Introduction
High mobility group box 1 (HMGB1) is not only a
non-histone nuclear protein that regulates gene transcription
(1), but can also be passively
released into the extracellular space as a proinflammatory mediator
by necrotic cells, or in an active manner by lipopolysaccharide
(LPS)-, tumor necrosis factor-α (TNF-α)-, interleukin-1β- and
interferon-γ-stimulated monocytes or macrophages (2). The release of HMGB1 is regulated by
elaborate mechanisms, including HMGB1 translocation from the
nucleus to the cytoplasm with post-translational modifications of
acetylation, methylation or phosphorylation, and its release into
the extracellular environment with inflammasome activation upon
appropriate stimulation (3,4).
When accumulating in the cytoplasm, HMGB1 has been reported to
regulate autophagy (5). This
evidence suggests that the function of HMGB1 is associated with its
location. Previous studies have also reported that the
extracellular functions of HMGB1 are dependent on its redox state
and combination of target receptors, which mediates the responses
to inflammation, immunity, chemotaxis and tissue regeneration
(6–8). The biological roles of extracellular
HMGB1 mainly involve its effects on macrophage proinflammatory
function and dendritic cell (DC) antigen presentation (9,10).
As extracellular HMGB1 is a critical mediator of lethality in
inflammatory diseases (11),
understanding the complex extracellular functions of HMGB1 is
necessary for the development of potential treatments.
The acetylation of HMGB1 was first described in
1979, which revealed that there are two sites of acetylation in the
HMGB1 protein, constituting lysine residues at positions 2 and 11
(12). A previous study indicated
that acetylation of HMGB1 at lysine 2 (isolated from cells cultured
in the presence of sodium n-butyrate) exhibited enhanced binding
affinity to distorted DNA structures (13) and is required for their potential
function in DNA replication (14).
A recent study revealed via mass spectrometry, that 8 of 43 lysine
residues are frequently modified in HMGB1 (15). The nuclear localization of HMGB1 is
affected by lysine acetylation, particularly within the two major
clusters of lysine residues at positions 27, 28 and 29 of the
nuclear localization signal 1 (NLS1) and residues 181, 182 and 183
of NLS2 (15). It has been
demonstrated that mutations of all six lysine residues into
glutamines, as a mimic of acetyl-lysine within the two clusters,
results in partial cytosolic localization (15). HMGB1 acetylation has been reported
to regulate its interaction with DNA and repair DNA damage
(16–18). In addition, accumulating
experimental evidence has indicated that HMGB1 modified by
acetylation serves an essential role in its nuclear translocation
(15,19); however, the effect of the
post-translational modification of acetylation on the extracellular
functions of HMGB1 is unknown. Therefore, the present study
proposed that the extracellular functions of acetylated HMGB1 and
non-acetylated HMGB1 differ.
In the present study, an optimized method of gene
mutation was employed to obtain mimicked acetylated HMGB1 (HMGB1-M)
and the effects of HMGB1-M and nonacetylated HMGB1 (HMGB1) on
macrophages and DCs were compared.
Materials and methods
Animals
Six female 5-week-old C57BL/6 mice (18–20 g) were
purchased from the Shanghai SLAC Laboratory Animal Co., Ltd.
(Shanghai, China). All mice were housed in the Animal Laboratory
Center of Tongji Medical College (Wuhan, China) in microisolator
cages, at 22–23°C in 50–65% humidity under a 12:12 light-dark cycle
and in specific pathogen-free conditions. The mice had access to a
standard diet and water for one week prior to use in experiments.
Animal studies were approved by the Animal Care and Use Committee
of Tongji Medical College, HUST (Wuhan, China).
Construction of plasmids
The pET28a (Merck KGaA, Darmstadt, Germany)-HMGB1
plasmid (HMGB1 sequence from mouse) was used as a template to
generate three lysine mutations at residues 27, 28, and 29 via
site-directed mutagenesis by polymerase chain reaction (PCR) with
mutant primer 1. The mutant plasmids were then used as a template
to generate another three mutated lysine residues 181, 182 and 183,
again by site directed mutagenesis via PCR with mutant primer 2,
forming the mutant plasmid (pET28a-HMGB1-M). Site directed
mutagenesis PCR was performed using the Eppendorf PCR system
(Eppendorf, Hamburg, Germany) with a final reaction volume of 50 µl
PCR mix containing 2 ng template, all mutant primers at 0.3 µM, 25
l 2X PCR buffer, and each dNTP at 0.4 mM and 1 U KOD FX Taq DNA
polymerase (Toyobo Life Science). The thermocycling conditions were
as follows: Initial denaturation at 94°C for 2 min followed by 30
cycles, each at 98°C for 10 sec, annealing at 60 °C for 30 sec,
extension at 68°C for 6 min (1 kbp/min). The PCR products were
treated with the restriction enzyme, DpnI (Toyobo Life
Science) at 37°C for 3 h. A total of 100 µl DH5α competent cells
(cat. no. C1100; Beijing Solarbio Science & Technology Co.,
Ltd., Beijing, China) were transformed with 1 µl of the
aforementioned PCR products, according to the manufacturer's
protocols and inoculated on Luria-Bertani plates containing
kanamycin (50 µg/ml) at 37°C for 12 h. A total of 10 random
colonies were selected for screening mutants, and their plasmids
were isolated by using the TIANprep Mini Plasmid kit (Tiangen
Biotech Co., Ltd., Beijing, China), according to the manufacturer's
protocols and PCR amplification was performed with the identifying
primer. Identifying PCR was performed in a final volume of 20 µl
PCR mix containing 20 ng template, each primer at 0.2 µM, 10 µl mix
buffer and 2 U Taq DNA polymerase (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) to identify the plasmids. The thermocycling
conditions were as follows: Initial denaturation at 94°C for 5 min
followed by 30 cycles, each at 94°C for 30 sec, annealing at 58°C
for 30 sec, extension at 72°C for 1 min and a final extension at
72°C for 5 min. The PCR products were evaluated by 1–2% agarose gel
electrophoresis and the bands were visualized using GoldView I
Nuclear Staining Dyes (Beijing Solarbio Science & Technology
Co., Ltd.). Subsequently, the pET28a-HMGB1 and pET28a-HMGB1-M
plasmids were employed for PCR amplification with transferring
primers, according to the identifying PCR conditions above. The PCR
products and pEGFP-N1 plasmid (Clontech; Takara Bio, Inc., Otsu,
Japan) were digested with XhoI (New England BioLabs, Inc., Ipswich,
MA, USA) and BamHI (New England BioLabs, Inc.), then the PCR
products and pEGFP-N1 plasmid were linked with T4 DNA Ligase (New
England BioLabs, Inc.), according to the manufacturer's protocols.
Reconstructed pEGFP-N1-HMGB1 and pEGFP-N1-HMGB1-M plasmids were
first screened by PCR with transferring primers according to the
identifying PCR conditions above, and double enzyme digestion
identification was performed with XhoI and BamHI. All
constructs were immediately sent to company (Thermo Fisher
Scientific, Inc.) for direct DNA sequencing. The coding sequence of
mouse HMGB1 (GenBank: BC006586.1; http://www.ncbi.nlm.nih.gov/genbank/) was used for
sequencing blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The primers used were: Mutant primer 1, HMGB1,
forward 5′-CAGCAGCAGCACCCGGATGCTTCTGTCAAC-3′, reverse
5′-TGCTGCTGCTGGTGCTCCTCCCGGCAAG-3′; mutant primer 2, HMGB1, forward
5′-AGCCAGCAACAGAAGGAAGAGGAAGATGATGAG-3′, reverse,
5′-CTTCTGTTGCTGGCTCTTTTCAGCCTTGAC-3′; identifying primer 1, forward
5′-GGGAGGAGCACCAGCAGC-3′, reverse 5′-CAGGATGCTCGCCTTTGATT-3′;
identifying primer 2, forward 5′-GCCGGGAGGAGCACCAGC-3′, reverse
5′-TCATCTTCCTCTTCCTGCTGTTG-3′ and transferring primer, forward
5′-CCCTCGAGATGGGCAAAGGAGATCCTAAGA-3′ and reverse,
5′-CGGGATCCCGTTCATCATCATCATCTTCTTCTTCA-3′. The nucleotides in bold
font indicate mutant sites.
Expression and purification of
recombinant HMGB1 and HMGB1-M
The pET28a-HMGB1 and pET28a-HMGB1-M plasmids (50 ng)
were transformed into 100 µl protease-deficient Escherichia
coli, strain BL21 (Beijing Solarbio Science & Technology
Co., Ltd.) at 42 °C for 90 sec, according to the DH5α
transformation protocol, The bacteria were incubated in ZYP-5052
medium (Amresco, LLC, Solon, OH, USA) containing kanamycin (50
µg/ml) overnight at 37°C with vigorous agitation. The bacterial
solution was centrifuged at 4°C, 8,000 × g for 10 min. Then, cell
lysis and purification of the recombinant protein were performed
with 1 ml HisTrap FF Crude columns (GE Healthcare Life Sciences,
Little Chalfont, UK), according to the manufacturer's protocols.
The affinity-purified recombinant proteins were further desalted
using Zeba Spin Desalting Columns (Pierce; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols. The
recombinant proteins were further purified by filtration and stored
in aliquots at −80°C until use. Contaminating lipolysaccharide (LPS
from protein preparations) was detected by a Pierce LAL Chromogenic
Endotoxin Quantitation kit (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocols. The LPS content in
purified HMGB1 and HMGB1-M samples was 1.31 and 1.11 pg/µl,
respectively. For all DC or RAW264.7 stimulation experiments with
the recombinant proteins, polymyxin B (Sigma-Aldrich; Merck KGaA)
was added to the cell culture medium at 6 units of polymyxin B per
picogram of LPS. Proteins were detected by Coomassie blue staining
at room temperature overnight following SDS-PAGE gel and protein
purity was determined using BandScan software version 5.0 (Glyko,
Novato, CA, USA).
Western blot analysis
Equal volumes (4 µl) of purified HMGB1 (0.32 µg/µl)
or HMGB1-M (0.19 µg/µl) proteins were separated by 12% SDS-PAGE.
The protein concentration was determined in duplicate wells with a
BCA protein assay kit (Thermo Fisher Scientific, Inc.), according
to the manufacturer's protocols. Proteins were transferred to a
0.45-µm polyvinylidene difluoride membrane, the membranes were
incubated with 5% nonfat dry milk for 2 h at room temperature.
Then, blots were incubated with rabbit monoclonal anti-HMGB1
(1:10,000; cat. no. ab79823, Abcam, Cambridge, MA, USA) overnight
at 4°C. The protein bands were visualized with Pierce™ ECL Plus
Western Blotting Substrate (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocols following incubation with
goat anti-rabbit IgG-horseradish peroxidase-conjugated secondary
antibodies (1:5,000; cat. no. sc-2004; Santa Cruz Biotechnology,
Inc. Dallas, TX, USA) for 1 h at room temperature.
Transfection of the recombinant
plasmids into cells and GFP imaging
RAW 264.7 cells (American Type Culture Collection,
Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's
medium (DMEM; HyClone; GE Healthcare) with 10% heat-inactivated
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in
a humidified 5% CO2 incubator at 37°C. The day prior to
transfection, 5×105 cells were plated on glass
coverslips in 24-well plates. When 90% confluence was achieved,
cells were transfected with 1 µg pEGFP-N1, pEGFP-N1-HMGB1, or
pEGFP-N1-HMGB1-M plasmids, respectively using GenePORTER reagent
(Genlantis, San Diego, CA, USA) according to the manufacturer's
protocols; untransfected cells served as the negative control.
After 48 h following transfection, cells were washed twice with
ice-cold PBS. Then, cells were fixed in 4% paraformaldehyde at room
temperature for 30 min and incubated with DAPI (Invitrogen; Thermo
Fisher Scientific, Inc.) at room temperature for 5 min to stain the
cell nuclei. Images were captured at ×400 magnification using an
inverted fluorescence microscope (CKX41SF; Olympus Corporation,
Tokyo, Japan) with an excitation wavelength of 488 nm, and the
percentage of cells with HMGB1 cytoplasmic accumulation was
manually counted under several randomly selected microscopic
fields. Cell fluorescence intensity was quantified with ImageJ
version 1.4 (National Institutes of Health, Bethesda, MD, USA).
ELISA and phagocytosis assay. A total of
5×105 RAW 264.7 cells were cultured in 24-well chamber
slides in DMEM supplemented with 10% FBS and stimulated with
HMGB1-M (5 µg/ml) or HMGB1 (5 µg/ml) recombinant protein for 16 h
to induce TNF-α secretion in a humidified 5% CO2
incubator at 37°C; untreated cells served as the negative control
(MED group). Culture supernatants were collected and assayed for
TNF-α production with an ELISA kit (cat. no. 88-7324-88;
Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols. Then, the cells were washed twice with
PBS and incubated with fluorescein isothiocyanate (FITC)-dextran
(1,000 µg/ml; Sigma-Aldrich; Merck KGaA) for 2 h in a 5%
CO2 incubator at 37°C. Cells were fixed in 4%
paraformaldehyde at room temperature for 30 min and then incubated
with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature for 5 min to stain the cell nuclei. Images were
captured from ten randomly selected microscopic fields at
magnification ×400 using an inverted fluorescence microscope
(CKX41SF, Olympus Corporation) with an excitation wavelength of 488
nm. Cells were also assessed by flow cytometry (LSR II; BD
Biosciences) and analyzed using FlowJo software version 7.6.1 (Tree
Star Inc., Ashland, OR, USA). The % phagocytosis was the %
FITC+ cells of total mononuclear cells.
Treatment of bone marrow-derived DCs (BMDCs) and
flow cytometry. Mice were sacrificed and disinfected in 75% ethanol
for 5 min. The femur and tibia were harvested to acquire BMDCs from
mouse bone marrow, as described previously (20). After 7 days of culture in RPMI 1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS in a 5% CO2 incubator at 37°C, 1×106
cells were stimulated with HMGB1 (5 µg/ml) or HMGB1-M (5 µg/ml) for
48 h and analyzed with flow cytometry; untreated cells (MED group)
served as the negative control. The cells were incubated with
monoclonal antibodies (mAbs) for 30 min at room temperature, and
the fluorochrome-conjugated mAbs (BD Biosciences, Franklin Lakes,
NJ, USA) used were as follows: Integrin α X (CD11c)-phycoerythrin
(PE)-Cy7, cluster of differentiation (CD)80-FITC, major
histocompatibility complex (MHC)-II-allophycocyanin (eBioscience;
Thermo Fisher Scientific, Inc.), and chemokine receptor type 4-PE
(CXCR4; BD Biosciences). Cells were acquired on a flow cytometer
(LSR II; BD Biosciences) and the data was analyzed using FlowJo
software version 7.6.1 (Tree Star Inc.).
Statistical analysis
Data are presented as the mean ± standard error of
the mean of two or three independent experiments. Statistical
analysis was performed using GraphPad Prism version 5.0 (GraphPad
Software, Inc., La Jolla, CA, USA). The difference between two
groups was performed using two-tailed unpaired Student's t-test.
Comparisons between multiple groups were performed with one-way
analysis of variance and a Newman-Keuls post-hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Preparation of HMGB1 and HMGB1-M
expression plasmids and proteins
The pET28a-HMGB1 plasmid was prepared in the present
study; other plasmids were reconstructed according as presented in
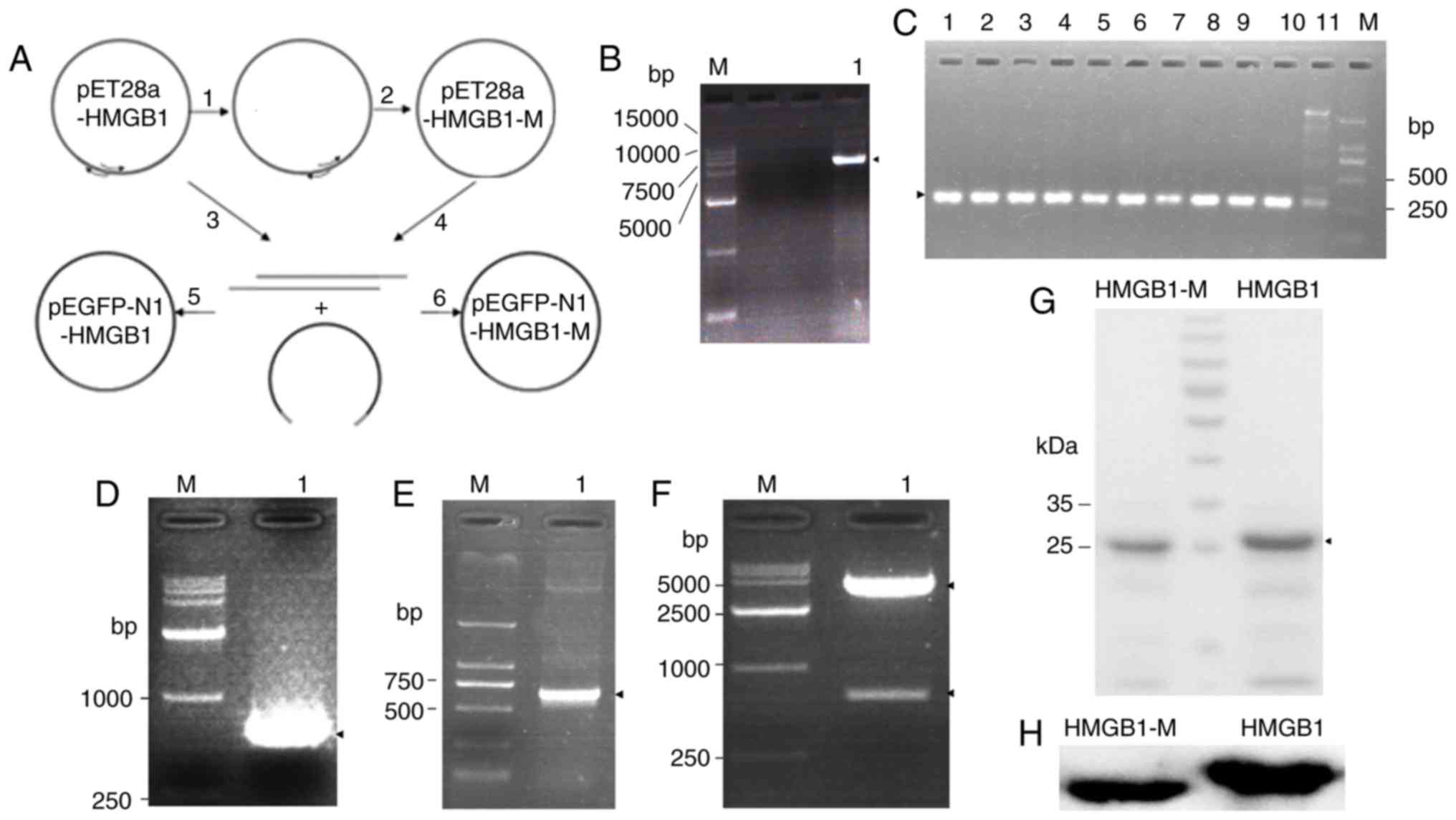
the flowchart (Fig. 1A). The
pET28a-HMGB plasmid was used as template for PCR amplification with
the mutant primer 1 to acquire partly mutant HMGB1 by mutating
lysine into glutamine at positions 27, 28 and 29. The PCR-amplified
circular products (~6 kb) were verified by agarose gel
electrophoresis (Fig. 1B). In the
present study, the aforementioned circular PCR products were
transfected into DH5α competent cells; 10 random transformed
colonies were selected to screen for mutants using PCR
amplification with identifying primer 1, which was designed against
the mutant gene but not the wild-type gene. The mutant samples only
had one bright band (285 bp), whereas the wild-type sample had
multiple bands (Fig. 1C).
Subsequently, one of the mutated plasmids was used as a template
for amplification with mutant primer 2 to acquire mutant HMGB1 via
the mutation of lysine residues 181, 182 and 183 into glutamines as
aforementioned. Random colonies were selected for screening mutants
to perform PCR amplification with identifying primer 2 as described
above (data not shown). Following sequencing analysis of the
plasmids, the pET28a-HMGB1-M plasmid was successfully constructed
(data not shown).
In the present study, two eukaryotic expression
plasmids were reconstructed by transferring HMGB1-M and HMGB1 DNA
fragments from pET28a to pEGFP-N1 plasmids by gene recombination.
Using the pET28a-HMGB1-M plasmid as a template, a HMGB1-M DNA
fragment was acquired by PCR amplification with transferring
primers. PCR products (648 bp) were verified by agarose gel
electrophoresis (Fig. 1D). Then,
PCR products were cloned into pEGFP-N1 plasmids by double enzyme
digestion and ligation reactions. Following the transformation of a
BL21 strain with the reaction mixtures, random colonies were
selected to acquire plasmids. All plasmids were verified by PCR
(Fig. 1E) and restriction enzyme
digestion (Fig. 1F). The
reconstruction process of the pEGFP-N1-HMGB1 plasmid was conducted
in the same manner as that for the pEGFP-N1-HMGB1-M plasmid (data
not shown). The pEGFP-N1-HMGB1-M/HMGB1 plasmids were finally
confirmed by sequencing analysis (data not shown).
To investigate the extracellular functions of the
HMGB1-M protein, pET28a-HMGB1-M/HMGB1-transformed strains were
employed to induce protein expression and the target proteins were
then purified. HMGB1 and HMGB1-M proteins were >80% pure, as
revealed by Coomassie blue staining of the SDS-PAGE gel (Fig. 1G). In addition, the purified
protein was identified by western blot analysis (Fig. 1H). These results indicated that
HMGB1 and HMGB1-M protein were successfully acquired.
HMGB1-M is expressed in the nucleus
and cytoplasm of RAW 264.7 cells
The HMGB1-M-GFP plasmid was transfected into RAW
264.7 cells to observe the intracellular distribution pattern of
this protein. HMGB1-M was expressed in the nucleus and cytoplasm,
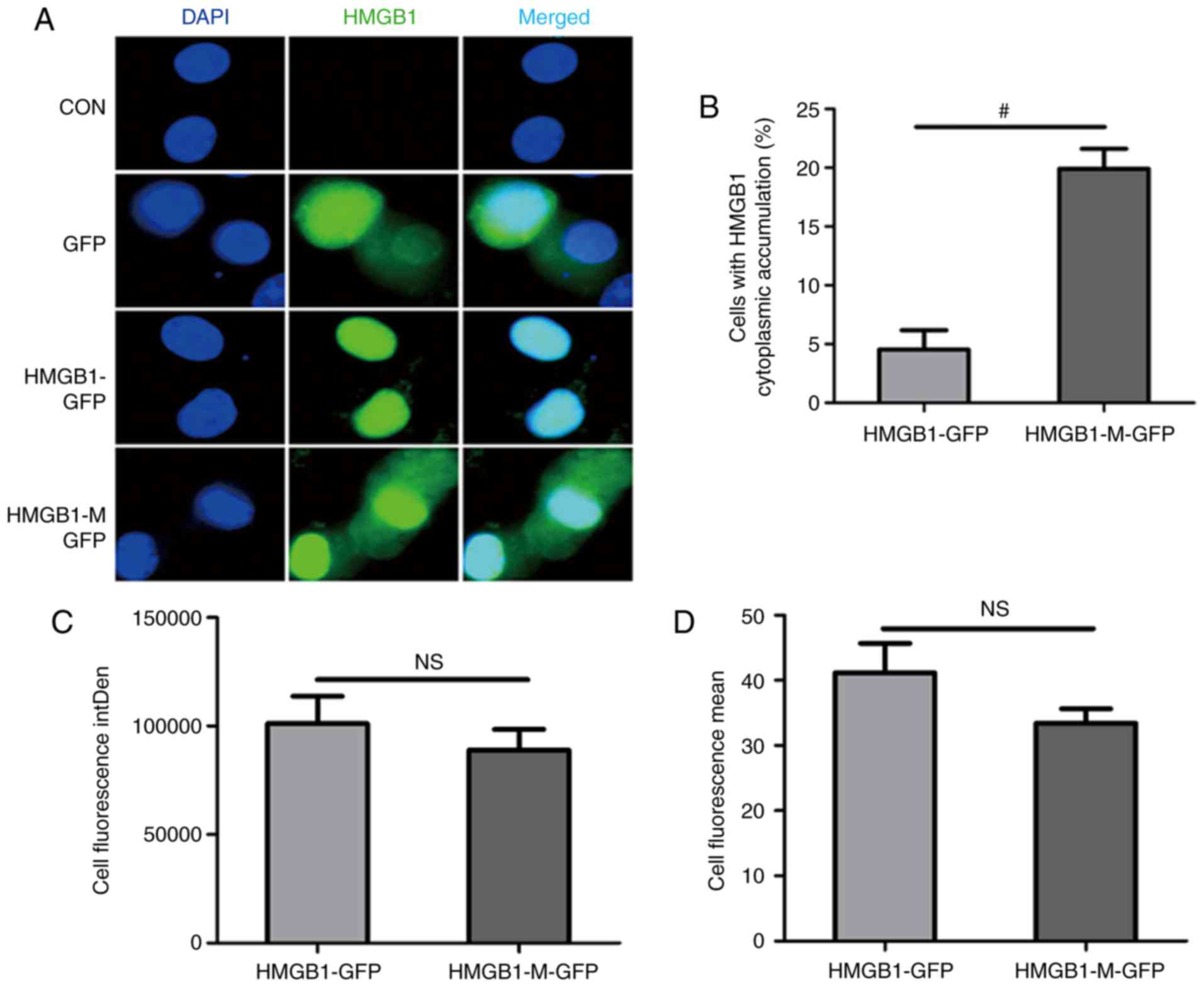
while HMGB1 was concentrated in the nucleus (Fig. 2A). Compared with HMGB1, HMGB1-M
expression significantly increased the percentage of HMGB1
cytoplasmic accumulation in RAW 264.7 cells (Fig. 2B). The integrated (Fig. 2C) and mean fluorescence intensity
(Fig. 2D) between the two groups
were not statistically significant.
HMGB1-M increases TNF-α production in
RAW 264.7 cells
To determine the potential role of HMGB1-M in
regulating the functions of macrophages, the phagocytotic function
and TNF-α secretion of RAW 264.7 cells treated with HMGB1-M or
HMGB1 were investigated. FITC-dextran was phagocytosed by RAW 264.7
cells and did not adhere to its surface (Fig. 3A). HMGB1-M and HMGB1 significantly
impaired the phagocytic activity of RAW 264.7 cells compared with
that in the MED group (Fig. 3B and
C); a significant decrease in the mean fluorescence intensity
(MFI) of FITC-dextran phagocytosis in RAW 264.7 cells was observed
in the HMGB1-M group compared with in the MED group (Fig. 3D). In addition, HMGB1 and HMGB1-M
were also significant inducers of TNF-α production in RAW 264.7
cells compared with in the MED group. Furthermore, HMGB1-M
significantly increased the production of TNF-α in RAW 264.7 cells
than HMGB1 (Fig. 3E). These
results demonstrated that HMGB1-M increased TNF-α production within
RAW 264.7 cells and exerted no significant effects on the
phagocytic potential of macrophages.
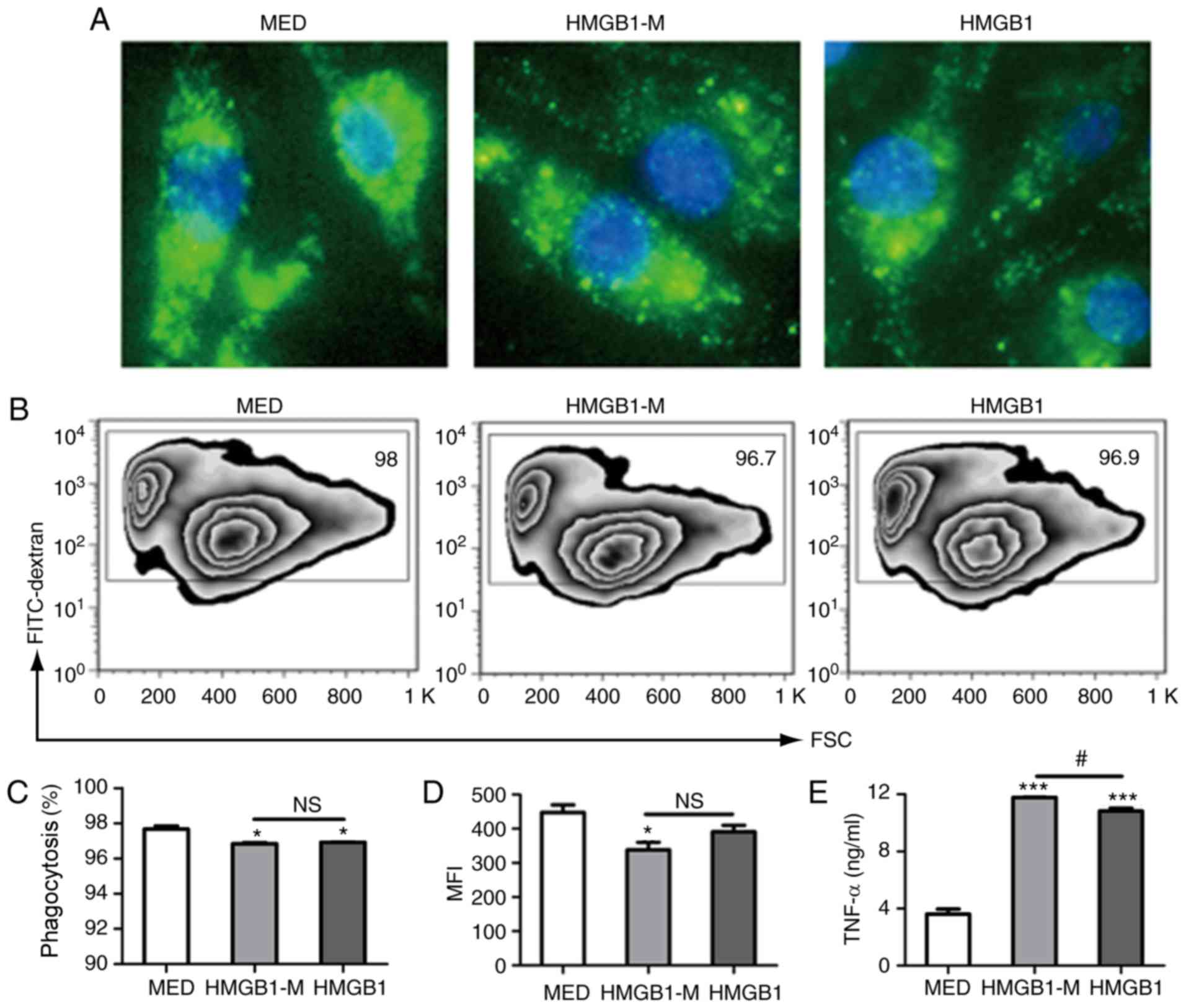 | Figure 3.HMGB1-M increases TNF-α secretion in
RAW 264.7 cells. The cells were stimulated for 16 h with HMGB1-M (5
µg/ml) or HMGB1 (5 µg/ml). Following the collection of culture
supernatants, the cells were incubated with FITC-dextran for 2 h.
(A) Morphological appearances of RAW 264.7 cells phagocytosing
FITC-dextran were assessed by fluorescence microscopy.
Magnification, ×400. (B) Phagocytic activity was analyzed by flow
cytometry. (C) Phagocytic ratio of FITC+ cells. (D) MFI
of cells is indicated. (E) TNF-α concentrations in culture
supernatants were measured by ELISA. *P<0.05, ***P<0.001 vs.
MED, and #P<0.05. The data are representative of
three independent experiments. FITC, fluorescein isothiocyanate;
HMGB1, high mobility group box 1; HMBG1-M, mimicked acetylated
HMGB1; MED, media control; MFI, mean fluorescence intensity; NS,
not significant; TNF-α, tumor-α. |
HMGB1-M decreases phenotypic
maturation of BMDCs
The present study evaluated the role of HMGB1-M in
BMDC function. HMGB1-M and HMGB1 was applied respectively to
stimulate immature BMDC for 48 h. The frequency of
CD11c+ cells among the total mononuclear cells and
percentage of CD11c+ cells expressing CD80, MHC-II or CXCR4 were
evaluated. Compared with the MED group, HMGB1-M and HMGB1 increased
the expression frequency of MHC-II and CD80 on BMDCs; however, the
frequency of CD11c expression was notably unaltered (Fig. 4A-C). Additionally, the present
study measured the expression of CXCR4 on BMDCs following
stimulation (Fig. 4D). Compared
with HMGB1, HMGB1-M significantly decreased the MFI of CD11c but
not the percentage of CD11c+ cells (Fig. 4E); the percentage and MFI of
MHC-II-expressing BMDCs was significantly decreased with HMGB1-M
compared with HMGB (Fig. 4F).
HMGB1-M exhibited no significant effects on the expression of CD80
compared with HMGB (Fig. 4G). It
has been demonstrated that HMGB1 is required for the migration of
maturing DCs due to upregulation of CXCR4 expression on BMDCs
(21). Compared with the MED
group, HMGB1-M and HMGB1 significantly promoted the percentage of
cells expressing CXCR4; however, no significant differences were
found between HMGB1-M and HMGB1 in the expression of CXCR4 on BMDCs
(Fig. 4H). These results
demonstrated that HMGB1-M downregulated phenotypic maturation of
BMDCs as demonstrated by the decreased MHC-II expression; however,
no notable effects on the expression of CD80 and CXCR4 on BMDCs
were observed.
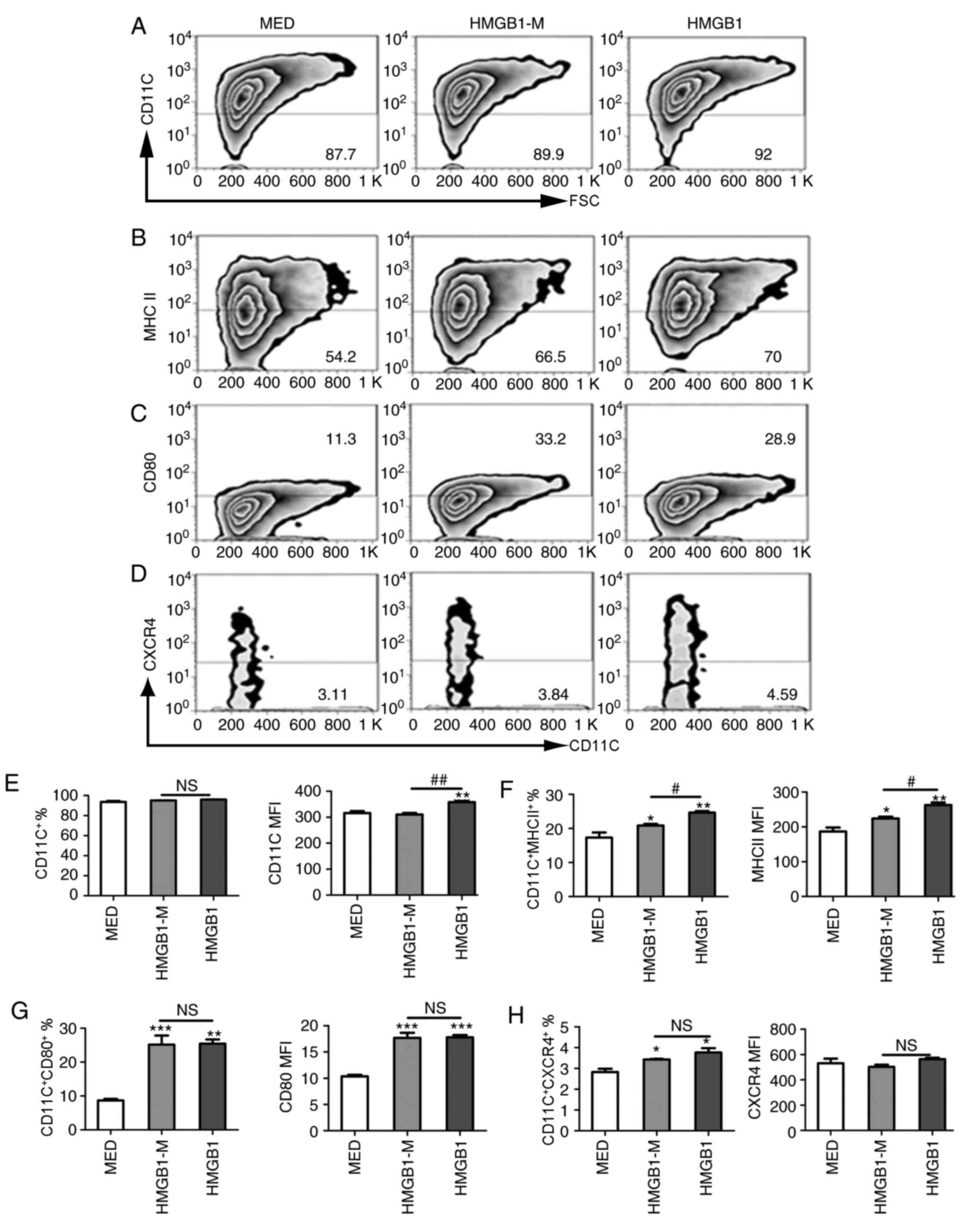 | Figure 4.HMGB1-M suppresses phenotypic
maturation of BMDCs. The cells were treated with HMGB1-M (5 µg/ml)
or HMGB1 (5 µg/ml) for 48 h. (A-D) BMDCs were collected and surface
molecule expression was assessed by flow cytometry. (E-H) Frequency
of CD11c+ among the total mononuclear cells and
percentage of CD80, MHC-II or CXCR4 expression of CD11c+ subsets
(left) and MFI is indicated (right). *P<0.05, **P<0.01 and
***P<0.001 vs. MED, and #P<0.05,
##P<0.01. The data are representative of three
independent experiments. BMDCs, bone marrow-derived dendritic
cells; CD80, cluster of differentiation 80; HMGB1, high mobility
group box 1; HMBG1-M, mimicked acetylated HMGB1; MED, media
control; MFI, mean fluorescence intensity; NS, not significant. |
Discussion
In the present study, a HMGB1-M plasmid was
successfully constructed to mimic the acetylated lysine residues of
HMGB1. HMGB1, as an extracellular mediator, serves an important
role within macrophages and DCs to facilitate the immune response
(9,10). HMGB1-M, compared with HMGB1,
increased TNF-α production within RAW 264.7 cells and suppressed
BMDC maturation, which may respectively affect the responses to
inflammation and antigen presentation.
Point mutation technology is a universal research
method for studying the function of proteins (22). To obtain the HMGB1-M protein, the
pET28a-HMGB1-M plasmid was constructed, which expressed the HMGB1-M
protein with mutated lysine residues, which were converted into
glutamines at positions 27, 28, 29, 181, 182 and 183. This mutation
process involved a total of six point mutations, in which three
mutations were conducted each time. Of note, the final PCR products
were inevitably mixed with some parental strands and undesired
mutations following DpnI digestion. A few modifications
based on other mutation methods were performed in the present study
(23–25), which included reducing the
concentration of the parent template at the beginning of the PCR
mutation reaction and constructing a pair of identification primers
to perform the preliminary screening of mutants following
site-directed mutagenesis via PCR. The forward primer contained
three mutant base sites, with the first base at the 3′ terminus
starting from the mutated site, and the first base at the 5′
terminus of the reverse primer, which started from base 335 in the
HMGB1 coding sequence. The pair of identifying primers had the
following characteristics: At least one of the forward and reverse
primers contained three point mutation sites at the 3′ terminus.
These optimizations for mutation may aid the generation of
mutations in future investigations.
The modification of acetylated HMGB1 is important
for its subcellular relocation and release into the extracellular
space (4). The two major clusters
(NLS1 and NLS2) of HMGB1 lysine residues are regions of high
acetylation. Lysine hyperacetylation within two major clusters was
reported to affect the subcellular relocation of HMGB1 (15). The mutation of six lysine residues
(positions: 27, 28, 29, 181, 182 and 183) into glutamines within
two major clusters, which mimicked acetylated lysine, was
associated with the translocation of HMGB1 from the nucleus to the
cytoplasm (15). HMGB1-M acquired
by mutation does not possess lysine acetylation in the HMGB1
protein via post-translational modifications (15). Treatment with histone deacetylase
inhibitors was reported to increase HMGB1 translocation to the
cytoplasm (15). These data are
consistent with the results of the present study, which
demonstrated that the HMGB1-M protein with mutations of lysine
residues at positions 27, 28, 29, 181, 182 and 183 into glutamines
was associated with increased cytoplasmic localization of HMGB1-M
in RAW 264.7 cells than the HMGB1 protein. In addition, the
molecular weight of the HMGB1-M protein band was slightly different
from that of HMGB1 in the present study, as the molecular weight of
substitutable glutamine (146.1456 g/mol) is lower than lysine
(146.1888 g/mol). Therefore, the mimicked multisite-acetylated
HMGB1 was selected as an alternative approach to investigate
acetylated HMGB1 in the present study to determine its
extracellular function.
Previous studies have reported that actively
released HMGB1 from macrophages undergoes post-translational
modifications, such as acetylation (15,19);
however, HMGB1 released from necrotic cells is not
post-translationally modified (4,9,26).
Therefore, there are likely two forms of HMGB1 (acetylated and
nonacetylated HMGB1) present in the extracellular environment. In
the present study, the effects of HMGB1-M with on macrophages and
DCs were compared with that of HMGB1. Subsequent analyses on TNF-α
production and phagocytic potential in RAW 264.7 cells, BMDC
maturation, and CXCR4 expression were conducted. Few investigations
into the functions of HMGB1-M and HMGB1 were conducted in the
present study; however, notable variations were reported. Compared
with HMGB1, HMGB1-M increased TNF-α release from RAW 264.7 cells
and decreased BMDC maturation, with no marked effects on phagocytic
potential of macrophages, or CD80 and CXCR4 expression. These
results suggested that HMGB1-M may serve a proinflammatory role,
whereas HMGB1 is more likely to serve a role in antigen
presentation. The reported variations between HMGB1-M and HMGB1
were notable; but may serve different roles in the regulation of
the immune response. Therefore, further investigation into these
associations in vivo are required in the future.
In conclusion, the present study employed a simple
and efficient method to induce mutations to conduct functional
studies of genes and proteins, which may be used in future
investigations; identifying primers were generated in the present
study to screen mutants. The results of the present study suggested
that mimicked acetylation may partly affect the extracellular
response of HMGB1 to inflammation and antigen presentation.
Therefore, the findings of the present study may provide novel
insight into the role of acetylated HMGB1 for the regulation of
immune responses in inflammatory diseases.
Acknowledgements
The authors would like to thank Mr. Bin Zhang, Dr
Hongping Ba, Dr Yan Sun, Dr Huijuan Zou, Dr Bingxia Ming, Dr Ming
Gao and Miss Lifeng Shi from the Department of Immunology, School
of Basic Medicine, Tongji Medical College, Huazhong University of
Science and Technology (Wuhan, China) for their technical
assistance with this study.
Funding
This study was supported by the National Natural
Science Foundation of China (NSFC grant nos. 31670876 and
31470852), and the Major State Basic Research Development Program
of China (973 Program, grant no. 2013CB530505).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CX was responsible for the concept and design of the
study, acquisition of data, analysis and interpretation of data,
drafting the article. HX, XY and XP designed the study. TZ and GF
interpreted the data. ZF contributed to the concept and design of
the study, interpretation of data, and critical revision of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Use Committee of Tongji Medical College, HUST (Wuhan,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ueda T, Chou H, Kawase T, Shirakawa H and
Yoshida M: Acidic C-tail of HMGB1 is required for its target
binding to nucleosome linker DNA and transcription stimulation.
Biochemistry. 43:9901–9908. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang H, Bloom O, Zhang M, Vishnubhakat JM,
Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, et
al: HMG-1 as a late mediator of endotoxin lethality in mice.
Science. 285:248–251. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu B, Nakamura T, Inouye K, Li J, Tang Y,
Lundbäck P, Valdes-Ferrer SI, Olofsson PS, Kalb T, Roth J, et al:
Novel role of PKR in inflammasome activation and HMGB1 release.
Nature. 488:670–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Q and Wang Y: HMG modifications and
nuclear function. Biochim Biophys Acta. 1799:28–36. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang D, Kang R, Livesey KM, Cheh CW,
Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ III
and Lotze MT: Endogenous HMGB1 regulates autophagy. J Cell Biol.
190:881–892. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Janko C, Filipovic M, Munoz LE, Schorn C,
Schett G, Ivanović-Burmazović I and Herrmann M: Redox modulation of
HMGB1-related signaling. Antioxid Redox Signal. 20:1075–1085. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang D, Kang R, Zeh HJ III and Lotze MT:
High-mobility group box 1, oxidative stress, and disease. Antioxid
Redox Signal. 14:1315–1335. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Venereau E, Casalgrandi M, Schiraldi M,
Antoine DJ, Cattaneo A, De Marchis F, Liu J, Antonelli A, Preti A,
Raeli L, et al: Mutually exclusive redox forms of HMGB1 promote
cell recruitment or proinflammatory cytokine release. J Exp Med.
209:1519–1528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dumitriu IE, Baruah P, Manfredi AA,
Bianchi ME and Rovere-Querini P: HMGB1: Guiding immunity from
within. Trends Immunol. 26:381–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Messmer D, Yang H, Telusma G, Knoll F, Li
J, Messmer B, Tracey KJ and Chiorazzi N: High mobility group box
protein 1: An endogenous signal for dendritic cell maturation and
Th1 polarization. J Immunol. 173:307–313. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andersson U and Tracey KJ: HMGB1 is a
therapeutic target for sterile inflammation and infection. Annu Rev
Immunol. 29:139–162. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sterner R, Vidali G and Allfrey VG:
Studies of acetylation and deacetylation in high mobility group
proteins. Identification of the sites of acetylation in HMG-1. J
Biol Chem. 254:11577–11583. 1979.PubMed/NCBI
|
|
13
|
Ugrinova I, Pasheva EA, Armengaud J and
Pashev IG: In vivo acetylation of HMG1 protein enhances its binding
affinity to distorted DNA structures. Biochemistry. 40:14655–14660.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alexandrova EA and Beltchev BG: Acetylated
HMG1 protein interacts specifically with homologous DNA polymerase
alpha in vitro. Biochem Biophys Res Commun. 154:918–927. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bonaldi T, Talamo F, Scaffidi P, Ferrera
D, Porto A, Bachi A, Rubartelli A, Agresti A and Bianchi ME:
Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect
it towards secretion. EMBO J. 22:5551–5560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Assenberg R, Webb M, Connolly E, Stott K,
Watson M, Hobbs J and Thomas JO: A critical role in
structure-specific DNA binding for the acetylatable lysine residues
in HMGB1. Biochem J. 411:553–561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Elenkov I, Pelovsky P, Ugrinova I,
Takahashi M and Pasheva E: The DNA binding and bending activities
of truncated tail-less HMGB1 protein are differentially affected by
lys-2 and lys-81 residues and their acetylation. Int J Biol Sci.
7:691–699. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lange SS, Mitchell DL and Vasquez KM: High
mobility group protein B1 enhances DNA repair and chromatin
modification after DNA damage. Proc Natl Acad Sci USA.
105:10320–10325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu B, Antoine DJ, Kwan K, Lundbäck P,
Wähämaa H, Schierbeck H, Robinson M, Van Zoelen MA, Yang H, Li J,
et al: JAK/STAT1 signaling promotes HMGB1 hyperacetylation and
nuclear translocation. Proc Natl Acad Sci USA. 111:3068–3073. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lutz MB, Kukutsch N, Ogilvie AL, Rössner
S, Koch F, Romani N and Schuler G: An advanced culture method for
generating large quantities of highly pure dendritic cells from
mouse bone marrow. J Immunol Methods. 223:77–92. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dumitriu IE, Bianchi ME, Bacci M, Manfredi
AA and Rovere-Querini P: The secretion of HMGB1 is required for the
migration of maturing dendritic cells. J Leukoc Biol. 81:84–91.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsieh PC and Vaisvila R: Protein
engineering: Single or multiple site-directed mutagenesis. Methods
Mol Biol. 978:173–186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aiyar A, Xiang Y and Leis J: Site-directed
mutagenesis using overlap extension PCR. Methods Mol Biol.
57:177–191. 1996.PubMed/NCBI
|
|
24
|
Zheng L, Baumann U and Reymond JL: An
efficient one-step site-directed and site-saturation mutagenesis
protocol. Nucleic Acids Res. 32:e1152004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Wu T, Song J, Chen XL, Zhang Y and
Wan Y: A mutant screening method by critical annealing
temperature-PCR for site-directed mutagenesis. BMC Biotechnol.
13:212013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scaffidi P, Misteli T and Bianchi ME:
Release of chromatin protein HMGB1 by necrotic cells triggers
inflammation. Nature. 418:191–195. 2002. View Article : Google Scholar : PubMed/NCBI
|