Introduction
MicroRNAs (miRNA/miR) represent a cluster of small
non-coding RNA molecules involved in controlling gene expression,
translation and cellular biological behaviors. In recent years,
growing evidence suggests that miRNAs serve a crucial role in tumor
development by functioning as oncogenes or tumor suppressors,
depending on their target genes and downstream signaling pathways
(1). Of these, miR-124, first
identified to be highly expressed in the brain, has been disclosed
to be aberrantly downregulated or silenced in multiple tumors
including liver, breast, cervical, nasopharyngeal and lung cancer
(2–6), which arises at least partly from DNA
methylation (7–9). Furthermore, miR-124 has also been
demonstrated to suppress cell growth, invasion,
epithelial-mesenchymal transition, and even metastasis through
targeting a series of oncogenes, including rho-associated protein
kinase, EZH2, signal transducer and activator of transcription 3
(STAT3), Slug and forkhead box protein Q1 (FOXQ1) (3,5,10,11).
Given that miR-124 regulates various malignant phenotypes in tumor
development by repressing different target genes, it is helpful to
understand the role of miR-124 regulatory networks in tumors in
order to identify novel miR-124-targeting genes.
Colorectal cancer (CRC) represents one of the
leading causes of cancer-associated mortality worldwide (12). The expression of miR-124 is
negatively correlated with grade of CRC differentiation, but
positively associated with poor prognosis, therefore represents an
independent prognostic factor for CRC (13,14).
Previous studies have demonstrated the tumor-suppressive properties
of miR-124 in the regulation of tumor growth and metastasis
(15–17). In addition, miR-124 has been
demonstrated to sensitize the response of tumor cells to
radiotherapy in CRC, through inhibiting paired mesoderm homeobox
protein 1 (18). Notably, an
integrating analysis demonstrated that miR-124 mediates crosstalk
within the Toll-like receptor signaling pathway in Crohn's disease,
ulcerative colitis and CRC (19),
which indicates that the involvement of miR-124 in CRC progression
begins at a very early stage of pathology.
IQ motif containing GTPase activating protein 1
(IQGAP1) is one of the largest known scaffold proteins,
participating in protein-protein interactions and integrating
diverse signaling pathways. Evidence implicates IQGAP1 as an
essential regulator of the mitogen activated protein kinase 1
(MAPK) (20–22) and Wnt/β-catenin signaling pathways
(23–25) that serve crucial roles in the
progression of multiple tumors. IQGAP1 is demonstrated to be
overexpressed in CRC tissues compared with the normal counterparts
(26) and tends to be expressed
more at the invasive front than at the upper portions within the
carcinoma tissues, which is associated with higher rates of distant
metastasis (26,27). Notably, in silico
predictions indicate that miR-124 has a conserved binding site
within the 3′untranslated region (UTR) of IQGAP1 mRNA. Therefore,
it was hypothesized that overexpression of IQGAP1 in CRC is, at
least partly, due to silencing of miR-124.
Materials and methods
Tissue samples and cell lines
A panel of 30 pairs of primary CRCs and their
matched adjacent normal mucosa, were obtained from patients (16
males and 14 females; aged 31–84 years) who underwent surgical
resections, between June 2016 to December 2016 at the First
People's Hospital of Yunnan province (Kunming, China). Tissues were
snap-frozen in liquid nitrogen immediately after resection and then
stored at −80°C for further use. The clinical features of all 30
CRC patients, including clinical stage, tumor location and
treatment prior to surgical resection are presented in Table I. All patients whose tissue samples
were collected for the study signed the informed consent. This
project was approved by the Ethics Committee of the First People's
Hospital of Yunnan Province.
 | Table I.Clinicopathological features of the
30 colorectal cancer patients analyzed in the study. |
Table I.
Clinicopathological features of the
30 colorectal cancer patients analyzed in the study.
|
Characteristics |
|
|---|
| Age |
|
| Mean
(SD) | 6.1 (14.1) |
|
Range | 31–84 |
| Sex |
|
|
Male | 16 |
|
Female | 14 |
| Stage |
|
| I | 3 |
| II | 7 |
|
III | 16 |
| IV | 4 |
| Location |
|
|
Rectum | 14 |
|
Colon | 16 |
| Histology |
|
|
Adenomatous carcinoma | 15 |
|
Mucinous carcinoma | 5 |
|
Differentiation |
|
|
Well | 6 |
|
Moderate | 17 |
|
Poor | 7 |
| Chemotherapy prior
to resection |
|
|
mFOLFOX6 | 2 |
|
Capecitabine | 2 |
| No
treatment | 26 |
Human CRC cell lines, including HT29, SW480, SW620,
DLD1, LoVo and HCT116, and 293T cells, were purchased from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China). CRC
cells were maintained routinely in RPMI-1640 medium (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin solution. The normal colonic
epithelial cell line FHC was kindly donated by Dr. Liang Peng from
Guangzhou Medical University (Guangzhou, China). FHC cells were
maintained in Dulbecco's modified Eagle's medium (DMEM): F12 medium
(Gibco; Thermo Fisher Scientific, Inc.) with supplements following
ATCC protocol. 293T cells were maintained in DMEM medium containing
high glucose supplemented with 10% FBS. All the cells mentioned
were cultured in a 37°C humidified atmosphere containing 5%
CO2.
Bioinformatics analyses
The analysis of the microarray dataset (accession
no. GSE20916) (28) was performed
using bioinformatics tool R2 (http://r2.amc.nl)
following the manufacturer's protocol. The 2 subgroups of normal
colon tissues differed in collection method. One consisted of 24
samples is collected by surgery, the other of 10 samples is
collected by colonoscopy. The colon tumor subgroup consisting of 30
samples were also from colonoscopy. Targetscan version 7.1
(http://targetscan.org), Pictar (http://pictar.mdc-berlin.de) and miRecords (http://c1.accurascience.com/miRecords/)
were used for prediction of miR-124 target genes.
Oligonucleotide and cell
transfection
Hsa-miR-124 mimics and its scrambled controls were
synthesized by Shanghai Genepharma Co., Ltd. (Shanghai, China). The
sequences of the hsa-miR-124 mimics were as follows: Sense
5′-UAAGGCACGCGGUGAAUGCC-3′ and antisense
5′-CAUUCACCGCGUGCCUUAUU-3′. The sequences of the scramble controls
were as follows: Sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense
5′-ACGUGACACGUUCGGAGAATT-3′. For transfection, cells were seeded
into 6-well clusters at a density of 3×106 cells/well
and transfected using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol, with 30 nM miR-124 mimics or scramble controls for 48 h
at 37°C before further experimentation in assays or RNA/protein
extraction.
Lentivirus packaging and stable cell
line establishment
IQGAP1-knockdown and negative control lentivirus
particles were packaged by co-transfecting 2.5 µg IQGAP1-short
hairpin (sh)RNA plasmids or shRNA scramble control plasmids
(GeneCopoeia, Inc., Rockville, MD, USA) using psi-LVRU6GP vector as
backbone (GeneCopoeia, Inc.) with lentiviral packaging plasmids
into 293T cells, using Lenti-Pac™ HIV Expression
Packaging Systems (GeneCopoeia, Inc.) according to the
manufacturer's protocol. The target sequence for IQGAP1-shRNA #1
and #2 are as follows: 5′-GGTTATCACCCTCATTCGTTC-3′ and
5′-GGCTTATGAGTACCTTTGTCA-3′. For lentivirus infection, cells were
incubated with viral supernatant in the presence of 8 µg/ml
polybrene for 24 h, followed by Puromycin (Invitrogen; Thermo
Fisher Scientific, Inc.) selection until drug-resistant colonies
became visible.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA was isolated from CRC tissues and cells
using the TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Mature miR-124 expression in cells was
determined using a Hairpin-it™ miRNAs qPCR kit (Shanghai
Genepharma, Co., Ltd.). RNU6B was used as an endogenous control.
IQGAP1 mRNA expression was determined by using SYBR green qPCR
assay (Takara Bio, Inc., Otsu, Japan). The thermocycling conditions
were as follows: Denaturation at 95°C for 3 min, followed by 40
cycles of amplification at 95°C for 12 sec and extension at 62°C
for 40 sec. GAPDH was used as the endogenous control. The sequence
of primers for IQGAP1 was as follows: Forward:
5′-GGGACCAACCAAAGTGTGTCAAC-3′, reverse:
5′-CTGCTCATTATTGCCTGTCTTGGA-3′. Primer sequence for GAPDH: Forward:
5′-TGACTTCAACAGCGACACCCA-3′, reverse: 5′-CACCCTGTTGCTGTAGCCAAA-3′.
Data was analyzed using the 2−ΔΔCq method (29). This experiment was repeated 3
separate times.
Cell growth assay and cell colony
formation assay
Cell growth was detected by using a Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) according to the manufacturer's protocol. For colony
formation assay, cells were trypsinized and plated on 6-well plates
at a density of 3×102 cells/well and cultured at 37°C
for 10 days. The colonies were stained with 0.1% crystal violet
solution containing 80% methanol for 5 min at room temperature. The
number of colonies defined as >50 cells/colony were counted at
×40 magnification by using a light microscope. The assays were
performed in triplicate of wells in 3 separate experiments.
Western blot analysis
Cultured cells were lysed in
radioimmunoprecipitation assay buffer (Cell Signaling Technology,
Inc., Danvers, MA, USA) with 1% PMSF, for 15 min on ice
Subsequently, cell lysates were centrifuged at 14,000 × g for 30
min and the supernatant was harvested. Protein concentration was
determined using a bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.). A total of 30 or 50 µg
protein/lane was loaded onto a 10% SDS-PAGE minigel and transferred
onto a polyvinylidene difluoride membrane. Membranes were blocked
using Tris-buffered saline containing 20% Tween-20 and 5% non-fat
milk at room temperature for 2 h. Following probing with 1:1,000
diluted anti-IQGAP1 (cat. no. 29016; Cell Signaling Technology,
Inc.), anti-phosphorylated (p)-extracellular signal regulated
kinase (ERK)1/2 (cat. no. 4370; Cell Signaling Technology, Inc.),
anti-ERK1/2 (cat. no. 4695; Cell Signaling Technology, Inc.) and
anti-β-catenin (cat. no. 9562; Cell Signaling Technology, Inc.)
respectively at 4°C overnight, the blots were subsequently
incubated with anti-rabbit horseradish peroxidase-conjugated
anti-immunoglobulin (Ig)G (cat. no. 7074; Cell Signaling
Technology, Inc.) for 1 h at room temperature. Signals were
visualized using ECL Substrates (Pierce; Thermo Fisher Scientific,
Inc.). β-actin (1:3,000; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) was used as an endogenous
protein for normalization. This experiment was repeated 3 separate
times.
Luciferase reporter assay
A fragment of wild type 3′UTR of IQGAP1 containing
the putative miR-124 binding site was amplified by PCR. The PCR
product was sub-cloned into a psiCHECK-2 vector (Promega
Corporation, Madison, WI, USA) immediately downstream of the
luciferase gene sequence. A corresponding psiCHECK-2 construct
containing the 3′UTR of IQGAP1 with a mutant seed sequence of
miR-124 was also synthesized.
293 cells were plated in triplicate wells in 24-well
clusters at a density of 1×105 cells/well, then
co-transfected with 0.1 µg psiCHECK-2 construct and 30 nM miR-124
mimic or scramble control using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation
for 48 h at 37°C, luciferase activity was detected using a
dual-luciferase reporter assay system (Promega Corporation) and
normalized by Renilla activity. The experiment was repeated
3 separate times.
Statistical analysis
Statistical analyses were performed using the SPSS
software, version 15.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad
Prism software version 6.01 (Graphpad Software, Inc., La Jolla, CA,
USA). Comparisons of miR-124 and IQGAP1 expression between CRC
tissues and paired adjacent colonic epithelial tissues were
performed using a Wilcoxon's paired test. All data are presented as
the mean ± standard deviation. Comparisons among multiple groups
were performed using one-way analysis of variance followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
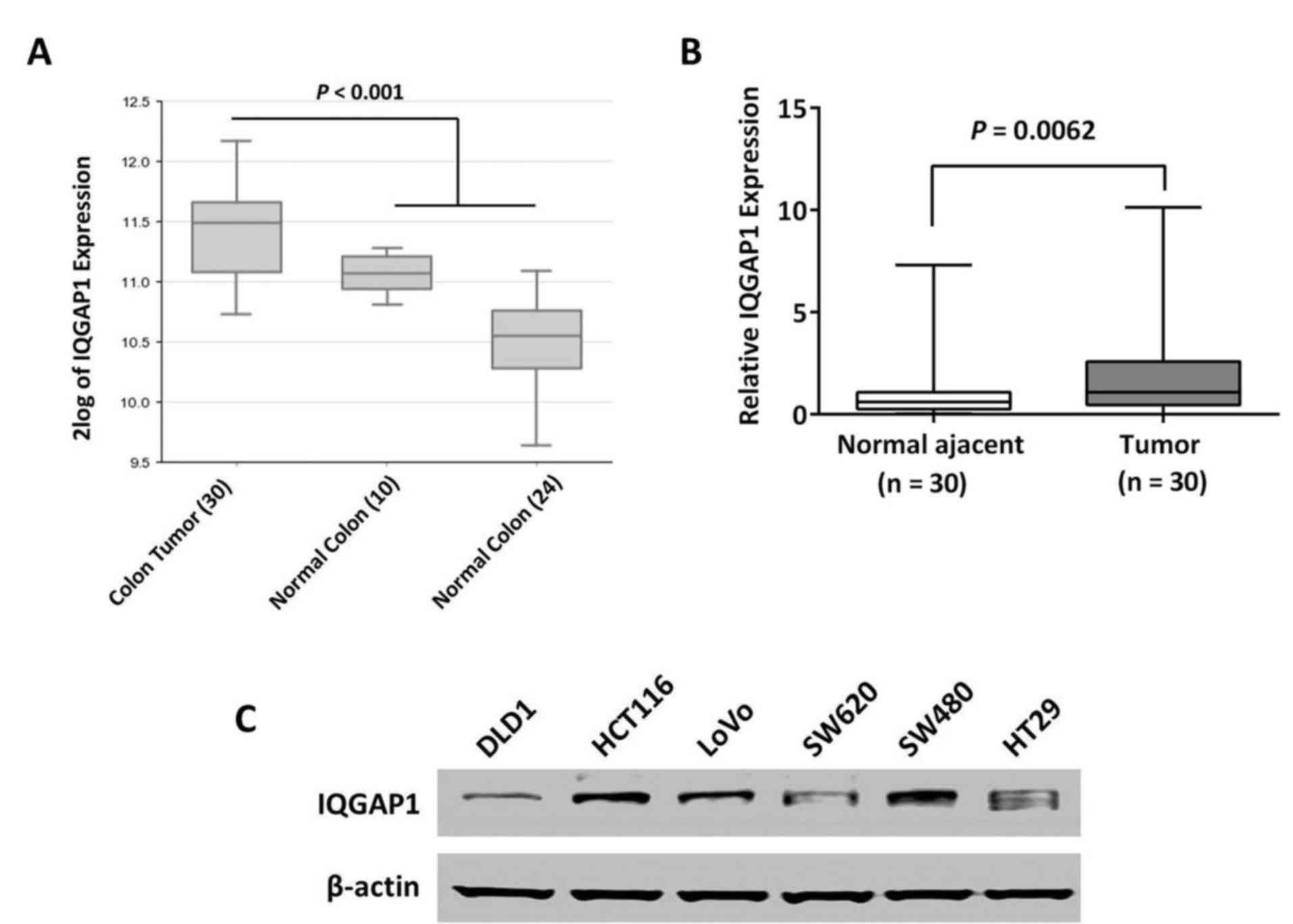
IQGAP1 expression is aberrantly
upregulated in CRC
IQGAP1 has been reported to be overexpressed in
multiple tumors, including CRC. Therefore gene expression profiles
of human CRC cohorts were first analyzed to observe the status of
IQGAP1 expression. As demonstrated in Fig. 1A, IQGAP1 was significantly
upregulated in the cohort of CRC tissues compared with the other 2
subgroups of normal colon tissues collected by surgery and
colonoscopy respectively (P<0.001). qPCR was performed to detect
IQGAP1 mRNA level in clinical specimens of CRC. In the cohort of 30
cases of paired CRC tissues and adjacent colon epithelia, it was
demonstrated that IQGAP1 was significantly upregulated in tumors
compared with the matched normal epithelia (P=0.0062; Fig. 1B). IQGAP1 protein levels in CRC
cell lines: HCT116, HT29, SW480, SW620, LoVo and DLD1, are
presented in Fig. 1C. Since HCT116
and SW480 cells expressed a high level of IQGAP1, they were
selected for establishing stable IQGAP1-knockdown cells.
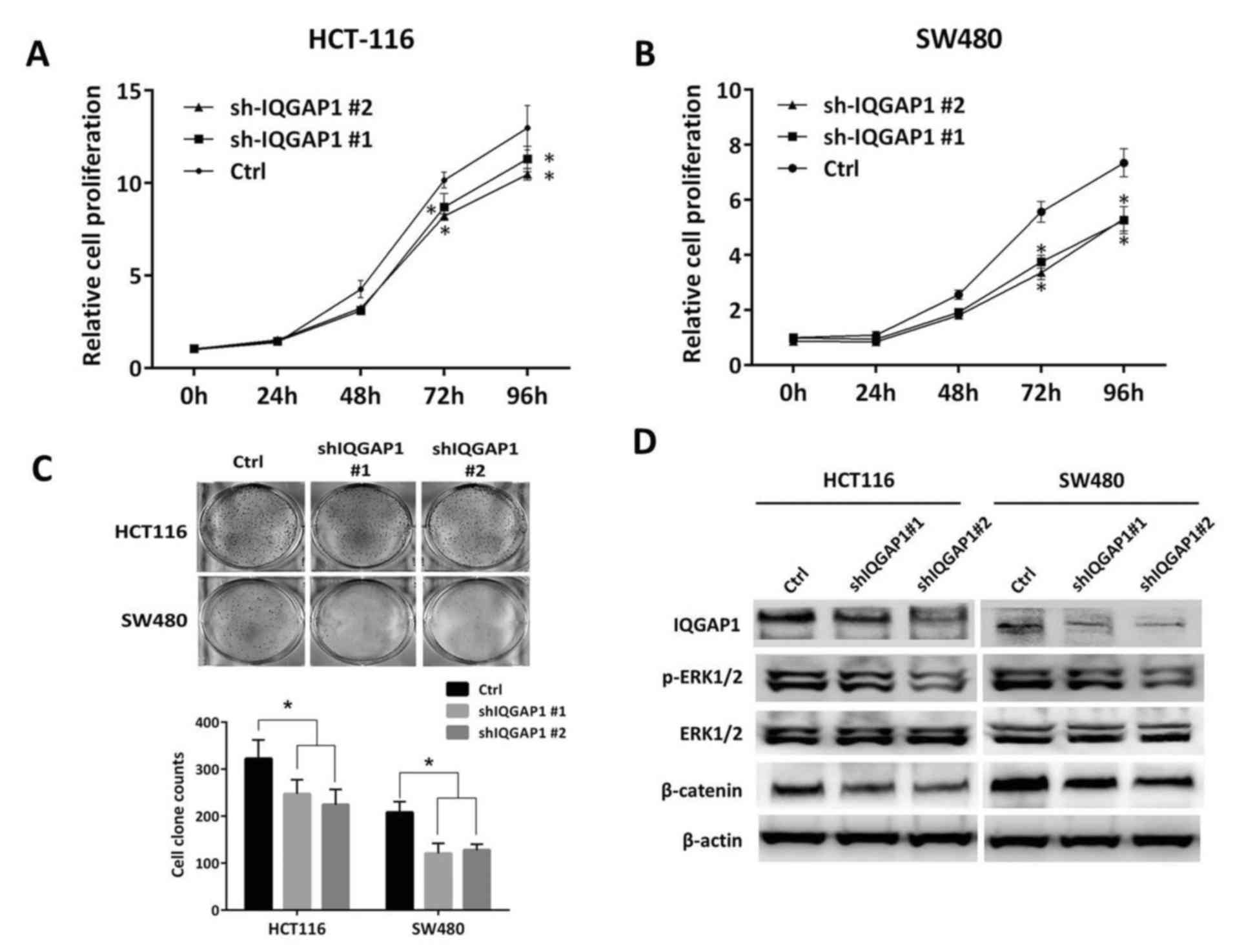
Knockdown of IQGAP1 inhibits CRC cell
growth and colony formation ability by suppressing phosphorylation
of ERK and antagonizing β-catenin activity
Next, the role of IQGAP1 in CRC growth and the
potential underlying mechanism was investigated. Stable IQGAP1
knockdown of CRC cells was generated by introducing IQGAP1
shRNA-packaged lentiviral particles into HCT116 and SW480 cells
which express relatively higher levels of IQGAP1 compared with the
other cell lines investigated in the present study. In the
loss-of-function assays, downregulation of IQGAP1 significantly
reduced cell growth (P<0.05) and colony formation ability
(Fig. 2A-C). Immunoblotting
demonstrated that in the IQGAP1-knockdown cells, phosphorylation of
ERK1/2 was impaired as well as β-catenin activity, (Fig. 2D), demonstrating the augmenting
effect of IQGAP1 on MAPK and β-catenin signaling.
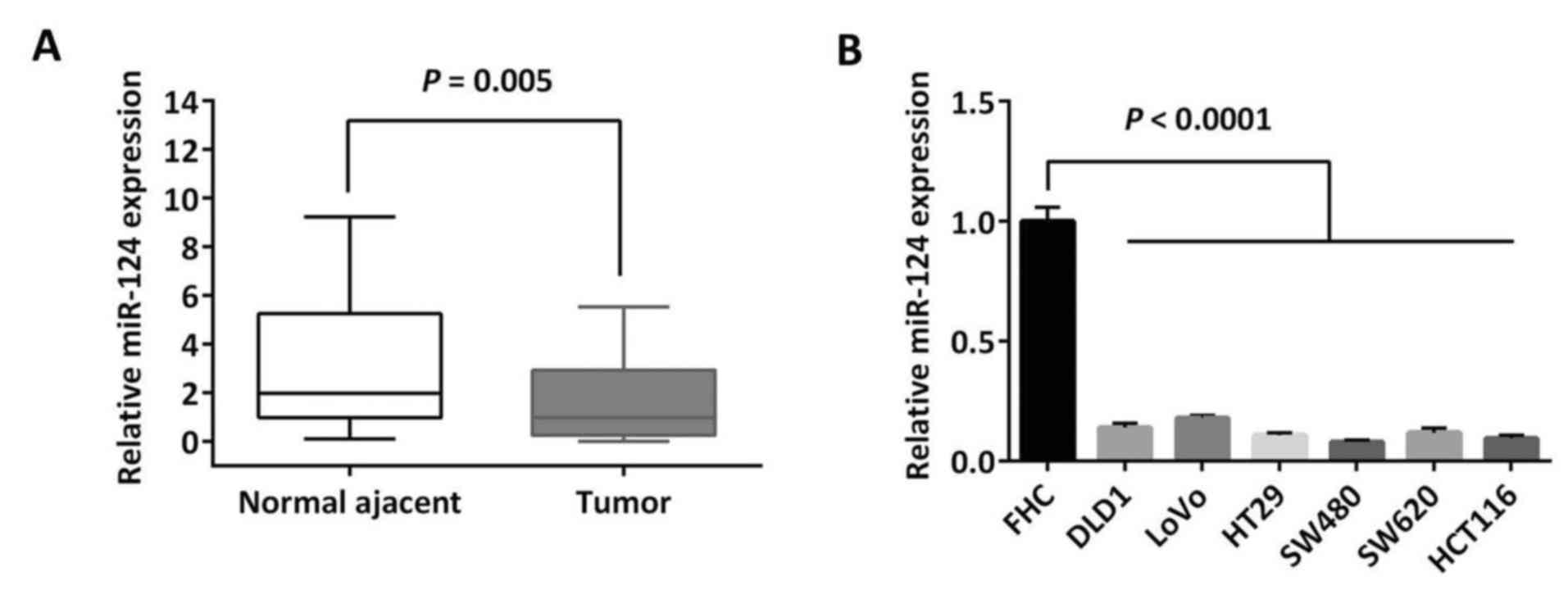
miR-124 is aberrantly downregulated in
CRC tissues
To demonstrate the expression pattern of miR-124 in
CRC, qPCR was performed to detect the miR-124 level in the clinical
specimens and cell lines. In the mentioned cohort of 30 cases of
paired CRC tissues and adjacent colon epithelia, it was
demonstrated that miR-124 was significantly downregulated in CRCs
compared with the adjacent normal tissues (P=0.005; Fig. 3A). The miR-124 expression level was
further examined in CRC cell lines and miR-124 expression in all 6
CRC cell lines mentioned was significantly decreased compared with
the normal colonic epithelial cell line FHC (P<0.0001; Fig. 3B). Notably, HCT116 and SW480 cells
that expressed relatively high IQGAP1 expression levels (Fig. 1C), were demonstrated to express
relatively low levels of miR-124 out of these CRC cell lines,
therefore were chosen for restoration of miR-124 in further assays
(Fig. 3B).
IQGAP1 is a direct target gene of
miR-124 in CRC
By utilizing bioinformatics algorithms
(Targetscan/Pictar/miRecords), it was identified that IQGAP1 was a
potential target gene of miR-124. The predicted binding site of
miR-124 within the 3′UTR of IQGAP1 mRNA is presented in Fig. 4A. It was hypothesized that miR-124
may target IQGAP1 in CRC. To investigate this further, an miR-124
mimic was introduced into HCT116 and SW480 cells. With the
restoration of miR-124 expression, IQGAP1 mRNA expression was
demonstrated to be significantly decreased by qPCR (P<0.05). In
addition, its protein level was demonstrated to be decreased by
immunoblotting (Fig. 4B and C),
while suppression of ERK1/2 phosphorylation and β-catenin was also
simultaneously observed (Fig. 4C).
To further investigate if the predicted binding site of miR-124 to
3′UTR of IQGAP1 mRNA is responsible for the downregulation of
IQGAP1, the 3′UTR of IQGAP1 was cloned into a luciferase reporter
vector wild-type (wt)-IQGAP1 and a corresponding mutant version
(mut-IQGAP1) was also constructed. wt-IQGAP1 and miR-124 mimic or
scrambled control were co-transfected into 293T and HCT116 cells to
perform a luciferase reporter assay. In 293T and HCT116 cells, the
luciferase activity of miR-124 transfected cells was significantly
reduced compared to the scrambled control cells (P<0.05;
Fig. 4D and E). Furthermore,
miR-124-mediated repression of luciferase activity was abolished by
the mutant putative binding site (Fig.
4D and E).
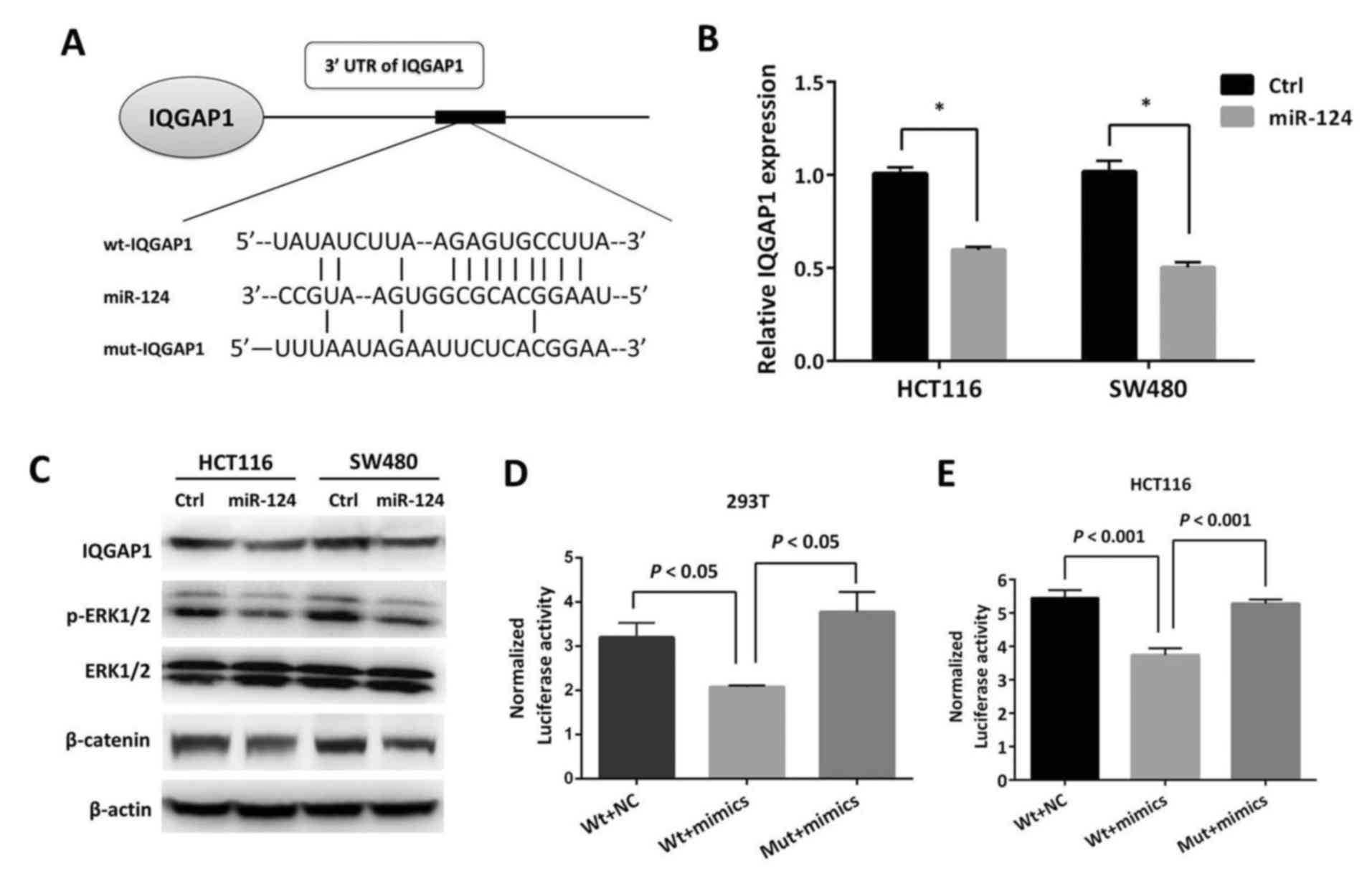 | Figure 4.IQGAP1 is a direct target gene of
miR-124 in CRC. (A) The predicted miR-124 binding site within
IQGAP1 3′UTR and its mutated version are as presented. (B)
Restoration of miR-124 in HCT116 and SW480 cells induced decreased
expression of IQGAP1 by quantitative polymerase chain reaction.
*P<0.05 vs. the Ctrl. (C) Restoration of miR-124 in HCT116 and
SW480 cells induced decreased phosphorylation of ERK1/2 and
β-catenin expression. The repression of luciferase activity by
wt-IQGAP1 3′UTR was dependent on miR-124, while mutated 3′UTR of
IQGAP1 abrogated miR-124-mediated repression of luciferase activity
in (D) 293T and (E) HCT116 cells. miR, microRNA; CRC, colorectal
cancer; IQGAP1, IQ motif containing GTPase activating protein 1;
Ctrl, control; UTR, untranslated region; p, phosphorylated; ERK,
extracellular signal regulated kinase; wt, wild-type; Mut, mutant;
NC, negative control. |
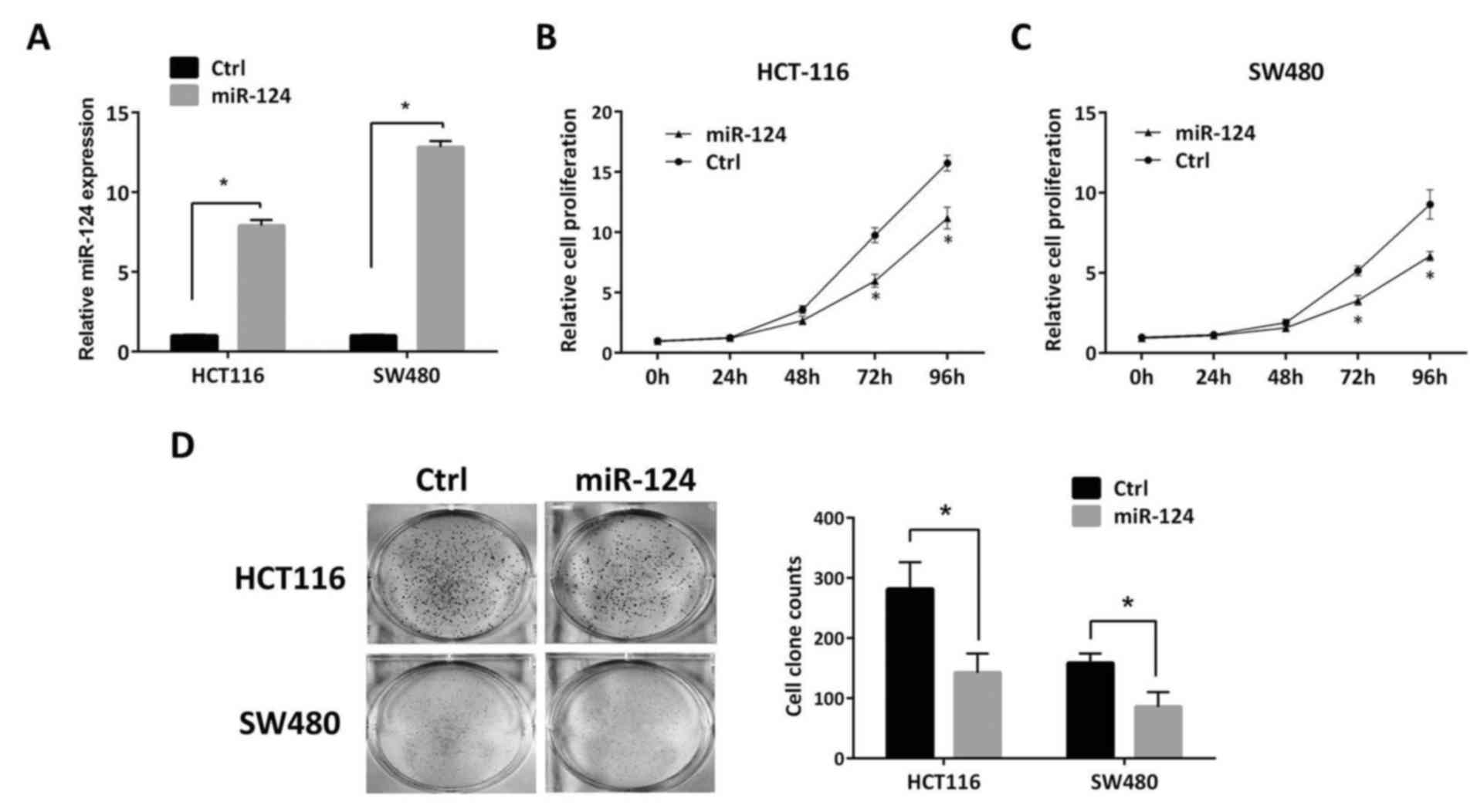
Restoration of miR-124 expression
suppresses cell growth and colony formation ability of CRC
To investigate the effects of miR-124 on cell growth
of CRC, the cell growth ability of miR-124-transfected HCT116 and
SW480 cells was measured. First of all, significant restoration of
miR-124 expression was performed by mimic transfection in the two
cell lines, HCT116 and SW480 (P<0.05; Fig. 5A). CCK-8 assays demonstrated that
restoration of miR-124 expression significantly repressed cell
growth ability in HCT116 and SW480 cells after 72 h (P<0.05;
Fig. 5B and C). Furthermore,
significantly decreased colony formation activity was also observed
in the two aforementioned cell lines (P<0.05; Fig. 5D). These results suggested that
miR-124 serves a suppressive role in tumor growth in CRC and its
absence may confer malignant potentials to CRC cells.
Discussion
IQGAP1, the best characterized member of the IQGAP
family, functions as a scaffold protein in the cytoplasm to curb,
compartmentalize and coordinate multiple signaling pathways in a
variety of cell types, and participates in cell-cell interaction,
cell adherence, and movement via actin/tubulin-based cytoskeletal
reorganization (30). In recent
years, studies demonstrated that IQGAP1 is frequently aberrantly
overexpressed in multiple tumors and integrates and mediates
several pro-oncogenic signaling pathways to promote tumorigenesis
and metastasis (31–34). IQGAP1 may bind directly to B-Raf,
dual specificity mitogen-activated protein kinase kinase 1 and ERK,
facilitating epidermal growth factor-induced activation of the MAPK
cascade. IQGAP1 binding to β-catenin enhances β-catenin nuclear
translocation, initiating transcription of cyclin D1 (35). Notably, a previous study by Jameson
et al (36) verified that
blockage of IQGAP1-ERK1/2 interactions by introducing a specific
IQGAP1 WW domain peptide, inhibits RAS- and RAF-driven
tumorigenesis and bypasses acquired resistance to the BRAF
inhibitor, which validates IQGAP1 inhibition as a promising
therapeutic strategy for tumors.
Dysregulation of miRNAs have been demonstrated to
account for malignant phenotypes in tumors. Growing evidence proves
that miR-124 is a miRNA, which exerts tumor suppressive effects but
is frequently absent in tumors (2–6).
Despite this miR-124 has been validated to target oncogenic genes
including STAT3, FOXQ1 and Slug (5,10,11)
and the present study aimed to identify the mechanistic link of
miR-124 to other potential target genes contributing to CRC
development. Since IQGAP1 is predicted as a potential target gene
of miR-124 by in silico prediction, it was hypothesized that
there is a regulative link between miR-124 and IQGAP1. In the
present study, IQGAP1 was demonstrated to be overexpressed in CRC
tissue. Knockdown of IQGAP1 in CRC cells could impede cell growth
and colony formation ability, which is considered to arise from
repression of phosphorylated (p)-ERK1/2 and β-catenin expression.
Similarly, restoration of miR-124 induced the inhibition of cell
growth and colony formation ability, and repressed p-ERK1/2 and
β-catenin. Given that miR-124 bound to the 3′UTR of IQGAP1 mRNA and
led to a decrease of IQGAP1 expression, it was demonstrated that
the absence of miR-124 was responsible, at least in part, for the
aberrant upregulation of IQGAP1 and subsequent enhanced activation
of MAPK and β-catenin signaling in CRC.
From the perspective of tumor treatment, it is
urgent to investigate promising therapeutic targets and invent
efficient approaches for tumor targeting. For example, the results
of the present study indicated that IQGAP1 is a potential target
for CRC treatment and further highlighted that miR-124 is in theory
a potential way to antagonize IQGAP1 similar to the WW domain
peptide of IQGAP1 mentioned. Recently, nanoparticles have emerged
as a popular concept for targeted delivery of therapeutic agents to
tumors (37–39). A type of combined polymeric
nanoparticle (PNP) entrapping phosphatidyl inositol 3 kinase
inhibitors or irinotecan suppresses or even eliminates lung
metastasis of CRC in a BALB/C mice model, but negligible
accumulation of PNPs is detected in the organs including the
spleen, liver and kidneys indicating good tissue selectivity of
PNPs and organ safety (40).
Notably, delivery of nanoparticles conjugated with miR-20a or
anti-miR-155 relieves aberrant expression of their protein targets
and eventually reduces tumor growth and liver metastasis in murine
colon cancer models (41,42). In addition, considering miR-124
suppresses tumor progression through targeting a list of
tumor-promoting proteins, miR-124-entrapped nanoparticles are
anticipated to be a novel therapeutic option for treating tumors
with absence of miR-124.
In conclusion, the present study provided evidence
that accounted for aberrant upregulation of IQGAP1 in CRC and newly
described a potential mechanism underlying dysregulated miR-124
indulging CRC progression. Therefore, it would be worthwhile to
estimate the implication of miR-124-targeted therapy for clinical
management of CRC.
Acknowledgements
The authors would like to thank Prof Liang Peng from
Guangzhou Medical University (Guangzhou, China) for his technical
assistance.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81502128)
and Joint Foundation of Kunming Medical University and Yunnan
Provincial Science and Technology Department (grant no.
2014FB085).
Availability of data and materials
The dataset GSE20916 analyzed during the present
study is publically available in GEO Datasets resource of NCBI
[https://www.ncbi.nlm.nih.gov/gds/?term=GSE20916].
Authors' contributions
QG and YZ designed the study. JF, WZ and YW
performed the experiments. PW analyzed the data and performed
statistical analysis. YZ wrote the paper. All authors read and
approved the manuscript.
Ethics approval and consent to
participate
All patients whose tissue samples were collected for
the study signed the informed consent. This project was approved by
the Ethics Committee of the First People's Hospital of Yunnan
Province.
Patient consent for publication
All patients whose tissue samples were collected for
the study signed an informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Esquela-Kerschera A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang YJ, Wang QY, Zhou CX, Yin QQ, He M,
Yu XT, Cao DX, Chen GQ, He JR and Zhao Q: MiR-124 targets Slug to
regulate epithelial-mesenchymal transition and metastasis of breast
cancer. Carcinogenesis. 34:713–722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wan HY, Li QQ, Zhang Y, Tian W, Li YN, Liu
M, Li X and Tang H: MiR-124 represses vasculogenic mimicry and cell
motility by targeting amotL1 in cervical cancer cells. Cancer Lett.
355:148–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Li H, Han J and Zhang Y:
Down-regulation of microRNA-124 is correlated with tumor metastasis
and poor prognosis in patients with lung cancer. Int J Clin Exp
Pathol. 8:1967–1972. 2015.PubMed/NCBI
|
|
7
|
Murray-Stewart T, Sierra JC, Piazuelo MB,
Mera RM, Chaturvedi R, Bravo LE, Correa P, Schneider BG, Wilson KT
and Casero RA: Epigenetic silencing of miR-124 prevents spermine
oxidase regulation: Implications for Helicobacter pylori-induced
gastric cancer. Oncogene. 35:5480–5488. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pronina IV, Loginov VI, Burdennyy AM,
Fridman MV, Senchenko VN, Kazubskaya TP, Kushlinskii NE, Dmitriev
AA and Braga EA: DNA methylation contributes to deregulation of 12
cancer-associated microRNAs and breast cancer progression. Gene.
604:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Zhang TT, Jin S, Liu H, Zhang X,
Ruan CG, Wu DP, Han Y and Wang XQ: Pyrosequencing quantified
methylation level of miR-124 predicts shorter survival for patients
with myelodysplastic syndrome. Clin Epigenetics. 9:912017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH,
Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al: The putative
tumour suppressor microRNA-124 modulates hepatocellular carcinoma
cell aggressiveness by repressing ROCK2 and EZH2. Gut. 61:278–289.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng Y, Li Y, Nian Y, Liu D, Dai F and
Zhang J: STAT3 is involved in miR-124-mediated suppressive effects
on esophageal cancer cells. BMC Cancer. 15:3062015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang MJ, Li Y, Wang R, Wang C, Yu YY, Yang
L, Zhang Y, Zhou B, Zhou ZG and Sun XF: Downregulation of
microRNA-124 is an independent prognostic factor in patients with
colorectal cancer. Int J Colorectal Dis. 28:183–189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jinushi T, Shibayama Y, Kinoshita I,
Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H
and Iseki K: Low expression levels of microRNA-124-5p correlated
with poor prognosis in colorectal cancer via targeting of SMC4.
Cancer Med. 3:1544–1552. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taniguchi K, Sugito N, Kumazaki M,
Shinohara H, Yamada N, Nakagawa Y, Ito Y, Otsuki Y, Uno B, Uchiyama
K and Akao Y: MicroRNA-124 inhibits cancer cell growth through
PTB1/PKM1/PKM2 feedback cascade incolorectal cancer. Cancer Lett.
363:17–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Z, Liu S, Tian L, Wu M, Ai F, Tang W,
Zhao L, Ding J, Zhang L and Tang A: miR-124 and miR-506 inhibit
colorectal cancer progression by targeting DNMT3B and DNMT1.
Oncotarget. 6:38139–38150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou L, Xu Z, Ren X, Chen K and Xin S:
MicroRNA-124 (MiR-124) inhibits cell proliferation, metastasis and
invasion in colorectal cancerby downregulating rho-associated
protein kinase 1(ROCK1). Cell Physiol Biochem. 38:1785–1795. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Zheng L, Huang J, Gao F, Lin X,
He L, Li D, Li Z, Ding Y and Chen L: MiR-124 Radiosensitizes human
colorectal cancer cells by targeting PRRX1. PLoS One. 9:e939172014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bai J, Li Y, Shao T, Zhao Z, Wang Y, Wu A,
Chen H, Li S, Jiang C, Xu J and Li X: Integrating analysis reveals
microRNA-mediated pathway crosstalk among Crohn's disease,
ulcerative colitis and colorectal cancer. Mol Biosyst.
10:2317–2328. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roy M, Li Z and Sacks DB: IQGAP1 is a
scaffold for mitogen-activated protein kinase signalling. Mol Cell
Biol. 25:7940–7952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roy M, Li Z and Sacks DB: IQGAP1 binds
ERK2 and modulates its activity. J Biol Chem. 279:17329–17337.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bourguignon LY, Gilad E, Rothman K and
Peyrollier K: Hyaluronan-CD44 interaction with IQGAP1 promotes
Cdc42 and ERK signaling, leading to actin binding, Elk-1/Estrogen
Receptor transcriptional activation, and ovarian cancer
progression. J Biol Chem. 280:11961–11972. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Briggs MW, Li Z and Sacks DB:
IQGAP1-mediated stimulation of transcriptional co-activation by
beta-catenin is modulated by calmodulin. J Biol Chem.
277:7453–7465. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Wang A, Wang F, Wang M, Zhu M, Ma
Y and Wu R: IQGAP1 activates Tcf signal independent of Rac1 and
Cdc42 in injury and repair of bronchial epithelial cells. Exp Mol
Pathol. 85:122–128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schmidt VA, Chiariello CS, Capilla E,
Miller F and Bahou WF: Development of hepatocellular carcinoma in
Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol.
28:1489–1502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nabeshima K, Shimao Y, Inoue T and Koono
M: Immunohistochemical analysis of IQGAP1 expression in human
colorectal carcinomas: Its overexpression in carcinomas and
association with invasion fronts. Cancer Lett. 176:101–109. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hayashi H, Nabeshima K, Aoki M, Hamasaki
M, Enatsu S, Yamauchi Y, Yamashita Y and Iwasaki H: Overexpression
of IQGAP1 in advanced colorectal cancer correlates with poor
prognosis-critical role in tumor invasion. Int J Cancer.
126:2563–2574. 2010.PubMed/NCBI
|
|
28
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5:e130912010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression using real time PCR and the 2 (-Delta
Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Abel AM, Schuldt KM, Rajasekaran K, Hwang
D, Riese MJ, Rao S, Thakar MS and Malarkannan S: IQGAP1: Insights
into the function of a molecular puppeteer. Mol Immunol.
65:336–349. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong P, Nabeshima K, Nishimura N, Kawakami
T, Hachisuga T, Kawarabayashi T and Iwasaki H: Overexpression and
diffuse expression pattern of IQGAP1 at invasion fronts are
independent prognostic parameters in ovarian carcinomas. Cancer
Lett. 243:120–127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen F, Zhu HH, Zhou LF, Wu SS, Wang J and
Chen Z: IQGAP1 is overexpressed in hepatocellular carcinoma and
promotes cell proliferation by Akt activation. Exp Mol Med.
42:477–483. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang XX, Li XZ, Zhai LQ, Liu ZR, Chen XJ
and Pei Y: Overexpression of IQGAP1 in human pancreatic cancer.
Hepatobiliary Pancreat Dis Int. 12:540–545. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Smith JM, Hedman AC and Sacks DB: IQGAPs
choreograph cellular signaling from the membrane to the nucleus.
Trends Cell Biol. 25:171–184. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
White CD, Brown MD and Sacks DB: IQGAPs in
Cancer: A family of scaffold proteins underlying tumorigenesis.
FEBS Lett. 583:1817–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jameson KL, Mazur PK, Zehnder AM, Zhang J,
Zarnegar B, Sage J and Khavari PA: IQGAP1 scaffold-kinase
interaction blockade selectively targets RAS-MAP kinase-driven
tumors. Nat Med. 19:626–630. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pi F, Binzel DW, Lee TJ, Li Z, Sun M,
Rychahou P, Li H, Haque F, Wang S, Croce CM, et al: Nanoparticle
orientation to control RNA loading and ligand display on
extracellular vesicles for cancer regression. Nat Nanotechnol.
13:82–89. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao CY, Cheng R, Yang Z and Tian ZM:
Nanotechnology for cancer therapy based on chemotherapy. Molecules.
23:E8262018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Awasthi R, Roseblade A, Hansbro PM,
Rathbone MJ, Dua K and Bebawy M: Nanoparticles in cancer treatment:
Opportunities and obstacles. Curr Drug Targets. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rychahou P, Bae Y, Reichel D, Zaytseva YY,
Lee EY, Napier D, Weiss HL, Roller N, Frohman H, Le AT and Mark
Evers B: Colorectal cancer lung metastasis treatment with
polymer-drug nanoparticles. J Control Release. 275:85–91. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Marquez J, Fernandez-Piñeiro I,
Araúzo-Bravo MJ, Poschmann G, Stühler K, Khatib AM, Sanchez A, Unda
F, Ibarretxe G, Bernales I and Badiola I: Targeting liver
sinusoidal endothelial cells with miR-20a-loaded nanoparticles
reduces murine coloncancer metastasis to the liver. Int J Cancer.
143:709–719. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Duo Y, Bi J, Zeng X, Mei L, Bao S,
He L, Shan A, Zhang Y and Yu X: Targeted delivery of anti-miR-155
by functionalized mesoporous silica nanoparticles for colorectal
cancer therapy. Int J Nanomedicine. 13:1241–1256. 2018. View Article : Google Scholar : PubMed/NCBI
|