divided into three groups: Normal, Nonrecurrent and
Recurrent. The differentially expressed genes between the three
groups were detected by Student's t-test with a P-value cutoff of
0.01. As a result, 793 (normal vs. recurrent), 871 (normal vs.
nonrecurrent) and 423 (recurrent vs. nonrecurrent) DEGs and their
corresponding t-scores were obtained.
Identification of MI-related protein
complexes
Protein complexes are groups of proteins that
interact with each other, which are fundamental functional units of
the macromolecular systems. 1,521 protein complexes were obtained
from the HPRD database with detailed protein annotations. By
following the work of Liu et al (16), a score Sc was
defined for each complex to measure its relevance to the
development of MI.
Sc=∑i=1NTiN
where N denotes the number of genes in the
complex c, and Ti represents the t-score
of gene i calculated by Student's t-test using the gene
expression data between two different groups.
To verify that the MI-related complexes were not
detected by chance, for each complex, a gene set was randomly
picked with the same number of genes as that in the complex, and
for each gene set, a score was calculated with the aforementioned
equation. This procedure was repeated 10,000 times and the P-value
was defined as the frequency of the gene set score larger than
Sc for the corresponding complex. Subsequently,
the complex was identified as related to MI if P<0.01. In
particular, only complexes that have at least 10 proteins were
considered in the present study.
Results and Discussion
Protein complexes comprising multiple proteins are
essential cellular functional units. Instead of focusing on a
single gene, the present study aimed to identify the protein
complexes that may serve important roles in MI. Therefore, protein
complexes comprising genes that were differentially expressed among
the normal, recurrent and nonrecurrent groups were regarded to
serve important roles in MI. On this basis, the protein complexes
that were significantly different among the three MI groups were
identified and the functions of those complexes were also
investigated (Fig. 1).
Identification of protein complexes
associated with MI
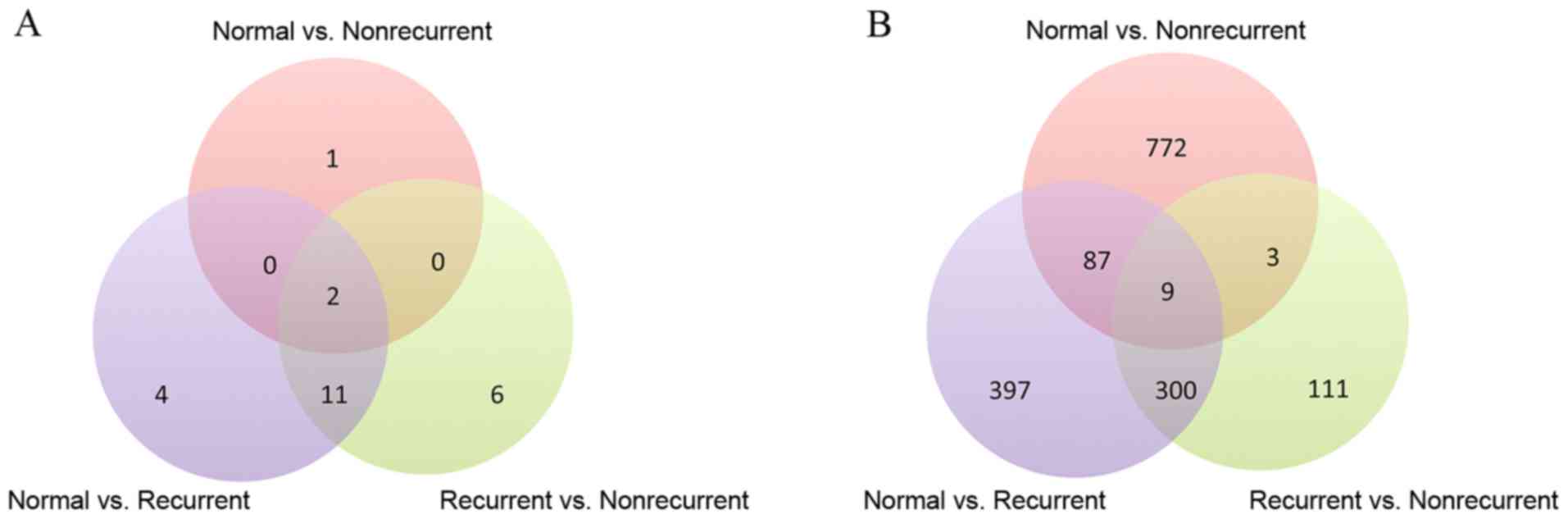
Gene expression data and protein complex annotations
were used to detect 17, 19 and 3 complexes as significantly
different between normal vs. recurrent, recurrent vs. nonrecurrent
and normal vs. nonrecurrent, respectively. Table I provides information about the
protein complexes that are different among the distinct groups;
protein complexes used in the present study were named and obtained
from the HPRD database. A Venn diagram of the three sets of protein
complexes detected for each of the three groups was created
(Fig. 2A), which revealed that
numerous complexes are shared among the three sets. In addition,
DEGs were identified by Student's t-test with a cutoff of
P<0.01. As a result, 793, 871 and 423 genes were detected to be
differentially expressed in the comparison of normal vs. recurrent,
normal vs. nonrecurrent and recurrent vs. nonrecurrent,
respectively (Fig. 2B). It was
noted that 31, 9 and 21 DEGs from the three above comparisons,
respectively, belonged to protein complexes. With the functional
annotations of protein complexes to which the DEGs belong, it was
possible to investigate the molecular mechanisms of MI from another
perspective.
 | Table I.Protein complexes that are
significantly different among the three myocardial infarction
groups. |
Table I.
Protein complexes that are
significantly different among the three myocardial infarction
groups.
|
Normal_Recurrent |
Recurrent_Nonrecurrent |
Normal_Nonrecurrent |
|---|
| COM_1553 | COM_1553 | COM_1553 |
| COM_2750 | COM_2750 | COM_2750 |
| COM_2322 | COM_1422 | COM_1648 |
| COM_1422 | COM_1427 |
|
| COM_1427 | COM_1426 |
|
| COM_970 | COM_970 |
|
| COM_2286 | COM_1505 |
|
| COM_2287 | COM_2286 |
|
| COM_2302 | COM_2287 |
|
| COM_2296 | COM_2322 |
|
| COM_2298 | COM_3000 |
|
| COM_1661 | COM_3014 |
|
| COM_1688 | COM_2296 |
|
| COM_1685 | COM_2998 |
|
| COM_33 | COM_242 |
|
| COM_2796 | COM_1661 |
|
| COM_2967 | COM_33 |
|
|
| COM_2796 |
|
|
| COM_2967 |
|
The results demonstrated that some protein complexes
are differentially expressed across different stages of MI; for
example, COM_1553 and COM_2750. Other protein complexes, such as
COM_1426 and COM_1505, are specifically differentially expressed
between recurrent and nonrecurrent groups. Only three protein
complexes were detected to be significantly different between
normal and nonrecurrent groups, two of which were also detected in
the comparisons of other groups. The small number of protein
complexes detected between normal and nonrecurrent groups may be
explained as no significant differences between these two groups
from the perspectives of molecules. This suggested that MI patients
in the absence of recurrence may restore to full health at greater
rates compared with MI patients with recurrence. The functions of
those proteins involved in the three complexes and their enriched
biological processes and pathways were also examined (Table II). The biological function
annotations were identified from the Gene Ontology database and the
pathway information is from the Kyoto Encyclopedia of Genes and
Genomes database; functional enrichment analysis was performed with
DAVID for each complex to determine the biological processes
associated with these protein complexes. For protein complex
COM_1648, which was differentially expressed only between normal
and nonrecurrent groups, several over-represented biological
processes have been detected to be associated with MI. For example,
it was reported that oxidized lipids and proteins, as well as
decreased antioxidant levels catalyzed by iron and copper, may be
detected in human atherosclerotic lesions (17). One previous study proposed that
zinc may displace iron and copper from oxidation-vulnerable sites
to limit the atherosclerotic damage (18). Another study suggested that the
polycomb-group complex serves a central role in the regulation of
heart development and functions (19). With an increased understanding of
ubiquitin ligases in cardiac disease, a number of studies have
emphasized the role of ubiquitin ligases in heart disease. For
instance, it was reported that specific ubiquitin ligases may serve
a role in the processes of cardiac hypertrophy and atrophy
(20), and the potential
therapeutic target roles of ubiquitin ligases in MI have also been
demonstrated (21). In addition,
it has been reported that microRNA (miRNA) miR-99a and let-7c
promoted cardiomyogenesis by upregulating their target genes,
whereas SWI/SNF related, matrix associated, actin dependent
regulator of chromatin, subfamily a, member identified in COM_1648
in the present study is a target gene of let-7c (22). These findings suggested that the
complexes detected by the present study may be associated with
MI.
 | Table II.GO function and KEGG pathway
enrichment analysis of genes from protein complexes that differ
between Normal and Nonrecurrent groups. |
Table II.
GO function and KEGG pathway
enrichment analysis of genes from protein complexes that differ
between Normal and Nonrecurrent groups.
| Category | Term | P-value |
|---|
| GOTERM_MF_FAT | GO:0008270~zinc ion
binding |
3.45×10−03 |
| GOTERM_CC_FAT | GO:0031519~PcG
protein complex |
4.69×10−03 |
| GOTERM_CC_FAT | GO:0001739~sex
chromatin |
6.56×10−03 |
| GOTERM_CC_FAT | GO:0000803~sex
chromosome |
7.02×10−03 |
| GOTERM_CC_FAT |
GO:0000151~ubiquitin ligase complex |
4.15×10−02 |
| GOTERM_CC_FAT |
GO:0005829~cytosol |
1.10×10−12 |
| GOTERM_BP_FAT |
GO:0006096~glycolysis |
2.14×10−07 |
| GOTERM_BP_FAT | GO:0046164~alcohol
catabolic process |
3.33×10−06 |
| GOTERM_BP_FAT | GO:0051789~response
to protein stimulus |
3.58×10−03 |
| KEGG_PATHWAY | hsa04530:Tight
junction |
8.35×10−03 |
Identification of protein complexes
associated with MI recurrence
By investigating the protein complexes that were
detected between recurrent and nonrecurrent groups, the enriched
biological processes and pathways were identified (Table III), some of which were strongly
implicated in MI, such as systemic lupus erythematosus. In
addition, it has been reported that patients with systemic lupus
erythematosus have an increased risk of CVD (23,24).
It has been reported that the incidence rates of coronary heart
disease exhibit differences according to sex, whereas the potential
role of the sex chromosomes has been unexplored to date. Recent
studies have shown that the sex chromosomes serve critical roles in
the difference in myocardial injury between the sexes (25,26).
In the present study, the over-represented cellular components of
the protein complexes were significantly enriched in sex chromatin,
sex chromosomes and X chromosomes, which suggested an association
between these complexes and MI. As shown in Table III, the protein complexes are
also enriched in DNA repair and telomere dysfunction. It was
demonstrated that the DNA repair genes, such as nei-like DNA
glycosylase 3, were also associated with increased risk of MI
(27,28). Furthermore, telomere dysfunction
has also emerged as an important factor in the molecular mechanism
of heart failure and the telomere-associated proteins were reported
to be involved in cardiovascular pathobiology (29,30).
These findings confirm that the proteins we detected here are
indeed related to MI.
 | Table III.GO function and KEGG pathway
enrichment analysis of protein complexes that differ between
Recurrent and Nonrecurrent groups. |
Table III.
GO function and KEGG pathway
enrichment analysis of protein complexes that differ between
Recurrent and Nonrecurrent groups.
| Category | Term | P-value |
|---|
| KEGG_PATHWAY | hsa05322:Systemic
lupus erythematosus |
1.58×10−05 |
| GOTERM_CC_FAT | GO:0001739~sex
chromatin |
7.31×10−05 |
| GOTERM_CC_FAT | GO:0000803~sex
chromosome |
8.43×10−05 |
| GOTERM_BP_FAT | GO:0000723~telomere
maintenance |
3.18×10−04 |
| GOTERM_BP_FAT | GO:0032200~telomere
organization |
3.41×10−04 |
| GOTERM_BP_FAT |
GO:0006302~double-strand break repair |
1.56×10−03 |
| GOTERM_BP_FAT | GO:0006281~DNA
repair |
2.24×10−03 |
| GOTERM_BP_FAT | GO:0006310~DNA
recombination |
4.40×10−03 |
| GOTERM_CC_FAT | GO:0000805~X
chromosome |
4.69×10−03 |
| GOTERM_BP_FAT | GO:0033554~cellular
response to stress |
1.52×10−02 |
| GOTERM_CC_FAT |
GO:0005853~eukaryotic translation
elongation factor 1 complex |
3.91×10−03 |
| GOTERM_BP_FAT | GO:0008380~RNA
splicing |
1.43×10−02 |
| GOTERM_CC_FAT |
GO:0005829~cytosol |
1.47×10−02 |
| GOTERM_MF_FAT | GO:0003723~RNA
binding |
1.51×10−02 |
Protein complexes were also detected between normal
and recurrent groups, and their enriched functions are listed in
Table IV, in which only the
processes related to heart disease, particularly MI, are listed for
clarification. These data indicated that the protein complexes were
mainly enriched in RNA polymerase II-associated activities. A
recent study have revealed that many of the CVD-associated mutant
genes were involved in transcription regulation, whereas RNA
polymerase II serves a key role in catalyzing the transcription of
DNA to mRNA, small nuclear RNA and miRNA (31). It has also been determined that
vitamin D receptor was able to reduce oxidative stress and inhibit
apoptosis in the MI injury, which suggested that the receptor may
be an attractive target for the treatment of heart disease
(32). These results may help us
further understand the roles of protein complexes in heart disease
and gain more insights into MI.
 | Table IV.GO function and KEGG pathway
enrichment analysis of protein complexes that differ between Normal
and Recurrent groups. |
Table IV.
GO function and KEGG pathway
enrichment analysis of protein complexes that differ between Normal
and Recurrent groups.
| Category | Term | P-value |
|---|
| GOTERM_CC_FAT | GO:0016592~Mediator
complex |
7.25×10−29 |
| GOTERM_MF_FAT | GO:0016455~RNA
polymerase II transcription mediator activity |
2.47×10−22 |
| GOTERM_MF_FAT |
GO:0003713~transcription coactivator
activity |
3.78×10−21 |
| GOTERM_BP_FAT |
GO:0006367~transcription initiation from
RNA polymerase II promoter |
5.07×10−21 |
| GOTERM_MF_FAT | GO:0016251~general
RNA polymerase II transcription factor activity |
6.14×10−20 |
| GOTERM_MF_FAT | GO:0042809~vitamin
D receptor binding |
3.39×10−17 |
| GOTERM_MF_FAT | GO:0046966~thyroid
hormone receptor binding |
4.64×10−16 |
| GOTERM_MF_FAT | GO:0003702~RNA
polymerase II transcription factor activity |
1.24×10−15 |
| GOTERM_BP_FAT | GO:0030521~androgen
receptor signaling pathway |
1.72×10−15 |
| GOTERM_BP_FAT |
GO:0006366~transcription from RNA
polymerase II promoter |
1.85×10−15 |
| GOTERM_MF_FAT | GO:0051427~hormone
receptor binding |
8.56×10−13 |
| GOTERM_BP_FAT | GO:0006461~
transcription regulator activity |
6.95×10−11 |
The overlapped protein complexes identified in both
normal vs. recurrent and nonrecurrent vs. recurrent comparison were
investigated further, as these complexes were considered to be
important in the recurrence of MI. Examination of the enriched
functions of these protein complexes revealed that some of them
were related to MI or heart disease (Table V). For example, aberrant RNA
splicing has been reported in heart diseases (33,34);
DNA damage and repair in atherosclerosis, which may lead to MI,
have also been reported, which suggested a novel mechanism of MI
(33). The release of DNA
resulting from damaged binding has also been reported to be
associated with MI (35), and
DNA-binding dyes, such as Hoechst, have been used to bind exposed
DNA and target injured myocardium (34,36).
Furthermore, previous studies have reported that cardiomyocyte cell
cycle activation may restore and enhance functions in injured
hearts after MI (37,38).
 | Table V.GO Functional enrichment analysis of
overlapped protein complexes between normal vs. recurrent and
nonrecurrent vs. recurrent. |
Table V.
GO Functional enrichment analysis of
overlapped protein complexes between normal vs. recurrent and
nonrecurrent vs. recurrent.
| Category | Term | P-value |
|---|
| GOTERM_BP_FAT | GO:0008380~RNA
splicing |
5.90×10−29 |
| GOTERM_CC_FAT |
GO:0030529~ribonucleoprotein complex |
3.66×10−22 |
| GOTERM_MF_FAT | GO:0003723~RNA
binding |
2.80×10−08 |
| GOTERM_CC_FAT | GO:0005686~snRNP
U2 |
8.48×10−06 |
| GOTERM_CC_FAT |
GO:0005829~cytosol |
1.40×10−09 |
| GOTERM_BP_FAT |
GO:0032268~regulation of cellular protein
metabolic process |
3.75×10−02 |
| GOTERM_CC_FAT |
GO:0005730~nucleolus |
1.71×10−03 |
| GOTERM_BP_FAT | GO:0006281~DNA
repair |
4.85×10−02 |
| GOTERM_BP_FAT | GO:0042254~ribosome
biogenesis |
2.26×10−11 |
| GOTERM_CC_FAT | GO:0015934~large
ribosomal subunit |
4.79×10−11 |
| GOTERM_CC_FAT |
GO:0022625~cytosolic large ribosomal
subunit |
1.01×10−10 |
| GOTERM_BP_FAT |
GO:0042273~ribosomal large subunit
biogenesis |
1.35×10−04 |
| GOTERM_CC_FAT |
GO:0044452~nucleolar part |
1.35×10−03 |
| GOTERM_MF_FAT | GO:0003684~damaged
DNA binding |
2.38×10−03 |
| GOTERM_CC_FAT | GO:0015935~small
ribosomal subunit |
7.20×10−03 |
| GOTERM_BP_FAT | GO:0006974~response
to DNA damage stimulus |
4.48×10−02 |
| GOTERM_BP_FAT | GO:0007049~cell
cycle |
6.93×10−24 |
| GOTERM_MF_FAT | GO:0016887~ATPase
activity |
5.67×10−06 |
Results from these functional enrichment analyses
indicated that the protein complexes detected by the present study
may serve important roles in promoting the development and
recurrence of MI. Although a number of enriched pathways were
identified, only a partial list of the known pathways related to MI
are presented in this study for clarification, where those novel
pathways may provide new insights into the molecular mechanism
underlying MI.
Identification of MI-associated
genes
In addition to the functional enrichment analyses of
the protein complexes aforementioned, the genes encoding proteins
from each complex were investigated to determine if they are
related to MI. Previous studies have revealed that several genes
were identified in the complexes that have been previously reported
to be relevant to cardiac disease. For example, it was reported
that the genes in the mediator of RNA polymerase II transcription
(MED) complex, such as MED1, MED12, MED13, MED14, MED23 and MED30,
serve important roles in CVD initiation and progression (39). In addition, mutations in MED13 have
been reported to be linked to CVDs and mutations in MED30 were
suggested to be important in CVD progression (31).
The present study identified 114 MI-associated genes
ranked by their relevant score from the GeneCards database
(40). Examination of these genes
in the protein complexes identified by the present study revealed
that numerous genes have been previously reported related to MI.
For example, mutations in proteasome subunit α6, a proteasome that
regulates the inflammation processes, have been demonstrated to be
a risk factor of MI (41,42). In addition, the
chaperonin-containing TCP1 subunit 7 (CCT7) gene may severely
impair soluble guanylyl cyclase activity (43). A recent study determined the
relationship between impaired soluble guanylyl cyclase-dependent
nitric oxide signaling and MI risk, and suggested that CCT7 may be
a new therapeutic target for MI (44).
The verification of the above genes in the protein
complexes related to MI as described above implied that the protein
complexes detected may be related to MI disease. In addition, the
complexes that were identified as significantly different among the
three groups, but in which no MI-related genes were found, were
also examined to determine whether the proteins from these
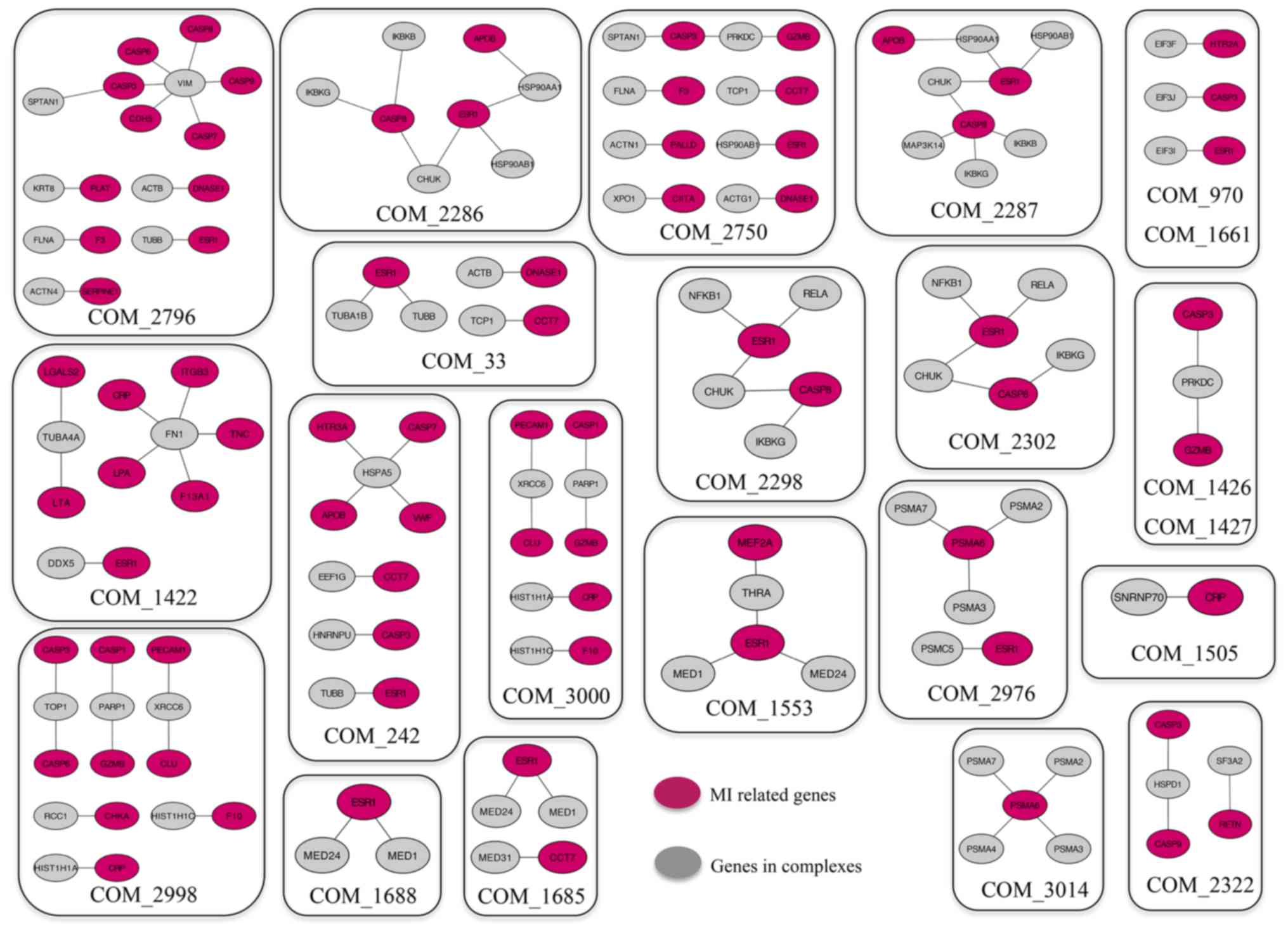
complexes interacted with known MI-related genes. Among the 24
complexes identified, proteins from 22 complexes exhibited
interactions with MI-related genes (Fig. 3), which indicated that these
proteins may also be related to MI and the corresponding complexes
may be important in MI development. For example, caspases have been
implicated in the molecular mechanism of MI (45,46);
heat shock protein family D, member, conserved helix-loop-helix
ubiquitous kinase and eukaryotic translation initiation factor 3
subunit J have been reported to interact with caspases, such as
caspase (CASP)3, CASP8 and CASP9, which suggested that these genes
may also be related to MI (43).
In summary, several proteins from the complexes identified by the
present study were validated to be related to MI, implying that the
corresponding complexes may be related to the development of
MI.
In conclusion, MI is the leading cause of mortality
worldwide. Elucidation of the molecular mechanisms underlying MI
may aid in better prevention of development of the disease and in
designing more effective treatments. Protein complexes composed of
multiple proteins are functional units of complex biological
systems, the dysfunction of which often leads to diseases. In the
present study, a bioinformatics approach was used to identify the
protein complexes associated with MI. Functional enrichment
analysis demonstrated that the protein complexes detected may serve
important roles in MI. Investigations of the proteins in the
detected complexes implied that some of the proteins have already
been reported as related to MI, whereas other proteins interacted
with known MI genes, which indicated that the protein complexes
detected are indeed important in MI. The protein complexes detected
here may improve our understanding of the molecular mechanisms
underlying MI, and may be used as biomarkers in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by Medical and
Health Technology Development Plan of Shandong Province
(2017ws137).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The public dataset GSE48060 can be obtain from Gene
Expression Omnibus (GEO) database.
Authors' contributions
NJ, FY, YQ, CL and HL analyzed and interpreted the
patient data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Go AS, Mozaffarian D, Roger VL, Benjamin
EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al:
Heart disease and stroke statistics-2013 update: A report from the
american heart association. Circulation. 127:e6–e245. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ge Y and Wang TJ: Identifying novel
biomarkers for cardiovascular disease risk prediction. J Intern
Med. 272:430–439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gerszten RE and Wang TJ: The search for
new cardiovascular biomarkers. Nature. 451:949–952. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pedrotty DM, Morley MP and Cappola TP:
Transcriptomic biomarkers of cardiovascular disease. Prog
Cardiovasc Dis. 55:64–69. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ishii N, Ozaki K, Sato H, Mizuno H, Saito
S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al:
Identification of a novel non-coding RNA, MIAT, that confers risk
of myocardial infarction. J Hum Genet. 51:1087–1099. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tatsuguchi M, Furutani M, Hinagata J,
Tanaka T, Furutani Y, Imamura S, Kawana M, Masaki T, Kasanuki H,
Sawamura T and Matsuoka R: Oxidized LDL receptor gene (OLR1) is
associated with the risk of myocardial infarction. Biochem Biophys
Res Commun. 303:247–250. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lage K, Karlberg EO, Storling ZM, Olason
PI, Pedersen AG, Rigina O, Hinsby AM, Tümer Z, Pociot F, Tommerup
N, et al: A human phenome-interactome network of protein complexes
implicated in genetic disorders. Nat Biotechnol. 25:309–316. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spirin V and Mirny LA: Protein complexes
and functional modules in molecular networks. Proc Natl Acad Sci
USA. 100:pp. 12123–12128. 2003; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meetei AR, Sechi S, Wallisch M, Yang D,
Young MK, Joenje H, Hoatlin ME and Wang W: A multiprotein nuclear
complex connects fanconi anemia and bloom syndrome. Mol Cell Biol.
23:3417–3426. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kalaitzidis D, Sykes SM, Wang Z, Punt N,
Tang Y, Ragu C, Sinha AU, Lane SW, Souza AL, Clish CB, et al: mTOR
complex 1 plays critical roles in hematopoiesis and
pten-loss-evoked leukemogenesis. Cell Stem Cell. 11:429–439. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fraser HB and Plotkin JB: Using protein
complexes to predict phenotypic effects of gene mutation. Genome
Biol. 8:R2522007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peri S, Navarro JD, Kristiansen TZ,
Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN,
Deshpande N, Suresh S, et al: Human protein reference database as a
discovery resource for proteomics. Nucleic Acids Res. 32:D497–D501.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu KQ, Liu ZP, Hao JK, Chen L and Zhao
XM: Identifying dysregulated pathways in cancers from pathway
interaction networks. BMC Bioinformatics. 13:1262012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Heinecke JW: Oxidants and antioxidants in
the pathogenesis of atherosclerosis: Implications for the oxidized
low density lipoprotein hypothesis. Atherosclerosis. 141:1–15.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stadler N, Stanley N, Heeneman S, Vacata
V, Daemen MJ, Bannon PG, Waltenberger J and Davies MJ: Accumulation
of zinc in human atherosclerotic lesions correlates with calcium
levels but does not protect against protein oxidation. Arterioscler
Thromb Vasc Biol. 28:1024–1030. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang QT: Epigenetic regulation of cardiac
development and function by polycomb group and trithorax group
proteins. Dev Dyn. 241:1021–1033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Willis MS, Bevilacqua A, Pulinilkunnil T,
Kienesberger P, Tannu M and Patterson C: The role of ubiquitin
ligases in cardiac disease. J Mol Cell Cardiol. 71:43–53. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Portbury AL, Ronnebaum SM, Zungu M,
Patterson C and Willis MS: Back to your heart: Ubiquitin proteasome
system-regulated signal transduction. J Mol Cell Cardiol.
52:526–537. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coppola A, Romito A, Borel C, Gehrig C,
Gagnebin M, Falconnet E, Izzo A, Altucci L, Banfi S, Antonarakis
SE, et al: Cardiomyogenesis is controlled by the miR-99a/let-7c
cluster and epigenetic modifications. Stem Cell Res. 12:323–337.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manzi S, Meilahn EN, Rairie JE, Conte CG,
Medsger TA Jr, Jansen-McWilliams L, D'Agostino RB and Kuller LH:
Age-specific incidence rates of myocardial infarction and angina in
women with systemic lupus erythematosus: Comparison with the
framingham study. Am J Epidemiol. 145:408–415. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hak AE, Karlson EW, Feskanich D, Stampfer
MJ and Costenbader KH: Systemic lupus erythematosus and the risk of
cardiovascular disease: Results from the nurses' health study.
Arthritis Rheum. 61:1396–1402. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang W, Huang W, Su S, Li B, Zhao W, Chen
S and Gu D: Association study of ACE2 (angiotensin I-converting
enzyme 2) gene polymorphisms with coronary heart disease and
myocardial infarction in a chinese han population. Clin Sci (Lond).
111:333–340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Chen X, McClusky R, Ruiz-Sundstrom
M, Itoh Y, Umar S, Arnold AP and Eghbali M: The number of X
chromosomes influences protection from cardiac
ischaemia/reperfusion injury in mice: One X is better than two.
Cardiovasc Res. 102:375–384. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Verschuren JJ, Trompet S, Deelen J, Stott
DJ, Sattar N, Buckley BM, Ford I, Heijmans BT, Guchelaar HJ,
Houwing-Duistermaat JJ, et al: Non-homologous end-joining pathway
associated with occurrence of myocardial infarction: Gene set
analysis of genome-wide association study data. PLoS One.
8:e562622013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Skarpengland T, Laugsand LE, Janszky I,
Luna L, Halvorsen B, Platou CG, Wang W, Vatten LJ, Damås JK,
Aukrust P, et al: Genetic variants in the DNA repair gene NEIL3 and
the risk of myocardial infarction in a nested case-control study.
The HUNT study. DNA Repair (Amst). 28:21–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wong JM and Collins K: Telomere
maintenance and disease. Lancet. 362:983–988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boccardi V, Esposito A, Rizzo MR, Marfella
R, Barbieri M and Paolisso G: Mediterranean diet, telomere
maintenance and health status among elderly. PLoS One.
8:e627812013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grueter CE: Mediator complex dependent
regulation of cardiac development and disease. Genomics Proteomics
Bioinformatics. 11:151–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yao T, Ying X, Zhao Y, Yuan A, He Q, Tong
H, Ding S, Liu J, Peng X, Gao E, Pu J and He B: Vitamin D receptor
activation protects against myocardial reperfusion injury through
inhibition of apoptosis and modulation of autophagy. Antioxid Redox
Signal. 22:633–650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mahmoudi M, Mercer J and Bennett M: DNA
damage and repair in atherosclerosis. Cardiovasc Res. 71:259–268.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baraldi PG, Bovero A, Fruttarolo F, Preti
D, Tabrizi MA, Pavani MG and Romagnoli R: DNA minor groove binders
as potential antitumor and antimicrobial agents. Med Res Rev.
24:475–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nguyen NT, Lindsey ML and Jin YF: Systems
analysis of gene ontology and biological pathways involved in
post-myocardial infarction responses. BMC Genomics. 16 Suppl
7:S182015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang S, Chen HH, Yuan H, Dai G, Schuhle
DT, Mekkaoui C, Ngoy S, Liao R, Caravan P, Josephson L and Sosnovik
DE: Molecular MRI of acute necrosis with a novel DNA-binding
gadolinium chelate: Kinetics of cell death and clearance in
infarcted myocardium. Circ Cardiovasc Imaging. 4:729–737. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Hu S, Ma G, Yao Y, Yan G, Chen J, Li
Y and Zhang Z: Acute myocardial infarction induced functional
cardiomyocytes to re-enter the cell cycle. Am J Transl Res.
5:327–335. 2013.PubMed/NCBI
|
|
38
|
Hassink RJ, Pasumarthi KB, Nakajima H,
Rubart M, Soonpaa MH, de la Rivière AB, Doevendans PA and Field LJ:
Cardiomyocyte cell cycle activation improves cardiac function after
myocardial infarction. Cardiovasc Res. 78:18–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schiano C, Casamassimi A, Vietri MT,
Rienzo M and Napoli C: The roles of mediator complex in
cardiovascular diseases. Biochim Biophys Acta. 1839:444–451. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rebhan M, Chalifa-Caspi V, Prilusky J and
Lancet D: GeneCards: A novel functional genomics compendium with
automated data mining and query reformulation support.
Bioinformatics. 14:656–664. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ozaki K, Sato H, Iida A, Mizuno H,
Nakamura T, Miyamoto Y, Takahashi A, Tsunoda T, Ikegawa S, Kamatani
N, et al: A functional SNP in PSMA6 confers risk of myocardial
infarction in the japanese population. Nat Genet. 38:921–925. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sjakste T, Poudziunas I, Ninio E, Perret
C, Pirags V, Nicaud V, Lazdins M, Evanss A, Morrison C, Cambien F
and Sjakste N: SNPs of PSMA6 gene-investigation of possible
association with myocardial infarction and type 2 diabetes
mellitus. Genetika. 43:553–559. 2007.PubMed/NCBI
|
|
43
|
Hanafy KA, Martin E and Murad F: CCTeta, a
Novel soluble guanylyl cyclase-interacting protein. J Biol Chem.
279:46946–46953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Erdmann J, Stark K, Esslinger UB, Rumpf
PM, Koesling D, de Wit C, Kaiser FJ, Braunholz D, Medack A, Fischer
M, et al: Dysfunctional nitric oxide signalling increases risk of
myocardial infarction. Nature. 504:432–436. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zidar N, Jera J, Maja J and Dusan S:
Caspases in myocardial infarction. Adv Clin Chem. 44:1–33. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mocanu MM, Baxter GF and Yellon DM:
Caspase inhibition and limitation of myocardial infarct size:
Protection against lethal reperfusion injury. Br J Pharmacol.
130:197–200. 2000. View Article : Google Scholar : PubMed/NCBI
|