Introduction
Cardiovascular disease (CVD) accounts for 16.7
million deaths every year, and is one of the leading causes of
deaths worldwide (1,2). Hyperglycemia is an important factor
in the induction of myocardial dysfunction and heart failure in
patients with diabetes (3).
Clinical evidence has demonstrated that elevated blood glucose
levels may result in the development of diabetic cardiomyopathy
(DC) (4). DC is a major
complication that increases the incidence and mortality of patients
with diabetes, and is a unique myocardial disease occurring
independently of hypertension and coronary atherosclerosis
(5). DC induces ischemic
myocardial injury and cardiac hypertrophy, which contributes to the
cardiac failure of patients with diabetes (6), and it is a complication of
hypertension and coronary artery disease (7). In addition, as an independent cause
of heart failure, it may be possible to reduce the incidence and
mortality of heart failure by preventing cardiac hypertrophy. There
are currently no effective treatments for DC; therefore,
investigation into the molecular mechanisms of DC and cardiac
hypertrophy may identify novel therapeutic strategies for heart
failure in patients with diabetes.
The majority of cardiomyocytes stop differentiating
and proliferating soon after birth. However, certain cardiomyocytes
re-enter into the cell cycle when stimulated by stress (8), thereby inducing the excessive
increase of nucleic acids and proteins. Cell sizes enlarge without
increasing cell numbers, eventually inducing cardiac hypertrophy
and heart failure (2,9). Apoptosis is a major mechanism of cell
death that consists of a series of tightly regulated cascades of
molecular processes (10).
Sustained hyperglycemia may induce the apoptosis of cardiomyocytes
in patients with diabetes (11,12).
It has also been demonstrated that cardiomyocyte apoptosis serves a
prominent role in the pathogenesis of DC (13).
Small ubiquitin-like modifiers (SUMOs) are highly
conserved ubiquitin-like proteins and 18% of their sequence is
homologous with ubiquitin. There are four distinct SUMO isoforms
(SUMO1, SUMO2, SUMO3 and SUMO4) in mammals (14). SUMOs primarily function in protein
post-translational modification, modify the stability and
interactions of proteins and regulate signal transduction (15,16).
SUMO1 primarily modifies physiological proteins, while SUMO2 and
SUMO3 have similar amino acid sequences and primarily modify stress
proteins associated with oxidative stress, heat shock and osmotic
pressure (17). The function of
SUMO4 remains unclear and its expression is detected in a limited
number of tissues, including the kidney, spleen and lymph node
(18,19). SUMOylation serves important roles
in physiological processes and the development of various diseases,
including inflammation, cancer and nervous lesions (20–23).
It has also been demonstrated that SUMOylation serves important
roles in the regulation of apoptosis in DC (24,25).
Transforming growth factor (TGF)-β is associated
with organ fibrosis and hypertrophy (12–14).
The TGF-β/Smad pathway regulates cell differentiation,
proliferation, migration and apoptosis and maybe adjusted by
post-translational modifications such as phosphorylation,
acetylation and ubiquitylation. It has been demonstrated that the
TGF-β/Smad pathway may be activated by high glucose (HG) via
regulation of the expression levels of Smad2 and Smad3, and the
pathway contributes to the fibrotic interstitium in DC (13,26,27).
Previous studies have also indicated that SUMO is involved in the
regulation of the TGF-β/Smad pathway (28,29).
However, the role served by the modifying effect of SUMOylation on
the TGF-β/Smad pathway in DC remains unclear.
In the present study, the effect of SUMO2
overexpression on HG-injured cardiomyocyte cell cycle and apoptosis
was investigated. The effect of SUMO2 overexpression on the
TGF-β/Smad pathway was subsequently evaluated. The results may
provide novel insights into SUMO2 as a potential biomarker for the
treatment of DC.
Materials and methods
Cell culture
H9c2 rat embryo cardiomyocytes purchased from the
American Type Culture Collection (Manassas, VA, USA) were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C with 5% CO2. Cells at logarithmic phase
were used in the current study. H9c2 cell morphology was identified
under an Olympus DSX100 optical microscope (Olympus Corporation,
Tokyo, Japan) at 48 h following treatment (magnification, ×100 or
200).
Cell viability assay
H9c2 cells were randomly divided into the following
five groups: HG groups treated with HG (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) under normal glucose concentration (5.6 mmol/l)
and three different HG concentrations (10, 20 and 30 mmol/l); and a
control group without any treatment (n=5 each group). Cell
viabilities were measured using a Cell Counting Kit (CCK)-8 assay
(Beyotime Institute of Biotechnology, Haimen, China) following HG
administration for different durations (6, 12 and 24 h). Following
treatment, cells were seeded in 96-well plates at an initial
density of 5×103 cells/well and incubated for the
indicated durations at 37°C. A total of 20 µl CCK-8 reagent was
subsequently added into each well of the plate and plates were
incubated at 37°C for 1 h. The optical density values were read at
450 nm by a microplate reader (Bio-Tek Instruments, Inc., Winooski,
VT, USA). Data were expressed as the percentage of viable cells as
follows: Relative viability (%) =
[A450(treated)-A450(blank)]/[A450(control)-A450(blank)]
×100.
Cell transfection
Cell transfection was performed following
construction of a SUMO2 overexpression plasmid with the
pGEM-T/pFLAGvector (Promega Corporation, Madison, WI, USA) and
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was
used as the transfection reagent. The empty vector was transfected
respectively at the same time as a transfection negative control
group. Cells were inoculated in DMEM culture media without
antibiotics. SUMO2 plasmid (4 µg) and Lipofectamine 2000 (10 µl)
were added into DMEM culture media without serum (or opti-MEM
media) when cell density reached 90–95%, and mixed gently with the
dilution of 1:2 (DNA: Lipofectamine 2000). The culture media was
changed into DMEM with 10% fetal bovine serum following culture for
6 h at 37°C with 5% CO2. The cell transfection rates
were detected by reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and western blot analysis following
culture for 48 h.
Cell group division
Cells were divided into five experimental groups to
perform the subsequent experiments: SUMO+HG group, consisting of
H9c2 cells treated with 20 mmol/l HG for 24 h following
transfection with SUMO2 overexpression plasmid for 12 h; Vect+HG
group, consisting of H9c2 cells treated with 20 mmol/l HG for 24 h
following transfection with empty plasmid vector for 12 h; Vect
group, consisting of H9c2 cells only transfected with empty plasmid
vector for 12 h; HG group, consisting of H9c2 cells treated with 20
mmol/l HG for 24 h only; and control group, consisting of H9c2
cells without any treatment.
RT-qPCR
mRNA expression levels were measured using RT-qPCR.
Total RNA was extracted from cells in each experimental group using
an RNeasy kit (Qiagen, Inc., Valencia, CA, USA) and cDNA was
reverse transcribed with 1 µg RNA at 42°C, for 60 min, using a
QuantiTect Reverse Transcription kit (Qiagen, Inc.), according to
the manufacturer's protocol. The qPCR amplification was performed
for 15 sec at 95°C, followed by 40 cycles of denaturation at 95°C
for 15 sec and annealing/extension at 60°C for 15 sec in an ABI
7300 Thermocycler (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using a Fast SYBR Green Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The quantification was identified
by 2−ΔΔCq (30).
Expression levels were normalized to that of GAPDH; the
oligonucleotide primer sequences use dare presented in Table I.
 | Table I.Primers used in reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primers used in reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Orientation | Sequence |
|---|
| SUMO2 | Forward |
GACGAGAAACCCAAGGA |
|
| Reverse |
CTGCCGTTCACAATAGG |
| CyclinA2 | Forward |
TGATGAAACTATGACCATGATGTCC |
|
| Reverse |
TTCACAGAACGCAGACCACC |
| CDKN1a | Forward |
TTGTCGCTGTCTTGCACTCT |
|
| Reverse |
GGCACTTCAGGGCTTTCTC |
| C-Myc | Forward |
GGTGGAAAACCCGACAGTCA |
|
| Reverse |
GCAACATAGGACGGAGAGCA |
| Bcl-2 | Forward |
CCCCTGGCATCTTCTCCTTCC |
|
| Reverse |
GGGTGACATCTCCCTGTGACG |
| Bax | Forward |
GGATGCGTCCACCAAGAA |
|
| Reverse |
ACGGAGGAAGTCCAGTGT |
| Caspase-3 | Forward |
GCCTCTGCCCGGTTAAGAAA |
|
| Reverse |
CATCTGTACCAGACCGAGCG |
| TGF-β1 | Forward |
CGCCTGCAGAGATTCAAGTC |
|
| Reverse |
GCCCTGTATTCCGTCTCCTT |
| Smad3 | Forward |
GTCATCTACTGCCGCTTGTG |
|
| Reverse |
GGGGATGGAATGGCTGTAGT |
| GAPDH | Forward |
GGTCATGAGTCCTTCCACGATA |
|
| Reverse |
ATGCTGGCGCTGAGTACGTC |
Western blot analysis
Cells were lysed by protein lysis reagent P0013 from
Beyotime Institute of Biotechnology (Haimen, China), followed by
centrifugation at 10,000 × g for 5 min at 4°C, and the supernatants
containing proteins were collected. Protein concentration was
determined by a BCA assay (Beyotime Institute of Biotechnology).
Proteins (10 µg) were subsequently subjected to each lane of 12%
SDS-PAGE and electro blotted onto polyvinylidene difluoride (PVDF)
membranes (GE Healthcare, Chicago, IL, USA). Following blocking
with 5% nonfat dry milk in PBS for 1 h at 37°C, the blotting
membranes were probed overnight at 4°C with primary antibodies
including rabbit anti-SUMO2 (1:2,000; ab209822), CyclinA2 (1:2,000;
ab137769), CDKN1a (1:1,000; ab109199), C-Myc (1:1,000; ab39688),
anti-B-cell lymphoma-2 (Bcl-2; 1:1,000; ab196495),
anti-Bcl-2-associated X (Bax; 1:1,000; ab53154),
anti-active-caspase-3 (1:200; ab2302), Smad3 (1:1,000; ab84177),
TGF-β1 (1:1,000; ab92486) and anti-GAPDH (1:2,500; ab9485) (all
from Abcam, Cambridge, UK). Then the membranes were subsequently
probed with the appropriate horseradish peroxidase (HRP)-conjugated
secondary antibodies: Goat anti-rabbit IgG H&L HRP (1:5,000;
ab6721; Abcam). The PVDF membrane was exposed to X-ray film and
immunoreactive bands were detected by reaction with enhanced
chemiluminescence (ECL) detection system reagent, GE ECL Start (GE
Healthcare). The membrane was probed with a monoclonal antibody for
GAPDH as the loading control. Band densities were quantified by
densitometry with Bio-Rad ChemiDoc XRS+ (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Cell cycle analysis
Cell cycle progression was evaluated by propidium
iodide (PI) staining. Cells (5.0×105/ml) from each group
were trypsinized, washed twice using PBS and fixed overnight at 4°C
in ice-cold 70% ethanol. Following two washes with PBS, cells were
incubated in 50 µg/ml PI (Invitrogen; Thermo Fisher Scientific,
Inc.) and 100 µg/ml RNase (Thermo Fisher Scientific, Inc.) for 30
min at room temperature. Thereafter, analysis was immediately
performed using FACSCalibur flow cytometer and BD
CellQuest™ Pro Software (BD Biosciences, Franklin Lakes,
NJ, USA). The proportion of cells in G0/G1, S
and G2/M phases was subsequently detected.
Apoptosis detection
The apoptosis status of the cells in each group was
determined by an Annexin V-fluorescein isothiocyanate (FITC)
Apoptosis kit (BioVision, Inc., Milpitas, CA, USA), according to
the manufacturer's protocols. Briefly, floating and trypsinized
adherent cells (5×105) from each group were collected
and re suspended in 500 µl PBS containing 5 µl Annexin V-FITC and 5
µl PI, and incubated for 5 min in the dark at room temperature.
Analysis was subsequently performed using a flow cytometer (BD
Biosciences). Cell Quest Pro software (BD Biosciences) was used to
analyze the apoptosis rate.
Statistical analysis
Data are presented as the mean ± standard deviation
of five independent experiments. Statistical analysis was performed
using SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) and data were
subjected to one-way analysis of variance followed by a Dunnett's
post-hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
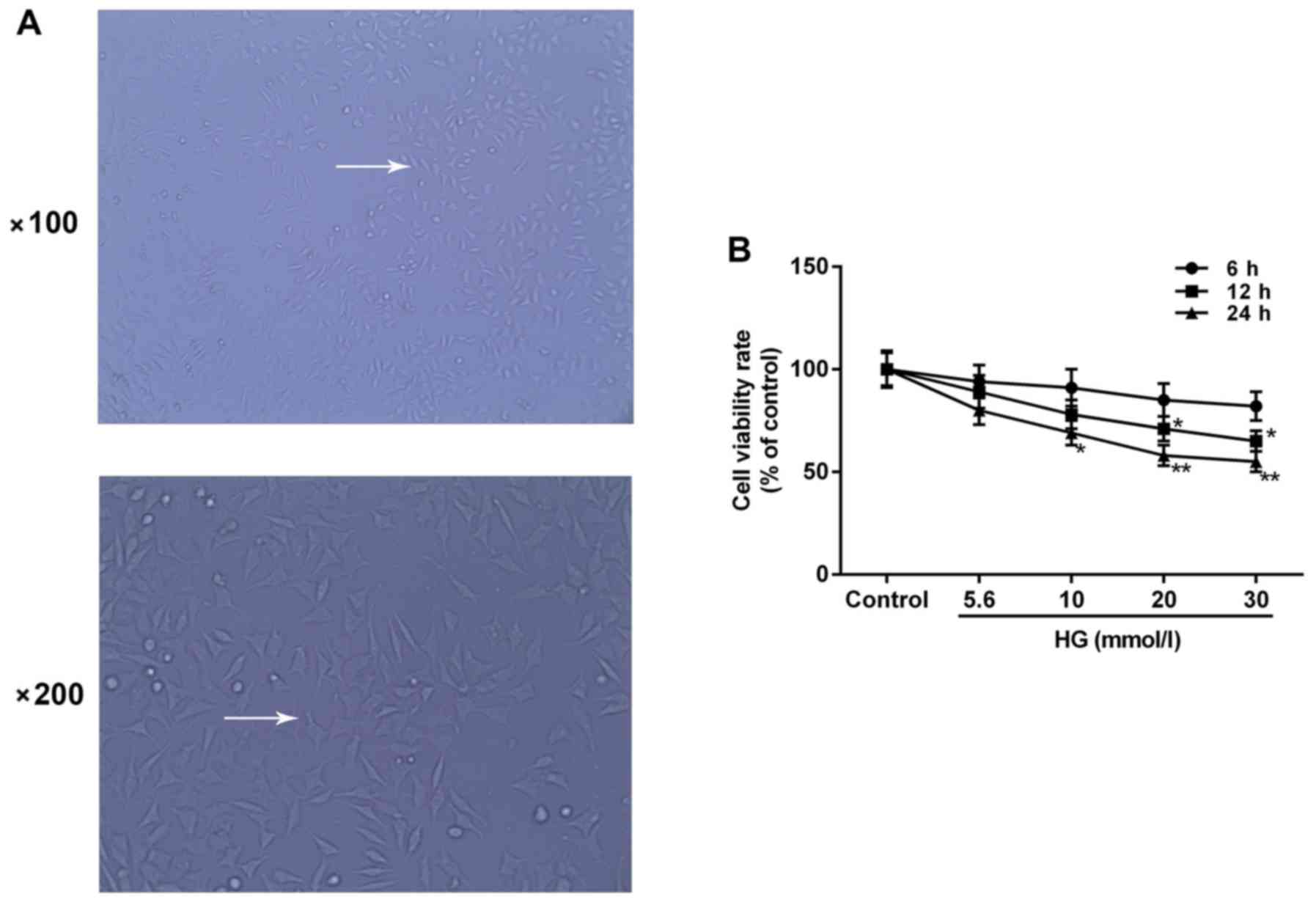
Cell morphology
H9c2 cells are rat embryo myocardial cells. Cell
morphology was identified under an optical microscope. Cells grew
well and were adherent to the bottom of culture flasks as a
monolayer at 48 h following treatment. The majority of cells
appeared to exhibit long-shuttle morphology and certain cells had a
triangular or irregular morphology (Fig. 1A).
Inhibitory effect of HG on H9c2 cell
viability
Cell viability was evaluated by a CCK-8 assay
following HG treatment of H9c2 cells at different concentrations
(0, 5.6, 10, 20 and 30 mmol/l) for different durations (6, 12 and
24 h) to indicate the damage induced by HG on H9c2 cells. The
results indicated that H9c2 cell viability decreased in a
dose-dependent and time-dependent manner. The viability was
significantly decreased by 42% compared with the control group when
treated with 20 mmol/l HG for 24 h (P<0.05) and was similar to
cells treated with 30 mmol/l HG for 24 h (Fig. 1B). Therefore, H9c2 cells treated
with 20 mmol/l HG for 24 h were selected for subsequent experiments
due to the extent of cytotoxicity at a low density (HG group).
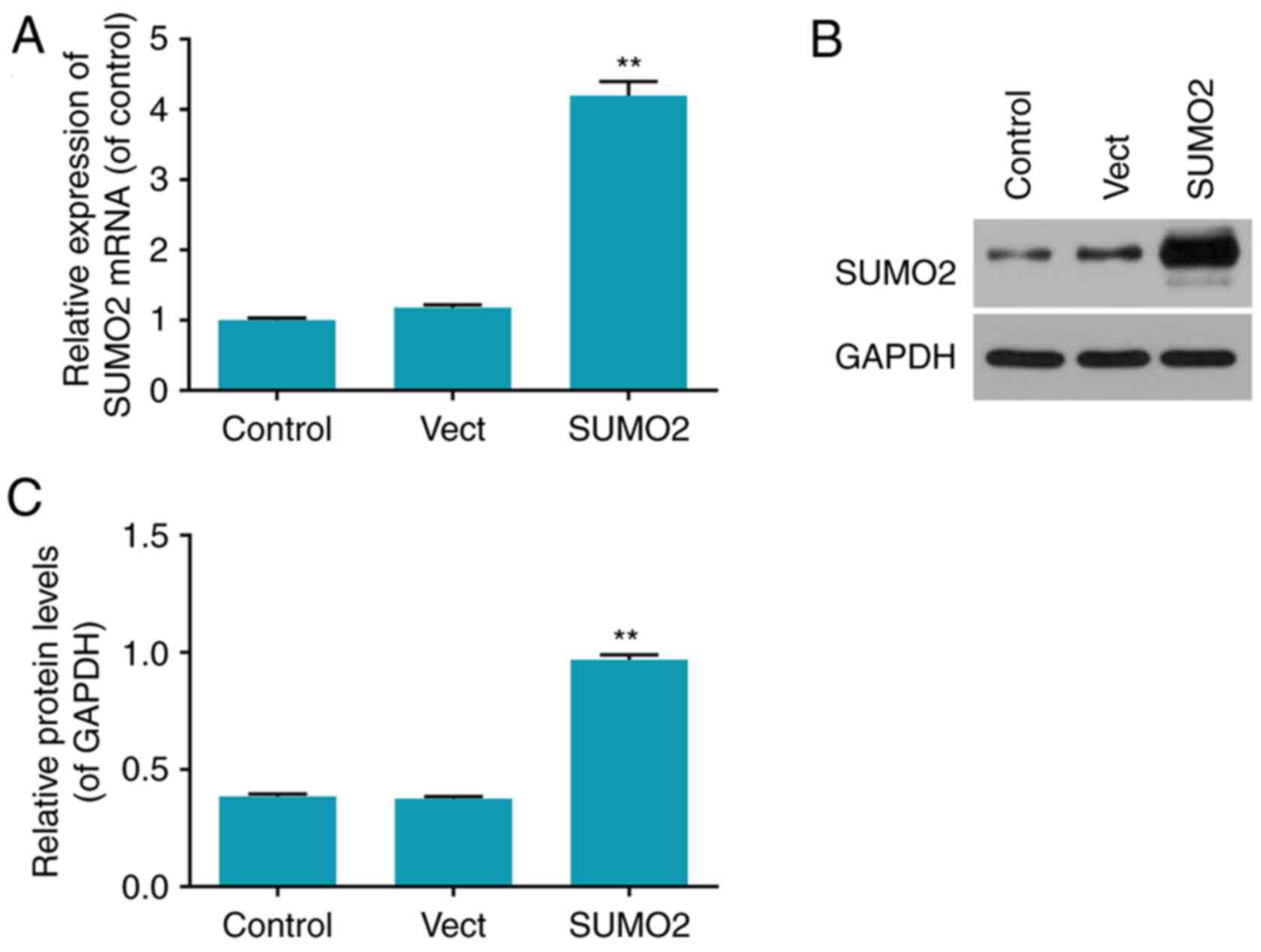
Transfection rates of overexpressed
SUMO2 in H9c2 cells
The transfection efficiency in each group was
determined by RT-qPCR and western blot analysis (Fig. 2). The mRNA and protein levels of
SUMO2 were significantly increased in the SUMO2 group compared with
the control (P<0.01; Fig. 2).
The mRNA and protein levels of SUMO2 in the Vect group transfected
with empty vector were not significantly different from the control
group (Fig. 2).
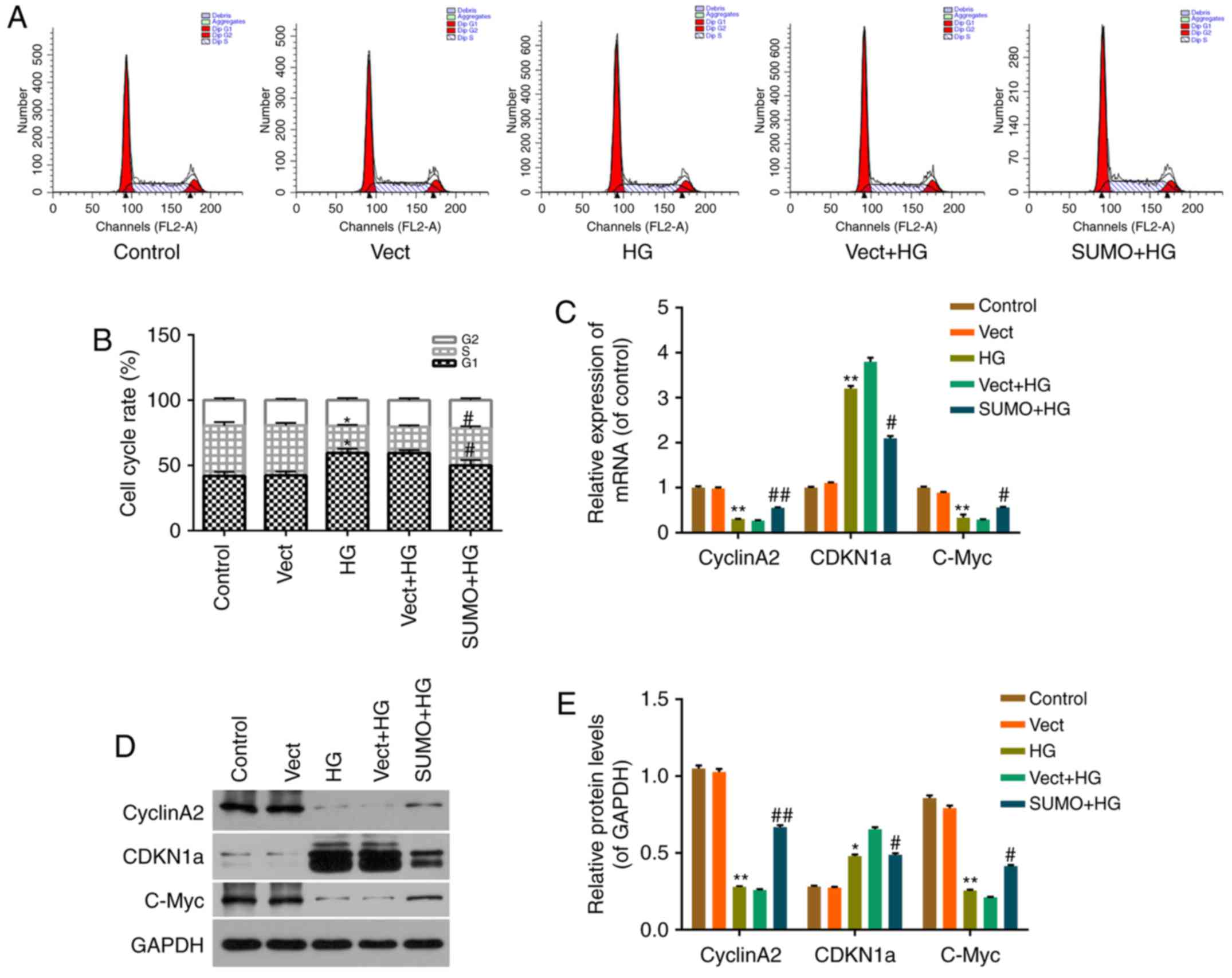
Effect of SUMO2 overexpression on
HG-induced H9c2 cell cycle arrest
The viability of HG-treated cells was decreased,
therefore, the subsequent effect of SUMO2 overexpression on H9c2
cell cycle progression was investigated by flow cytometry.
Treatment of H9c2 cells with HG blocked the cell cycle transition
from G1 to S phase (Fig. 3A
and B). The percentage of cells in G0/G1
phases significantly increased from 41.97 to 59.58% (P<0.05),
while the S phase fraction decreased from 38.45 to 29.60%
(P<0.05), in the HG group compared with the control group
(Fig. 3A and B). Following SUMO2
overexpression, the cell cycle transition from G1 to S phase
recovered significantly, the percentage of cells in
G0/G1 phases significantly decreased from
59.44 to 50.06% (P<0.05), while the S phase fraction
significantly increased from 20.06 to 28.44% (P<0.05) in the
SUMO + HG group compared with the Vect+HG group (Fig. 3A and B).
Effect of SUMO2 on cell
cycle-associated factors in H9c2 cells treated with HG
To investigate the mechanism by which SUMO2
overexpression protects H9c2 cells from HG-induced cell viability
inhibition and cell cycle arrest, RT-qPCR and western blot analysis
were performed. mRNA and protein levels of cell cycle-associated
factors, including CyclinA2, CDKN1a and C-Myc, were detected in
each group. The results indicated that the mRNA and protein levels
of CDKN1a were significantly increased in the HG group compared
with the control group (P<0.01 and P<0.05, respectively) and
significantly decreased in the SUMO+HG group compared with the
Vect+HG group (both P<0.05; Fig.
3C-E). By contrast, the mRNA and protein levels of CyclinA2 and
C-Myc were significantly decreased in the HG group compared with
the control group (all P<0.01) and significantly increased in
the SUMO+HG group compared with the Vect+HG group (P<0.05;
Fig. 3C-E).
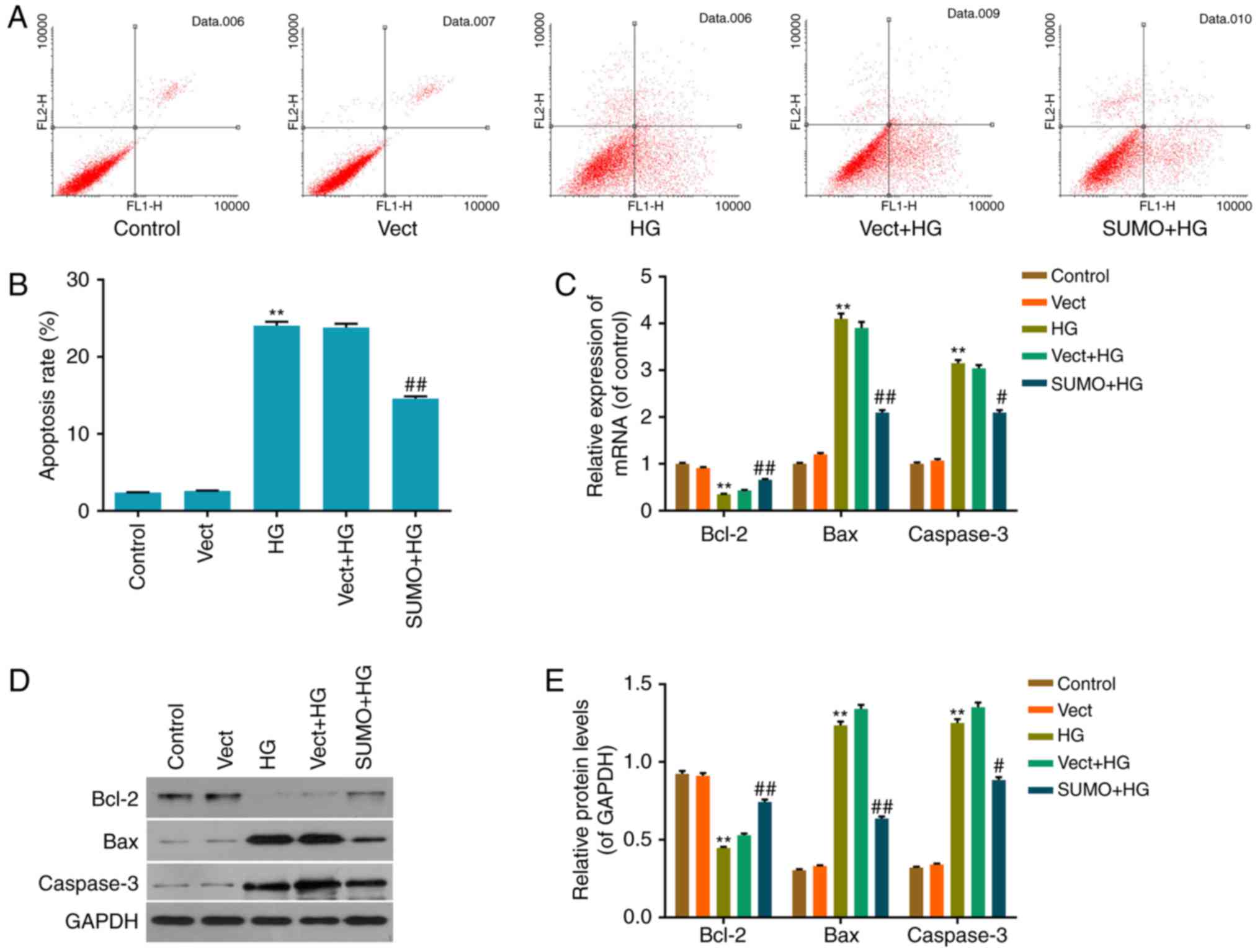
Inhibitory effect of SUMO2
overexpression on HG-induced H9c2 cell apoptosis
The inhibition of apoptosis by SUMO2 following HG
treatment was investigated by performing an Annexin V/PI
double-stain assay to detect the apoptosis status in each group.
The apoptosis rate of the HG group was significantly increased
compared with the control group (P<0.01) and the apoptosis rate
of the SUMO+HG group was significantly decreased by ~42% of that of
the Vect+HG group (P<0.01; Fig. 4A
and B). Thus, the results indicated that SUMO2 overexpression
may inhibit the promotion of H9c2 cell apoptosis by HG.
Effect of SUMO2 overexpression on
apoptosis-associated factors in H9c2 cells treated with HG
To investigate the mechanism by which SUMO2
overexpression protects H9c2 cells from HG-induced inhibition of
cell viability inhibition and apoptosis promotion, RT-qPCR and
western blot analysis were performed to detect mRNA and protein
levels of apoptosis-associated factors, including Bax, Bcl-2 and
Caspase-3, in each group. The results indicated that the mRNA and
protein levels of the apoptosis activating factors Bax and
Caspase-3 were significantly increased in the HG group compared
with the control group (P<0.01) and significantly decreased in
the SUMO+HG group compared with the Vect+HG group (P<0.05;
Fig. 4C-E). By contrast, the mRNA
and protein levels of the apoptosis inhibitor Bcl-2 were
significantly decreased in the HG group compared with the control
group (P<0.01) and significantly increased in the SUMO+HG group
compared with the Vect+HG group (P<0.01; Fig. 4C-E).
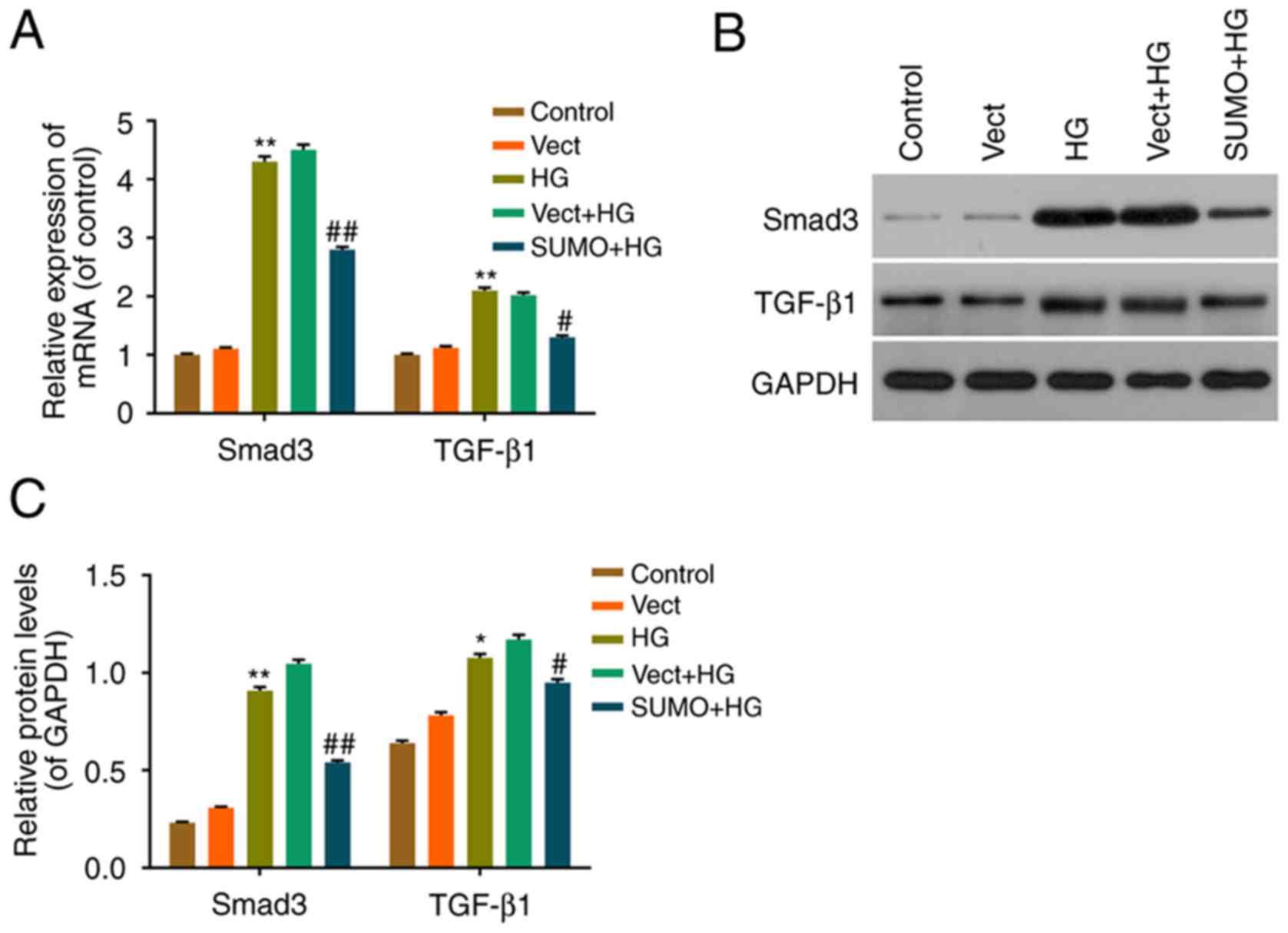
Effect of SUMO2 overexpression on the
TGF-β/Smad pathway in H9c2 cells treated with HG
The effect of SUMO2 overexpression on downstream
effectors in the TGF-β/Smad pathway was also investigated. RT-qPCR
and western blot analysis were performed to assess the mRNA and
protein levels of TGF-β1 and Smad3. The expression of TGF-β1 and
Smad3 mRNA and protein was significantly increased in the HG group
compared with the control group (P<0.05) and significantly
decreased in the SUMO+HG group compared with the Vect+HG group
(P<0.05) (Fig. 5). These
results therefore indicated that SUMO2 overexpression may inhibit
cell apoptosis by regulating the TGF-β/Smad pathway in H9c2 cells
treated with HG.
Discussion
Hyperglycemia is an inducing factor of DC, which
increases the mortality of patients with diabetes and is
characterized by cardiac hypertrophy (31,32).
Investigation of the molecular mechanisms of DC and cardiac
hypertrophy may uncover novel therapeutic strategies for heart
failure in patients with diabetes. It may be hypothesized that
SUMOs may aid in treating DC, as they have an important
modification function in physiological process and disease
development (17,33,34).
The current study investigated how overexpression of
SUMO2 affected HG-induced cardiomyocyte injury, with a focus on the
cell cycle and apoptosis. H9c2 rat embryo cardiomyocytes with a
classical long shuttle type morphology, identified by optical
microscopy, were employed in the current study. The viability of
HG-injured H9c2 cells was evaluated by a CCK-8 assay and the
results indicated that the viability was decreased in a
dose-dependent (0, 5.6, 10, 20 and 30 mmol/l) and time-dependent
(6, 12 and 24 h) manner. The degree of inhibition on cell viability
with 30 mmol/l HG was more than that of 20 mmol/l, however, the
increase was not significant. Therefore, H9c2 cells treated with 20
mmol/l HG for 24 h were selected for subsequent experiments.
The cell cycle is the process by which a cell
divides into two daughter cells and it consists of interphase
(G1, S and G2 phases) and mitotic (M) phase
(35). Cell cycle regulatory
factors, which include cyclin, CDKs and CDK inhibitors, serve
critical roles in the regulation of the cell cycle (36–38).
Different combinations of cyclins and CDKs control cell cycle
progression. Cardiac hypertrophy occurs when cardiomyocytes grow
without division, during which cell cycle regulatory factors may
serve prominent roles (39,40).
The results of the current study indicated that the cell cycle was
blocked between G1/S phases when H9c2 cells were treated
with 20 mmol/l HG for 24 h and the blocking effect was attenuated
by SUMO2 overexpression. RT-qPCR and western blot analysis were
performed to determine the underlying molecular mechanisms of this
effect of SUMO2 overexpression. The results demonstrated that the
expression of the cell cycle activating factors CyclinA2 and C-Myc
were downregulated by HG and upregulated by SUMO2 overexpression in
HG-injured H9c2 cells. By contrast, the expression of CDKN1a, also
termed p21, was upregulated by HG and downregulated by SUMO2
overexpression in HG-injured H9c2 cells. The combination of
CyclinA2 with CDKs serves critical roles in the transition of the
cell cycle from G1/S phase to G2/M phase,
which promotes cell mitosis (41,42),
and C-Myc encodes a phosphoprotein that facilitates cell
proliferation and differentiation (43,44).
CDKN1a is a critical negative regulator in the cell cycle that
inhibits the activation of Cyclin and CDK complexes (45,46).
Hence, SUMO2 overexpression may attenuate cell cycle arrest induced
by HG in H9c2 cells via regulation of cell cycle-associated
factors, by inhibiting the cyclin transition-promoting function of
CyclinA2, the cycle promoting function of C-Myc and inhibiting the
function of CDKN1a.
In addition to the cell cycle, there is evidence
indicating that apoptosis constitutes the prevailing form of
myocyte death (47–49). Cardiomyocyte apoptosis was reported
to be the pathophysiological basis of the development of DC and to
account for the high incidence of heart failure (11,13).
Therefore, it is critical to further investigate the molecular
mechanism of cardiomyocyte apoptosis. Annexin V/PI double-stain
assay and flow cytometry were performed in the current study to
determine whether SUMO2 overexpression affects cell apoptosis
stimulated by HG, and the results demonstrated that SUMO2
overexpression decreased the rate of apoptosis induced by HG in
H9c2 cells. The results of RT-qPCR and western blotting indicated
that the mechanism underlying the aforementioned effects was
associated with the regulation of apoptosis-associated factors,
including Bax, Bcl-2 and Caspase-3. Bcl-2 facilitates cell mitosis
and inhibits apoptosis by regulating the outer membrane
permeability of mitochondria (21,28).
By contrast, Bax facilitates apoptosis by forming a heterodimer
with Bcl-2 and inhibiting its function (33,41,50).
Caspase-3 is an important member of the caspase family. As a
conjunct activating factor in apoptosis signal transduction, it
directly participates in cell regulation, signal transduction and
late apoptosis (51,52). The results of the current study
indicated that SUMO2 overexpression activated the apoptosis
inhibitor Bcl-2 and inhibited the proapoptotic factors Bax and
Caspase-3 in HG-injured H9c2 cells, thereby leading to apoptosis
inhibition.
The effects of SUMO2 overexpression on the
downstream effectors in the TGF-β/Smad pathway in H9c2 cells was
also investigated. The TGF-β/Smad pathway is an effective signaling
pathway involved in the acceleration of oxidative stress, apoptosis
and inflammation, and is therefore implicated in various diseases,
including cardiac fibrosis (26,53,54).
Among eight Smad family members (Smad1-8), Smad2/3 and Smad4 are
the critical factors in the TGF-β pathway (55). The SUMOylation of Smad3 and Smad4
have been previously reported to inhibit TGF-β/Smad transcriptional
activity (56–58). The expression of Smad2/3 was also
demonstrated to be regulated by HG in rat renal tubular epithelial
or mesangial cells (59).
Therefore, the current study focused on Smad3. The results of the
current study indicated that SUMO2 overexpression significantly
decreased the expression of TGF-β1 and Smad3 mRNA and protein in
HG-injured H9c2 cells, indicating that SUMO2 may function in the
TGF-β/Smad pathway by inhibiting the activities of TGF-β and Smad3
in myocardial cells under HG stress conditions to inhibit cell
apoptosis.
In conclusion, a HG-injured H9c2 cardiomyocyte model
was established in the current study to investigate how SUMO2
overexpression alleviates cell cycle arrest and apoptosis promotion
induced by HG, and the results demonstrated that this may occur via
regulation of cell cycle- and apoptosis-associated factors, as well
as inhibition of the TGF-β/Smad pathway. SUMO2 is downregulated by
HG, therefore, it was over-expressed in the present study in order
to recover it. Future studies are required to knockdown SUMO2 and
to determine the effect of SUMO2 on the SUMOylation of associated
factors. However, the results of the current study indicate that
SUMO2 may be a potential biomarker of an important endogenous
protection factor in cardiomyocytes, which may provide novel
insights for the protection of cardiomyocytes and may aid in the
diagnosis and prognosis of patients with DC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CZ made substantial contributions to the design of
the present study and wrote the manuscript. QS made substantial
contributions to the conception of the present study.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Manyari DE: Prognostic implications of
echocardiographically determined left ventricular mass in the
Framingham Heart Study. N Engl J Med. 323:1706–1707. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rohini A, Agrawal N, Koyani CN and Singh
R: Molecular targets and regulators of cardiac hypertrophy.
Pharmacol Res. 61:269–280. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tate M, Deo M, Cao AH, Hood SG, Huynh K,
Kiriazis H, Du XJ, Julius TL, Figtree GA, Dusting GJ, et al:
Insulin replacement limits progression of diabetic cardiomyopathy
in the low-dose streptozotocin-induced diabetic rat. Diab Vasc Dis
Res. 14:423–433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu X, Bai T, Xu Z, Liu Q, Zheng Y and Cai
L: Pathophysiological fundamentals of diabetic cardiomyopathy.
Compr Physiol. 7:693–711. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mishra PK, Ying W, Nandi SS, Bandyopadhyay
GK, Patel KK and Mahata SK: Diabetic cardiomyopathy: An
immunometabolic perspective. Front endocrinol (Lausanne). 8:722017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prakoso D, DeBlasio MJ, Qin C, Rosli S,
Kiriazis H, Qian H, Du XJ, Weeks KL, Gregorevic P, McMullen JR and
Ritchie RH: Phosphoinositide 3-Kinase (p110α) gene delivery limits
diabetes-induced cardiac NADPH oxidase and cardiomyopathy in a
mouse model with established diastolic dysfunction. Clin Sci
(Lond). 131:1345–1360. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bell DS: Diabetic cardiomyopathy. A unique
entity or a complication of coronary artery disease? Diabetes Care.
18:708–714. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ounzain S and Pedrazzini T: The promise of
enhancer-associated long noncoding RNAs in cardiac regeneration.
Trends Cardiovasc Med. 25:592–602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dhiman M and Garg NJ: NADPH oxidase
inhibition ameliorates Trypanosoma cruzi-induced myocarditis during
Chagas disease. J Pathol. 225:583–596. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marushchak M, Lisnianska N, Krynytska
Capital IU and Chornomydz I: The mechanisms of apoptosis initiation
in rats with chronic enterocolitis combined with
streptozotocin-induced diabetes. Georgian Med News. 125–130.
2017.PubMed/NCBI
|
|
11
|
Hong YA, Lim JH, Kim MY, Kim Y, Park HS,
Kim HW, Choi BS, Chang YS, Kim HW, Kim TY, et al: Extracellular
superoxide dismutase attenuates renal oxidative stress through the
activation of AMPK in diabetic nephropathy. Antioxid Redox Signal.
Nov 14–2017.(Epub ahead of print). doi: 10.1089/ars.2017.7207.
|
|
12
|
Tabebi M, Khabou B, Boukadi H, Ben Hamad
M, Ben Rhouma B, Tounsi S, Maalej A, Kamoun H, Keskes-Ammar L, Abid
M, et al: Association study of apoptosis gene polymorphisms in
mitochondrial diabetes: A potential role in the pathogenicity of
MD. Gene. 639:18–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bugyei-Twum A, Advani A, Advani SL, Zhang
Y, Thai K, Kelly DJ and Connelly KA: High glucose induces Smad
activation via the transcriptional coregulator p300 and contributes
to cardiac fibrosis and hypertrophy. Cardiovasc Diabetol.
13:892014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wilkinson KA and Henley JM: Mechanisms,
regulation and consequences of protein SUMOylation. Biochem J.
428:133–145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Princz A and Tavernarakis N: The role of
SUMOylation in ageing and senescent decline. Mech Ageing Dev.
162:85–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hay RT: SUMO: A history of modification.
Mol Cell. 18:1–12. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li XC, Zeng Y, Sun RR, Liu M, Chen S and
Zhang PY: SUMOylation in cardiac disorders-a review. Eur Rev Med
Pharmacol Sci. 21:1583–1587. 2017.PubMed/NCBI
|
|
18
|
Guo B, Yang SH, Witty J and Sharrocks AD:
Signalling pathways and the regulation of SUMO modification.
Biochem Soc Trans. 35:1414–1418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Geiss-Friedlander R and Melchior F:
Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol.
8:947–956. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rosonina E, Akhter A, Dou Y, Babu J and
Theivakadadcham Sri VS: Regulation of transcription factors by
sumoylation. Transcription. 8:220–231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bedford L, Lowe J, Dick LR, Mayer RJ and
Brownell JE: Ubiquitin-like protein conjugation and the
ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov.
10:29–46. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paddibhatla I, Lee MJ, Kalamarz ME,
Ferrarese R and Govind S: Role for sumoylation in systemic
inflammation and immune homeostasis in Drosophila larvae.
PLoS Pathog. 6:e10012342010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wilson VG: Viral interplay with the host
sumoylation system. Adv Exp Med Biol. 963:359–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang E and Abe J: Kinase-SUMO networks in
diabetes-mediated cardiovascular disease. Metabolism Clin Exp.
65:623–633. 2016. View Article : Google Scholar
|
|
25
|
Shishido T, Woo CH, Ding B, McClain C,
Molina CA, Yan C, Yang J and Abe J: Effects of MEK5/ERK5
association on small ubiquitin-related modification of ERK5:
Implications for diabetic ventricular dysfunction after myocardial
infarction. Circ Res. 102:1416–1425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang L, Mao Y, Pan J, Wang S, Chen L and
Xiang J: Bamboo leaf extract ameliorates cardiac fibrosis possibly
via alleviating inflammation, oxidative stress and apoptosis.
Biomed Pharmacother. 95:808–817. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu H, Sun F, Li X and Sun L:
Downregulation of miR-23a inhibits high glucose-induced EMT and
renal fibrogenesis by upregulation of SnoN. Hum Cell. 31:22–32.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kang JS, Saunier EF, Akhurst RJ and
Derynck R: The type I TGF-beta receptor is covalently modified and
regulated by sumoylation. Nat Cell Biol. 10:654–664. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin X, Liang M, Liang YY, Brunicardi FC
and Feng XH: SUMO-1/Ubc9 promotes nuclear accumulation and
metabolic stability of tumor suppressor Smad4. J Biol Chem.
278:31043–31048. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haas AV and McDonnell ME: Pathogenesis of
cardiovascular disease in diabetes. Endocrinol Metab Clin North Am.
47:51–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singh RM, Waqar T, Howarth FC, Adeghate E,
Bidasee K and Singh J: Hyperglycemia-induced cardiac contractile
dysfunction in the diabetic heart. Heart Fail Rev. 23:37–54. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bernt A, Rangrez AY, Eden M, Jungmann A,
Katz S, Rohr C, Muller OJ, Katus HA, Sossalla ST, Williams T, et
al: Sumoylation-independent activation of
Calcineurin-NFAT-signaling via SUMO2 mediates cardiomyocyte
hypertrophy. Sci Rep. 6:357582016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim EY, Zhang Y, Ye B, Segura AM, Beketaev
I, Xi Y, Yu W, Chang J, Li F and Wang J: Involvement of activated
SUMO-2 conjugation in cardiomyopathy. Biochim Biophys Acta.
1852:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Valle-Casuso JC, Allouch A, David A, Lenzi
GM, Studdard L, Barre-Sinoussi F, Muller-Trutwin M, Kim B, Pancino
G and Saez-Cirion A: p21 restricts HIV-1 in monocyte-derived
dendritic cells through the reduction of dNTP biosynthesis and
regulation of SAMHD1 antiviral activity. J Virol. 91:e013242017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brooks G, Poolman RA and Li JM: Arresting
developments in the cardiac myocyte cell cycle: Role of
cyclin-dependent kinase inhibitors. Cardiovasc Res. 39:301–311.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hauck L, Harms C, Grothe D, An J, Gertz K,
Kronenberg G, Dietz R, Endres M and von Harsdorf R: Critical role
for FoxO3a-dependent regulation of p21CIP1/WAF1 in response to
statin signaling in cardiac myocytes. Circ Res. 100:50–60. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Koga K, Kenessey A and Ojamaa K:
Macrophage migration inhibitory factor antagonizes pressure
overload-induced cardiac hypertrophy. Am J Physiol Heart Circ
Physiol. 304:H282–H293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kaplan A, Abidi E, Ghali R, Booz GW,
Kobeissy F and Zouein FA: Functional, cellular and molecular
remodeling of the heart under influence of oxidative cigarette
tobacco smoke. Oxid Med Cell Longev. 2017:37591862017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang D, Li Y, Heims-Waldron D, Bezzerides
V, Guatimosim S, Guo Y, Gu F, Zhou P, Lin Z, Ma Q, et al:
Mitochondrial cardiomyopathy caused by elevated reactive oxygen
species and impaired cardiomyocyte proliferation. Circ Res.
122:74–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deng H, Cheng Y, Guo Z, Zhang F, Lu X,
Feng L, Wang X and Xu Z: Overexpression of cyclinA2 ameliorates
hypoxia-impaired proliferation of cardiomyocytes. Exp Ther Med.
8:1513–1517. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fu XJ, Li HX, Yang K, Chen D and Tang H:
The important tumor suppressor role of PER1 in regulating the
cyclin-CDK-CKI network in SCC15 human oral squamous cell carcinoma
cells. Onco Targets Ther. 9:2237–2245. 2016.PubMed/NCBI
|
|
43
|
Yang XH, Tang F, Shin J and Cunningham JM:
Incorporating genomic, transcriptomic and clinical data: a
prognostic and stem cell-like MYC and PRC imbalance in high-risk
neuroblastoma. BMC Syst Biol. 11 Suppl 5:922017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu P, Su J, Song X and Wang S: Activation
of nuclear β-catenin/c-Myc axis promotes oxidative stress injury in
streptozotocin-induced diabetic cardiomyopathy. Biochem Biophys Res
Commun. 493:1573–1580. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yoon MK, Mitrea DM, Ou L and Kriwacki RW:
Cell cycle regulation by the intrinsically disordered proteins p21
and p27. Biochem Soc Trans. 40:981–988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yanagi T, Nagai K, Shimizu H and Matsuzawa
SI: Melanoma antigen A12 regulates cell cycle via tumor suppressor
p21 expression. Oncotarget. 8:68448–68459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou C, Huang J, Li Q, Zhan C, Xu X, Zhang
X, Ai D, Zhu Y, Wen Z and Wang DW: CYP2J2-derived EETs attenuated
ethanol-induced myocardial dysfunction through inducing autophagy
and reducing apoptosis. Free Radic Biol Med. 117:168–179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen TS, Kuo CH, Battsengel S, Pan LF, Day
CH, Shen CY, Chung LC, Padma VV, Yao CK, Lin YM and Huang CY:
Adipose-derived stem cells decrease cardiomyocyte damage induced by
porphyromonas gingivalis endotoxin through suppressing hypertrophy,
apoptosis, fibrosis and MAPK markers. Environ Toxicol. 33:508–513.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wei X, Yang Y, Jiang YJ, Lei JM, Guo JW
and Xiao H: Relaxin ameliorates high glucose-induced cardiomyocyte
hypertrophy and apoptosis via the Notch1 pathway. Exp Ther Med.
15:691–698. 2018.PubMed/NCBI
|
|
50
|
Yao C, Cao X, Fu Z, Tian J, Dong W, Xu J,
An K, Zhai L and Yu J: Boschniakia rossica polysaccharide
triggers laryngeal carcinoma cell apoptosis by regulating
expression of Bcl-2, caspase-3 and P53. Med Sci Monit.
23:2059–2064. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lv L and Liu B: Antitumor effects of
bakuchiol on human gastric carcinoma cell lines are mediated
through PI3K/AKT and MAPK signaling pathways. Mol Med Rep.
16:8977–8982. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cui X, Jing X, Wu X, Bi X, Liu J, Long Z,
Zhang X, Zhang D, Jia H, Su D and Huo K: Abnormal expression levels
of BMP15/Smad1 are associated with granulosa cell apoptosis in
patients with polycystic ovary syndrome. Mol Med Rep. 16:8231–8236.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lustri AM, Di Matteo S, Fraveto A,
Costantini D, Cantafora A, Napoletano C, Bragazzi MC, Giuliante F,
De Rose AM, Berloco PB, et al: TGF-β signaling is an effective
target to impair survival and induce apoptosis of human
cholangiocarcinoma cells: A study on human primary cell cultures.
PLoS One. 12:e01839322017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Qiu M, Chen Y, Chen L, Zeng J and Liu J:
Transforming growth factor β1 and Fas ligand synergistically
enhance immune tolerance in dendritic cells in liver
transplantation. J Surg Res. 218:180–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen K, Cheng L, Qian W, Jiang Z, Sun L,
Zhao Y, Zhou Y, Zhao L, Wang P, Duan W, et al: Itraconazole
inhibits invasion and migration of pancreatic cancer cells by
suppressing TGF-beta/SMAD2/3 signaling. Oncol Rep. 39:1573–1582.
2018.PubMed/NCBI
|
|
56
|
Imoto S, Sugiyama K, Muromoto R, Sato N,
Yamamoto T and Matsuda T: Regulation of transforming growth
factor-beta signaling by protein inhibitor of activated STAT, PIASy
through Smad3. J Biol Chem. 278:34253–34258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Imoto S, Ohbayashi N, Ikeda O, Kamitani S,
Muromoto R, Sekine Y and Matsuda T: Sumoylation of Smad3 stimulates
its nuclear export during PIASy-mediated suppression of TGF-β
signaling. Biochem Biophys Res Commun. 370:359–365. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Long J, Wang G, He D and Liu F: Repression
of Smad4 transcriptional activity by SUMO modification. Biochem J.
379:23–29. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tang WB, Ling GH, Sun L, Zhang K, Zhu X,
Zhou X and Liu FY: Smad anchor for receptor activation regulates
high glucose-induced EMT via modulation of Smad2 and Smad3
activities in renal tubular epithelial cells. Nephron. 130:213–220.
2015. View Article : Google Scholar : PubMed/NCBI
|