Introduction
Owing to changes in lifestyle and dietary structure
in modern society, the incidence of obesity has gradually increased
worldwide (1). Obesity, a lipid
metabolism disorder, can lead to an elevated level of free fatty
acids (FFAs) (2), which have been
reported to be associated with an increased risk and severity of
heart failure (3). Furthermore,
our previous study (4) and other
researchers (5,6) have demonstrated that palmitic acid
(PA), a major component of FFAs, significantly reduces the
viability of cardiomyocytes and induces myocardial injury in
vitro and in vivo. Therefore, PA-induced myocardial
injury is considered to be one of the factors contributing to
obesity-induced impairment of the heart. However, the mechanism of
PA-induced myocardial injury has not been completely
elucidated.
A long noncoding RNA (lncRNA) is defined as an RNA
transcript of >200 nucleotides in length that regulates gene
expression at multiple levels (7).
For instance, the lncRNAs H19 (8)
and X-inactive specific transcript (Xist) (9) are involved in genomic imprinting,
which is a type of epigenetic regulation. Furthermore, the lncRNA
PANDA participated in transcriptional regulation via a molecular
decoy (10). In addition, an
increasing number of lncRNAs have been reported to regulate gene
expression at the post-transcriptional level as a competitive
endogenous RNA (ceRNA) (11–13).
Therefore, lncRNAs may provide novel insight into the underlying
mechanism of PA-induced myocardial injury.
Growth arrest-specific transcript 5 (GAS5) is a
well-known lncRNA that was initially identified in mouse NIH 3T3
cells (14). GAS5 has been
reported to be involved in multiple pathological processes,
including tumorigenesis and metastasis (15,16),
ischemic stroke (17), hepatic and
cardiac fibrosis (18,19), and the development of
osteoarthritis (20). Notably,
several lncRNAs have been reported to be involved in lipotoxicity.
For instance, metastasis-associated lung adenocarcinoma transcript
1 promotes PA-induced lipid accumulation in hepatocytes by
increasing the stability of the sterol regulatory element binding
transcription factor 1 protein (21), while growth arrested DNA-damage
inducible gene 7 is a key regulator of PA-induced oxidative stress
and cell death in cultured fibroblasts (22). However, whether GAS5 serves a role
in PA-induced myocardial injury remains elusive.
The aim of the present study was to explore the role
of GAS5 in PA-induced myocardial injury and its underlying
mechanism of action. The results demonstrated that GAS5 was
significantly upregulated in cardiomyocytes following treatment
with PA, while knockdown of GAS5 expression alleviated PA-induced
myocardial injury and inflammatory activation. Mechanistically,
GAS5 was identified to act as a ceRNA to regulate high mobility
group box 1 (HMGB1) expression by binding with microRNA (miR)-26a,
subsequently inhibiting nuclear factor (NF)-κB activation.
Materials and methods
Cell culture and PA treatment
The rat myocardial H9c2 cell line was obtained from
the Shanghai Institutes for Biological Sciences (Shanghai, China).
Cells were grown in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (TBD Science, Tianjin, China), 100 units of
penicillin/ml and 100 µg of streptomycin/ml in a humidified
atmosphere at 37°C with 5% CO2. H9c2 cells were treated
with 400 µM PA (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for
consecutive 6, 12 or 24 h to induce myocardial lipotoxic injury
according to the previously published study (4).
Oil Red O staining
Following the aforementioned treatment with PA for
24 h, cells were fixed in 4% paraformaldehyde for 30 min at room
temperature. Following washing three times with phosphate-buffered
saline, cells were incubated with 3 mg/ml Oil Red O working
solution (Sigma-Aldrich; Merck KGaA) for 30 min at room
temperature. Cell nuclei were then stained with hematoxylin for 2
min at room temperature, followed by imaging with a light
microscope (Olympus Corporation, Tokyo, Japan).
Cell transfection
GAS5-specific small interfering RNA (siRNA) (sense,
5′-TCTCACAGGCAGTTCTGTGG-3′, antisense, 5′-ATCCATCCAGTCACCTCTGG-3′),
scrambled siRNA [serving as the negative control (NC)], (sense,
5′-UUCUCCGAACGUGUCACGUTT-3′, antisense,
5′-ACGUGACACGUUCGGAGAATT-3′) miR-26a mimics
(5′-UUCAAGUAAUCCAGGAUAGGCU-3′), mimics NC
(5′-UUCUCCGAACGUGUCACGUTT-3′), miR-26a inhibitor
(5′-AGCCTATCCTGGATTACTTGAA-3′) and inhibitor NC
(5′-CAGUACUUUUGUGUAGUACAA-3′) were designed and synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China). Briefly, cells
were transfected with the 5 µl siRNAs (20 µM) and 200 µl miRNA
nucleotide sequences (100 nM) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. At 24 h post-transfection, cells were then
washed and cultured in fresh medium, followed by treated with 400
µM PA for 24 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. RNA concentration and A260/280 was
measured using a NanoPhotometer P-Class (Implen GmbH, Munich,
Germany). To detect mRNA expression levels, the PrimeScript RT
reagent kit with gDNA Eraser (Takara Bio, Inc., Otsu, Japan) and
SYBR Premix Ex Taq II (Takara Bio, Inc.) were used for RT and qPCR,
respectively, according to the manufacturer's protocol. To detect
miR-26a expression, Mir-X™ miRNA First Strand Synthesis kit and
Mir-X™ miRNA RT-qPCR SYBR® kit (Takara Bio, Inc., Otsu,
Japan) were used for RT and qPCR, respectively, according to the
manufacturer's protocol. PCR was performed at 42°C for 30 min; 95°C
for 10 min, followed by 45 cycles of amplification at 95°C for 20
sec, 62°C for 30 sec, 72°C for 30 sec. The melt curve stage was
performed at 95°C for 15 sec, 60°C for 1 min, 95°C for 15 sec.
GAPDH and U6 were used as the internal controls in the detection of
mRNA and miRNA expression levels. All oligonucleotide primers were
designed by Shanghai Sangon Biotech Co., Ltd. (Table I). The relative expression was
analyzed using 2−ΔΔCq method (23).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Name | Sequences
(5′-3′) |
|---|
| GAS5 | Forward:
TCTCACAGGCAGTTCTGTGG |
|
| Reverse:
ATCCATCCAGTCACCTCTGG |
| TNF-α | Forward:
GAAGAGAACCTGGGAGTAGATAAGG |
|
| Reverse:
GTCGTAGCAAACCACCAAGC |
| IL-1β | Forward:
TCGTTGCTTGTCTCTCCTTG |
|
| Reverse:
AAAAATGCCTCGTGCTGTCT |
| HMGB1 | Forward:
CAAACCTGCCGGGAGGAGCA |
|
| Reverse:
TCTTTCATAACGAGCCTTGTCAGCC |
| GAPDH | Forward:
ATCATCAGCAATGCCTCC |
|
| Reverse:
CATCACGCCACAGTTTCC |
| miR-26a | RT:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCCAG |
|
| Forward:
AAGGAGAACCCGTAGATCCG |
|
| Reverse:
GTGCAGGGTCCGAGGTATTC |
Cell Counting Kit (CCK)-8 assay
Cells were seeded at 5×103 cells/well in
96-well plates. Following transfection for 24 h, the cell viability
was determined by CCK-8 assay (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) according to the manufacturer's protocol.
The absorbance value was detected at 450 nm by an Epoch 2
Microplate Spectrophotometer (BioTek Instruments, Inc., Winooski,
VT, USA).
Lactate dehydrogenase (LDH) assay
Cells were seeded at 1×105 cells/well in
24-well plates. Following transfection for 24 h, LDH released from
the cardiomyocytes into the culture medium was measured using an
LDH assay kit (cat. no. A020-1; Nanjing Jiancheng Bioengineering
Institute, Nanjing, China) according to the manufacturer's
protocol. The absorbance value was detected at 450 nm by an Epoch 2
Microplate Spectrophotometer (BioTek Instruments, Inc., Winooski,
VT, USA).
Enzyme-linked immunosorbent assay
(ELISA) for measuring cytokine and NF-κB activities
Cells were seeded at 1×105 cells/well in
24-well plates. Following transfection for 24 h, the concentrations
of tumor necrosis factor α (TNF-α) and interleukin-1β (IL-1β) in
the culture medium were detected using commercial rat TNF-α or
IL-1β ELISA kits (cat. no. EK3822/2 and EK301B2/2, respectively;
Hangzhou MultiSciences Biotech Co., Ltd., Hangzhou, China)
according to the manufacturer's protocol. To measure the activity
of NF-κB, cells were harvested after transfection and PA treatment,
and nuclear extract lysates were extracted from harvested cells
using Nuclear Extraction kit (cat. no. ab113474; Abcam, Cambridge,
UK), according to the manufacturer's protocol. The activity of
NF-κB in nuclear extract lysates was detected using NF-κB p65
transcription factor assay kit (cat. no. ab133112; Abcam) according
to the recommended experimental protocol.
Bioinformatics analysis
starBase v2.0 (http://starbase.sysu.edu.cn/index.php) and LncBase
Predicted v.2 tool (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted)
were employed to predict the binding cites between GAS5 and
miRNAs.
Luciferase activity assay
Luciferase reporter plasmids (pmirGLO-GAS5-wt and
pmirGLO-GAS5-mut) were obtained from Shanghai GenePharma Co., Ltd.
Briefly, 2×105 cells were plated into each well of a
24-well plates and transfected with pmirGLO-GAS5-wt or
pmirGLO-GAS5-mut together with miR-26a mimics or mimics NC, as
previously described (18). At 48
h post-transfection, luciferase activity was analyzed using the
Dual-Glo Luciferase assay system (Promega Corporation, Madison, WI,
USA) according to the manufacturer's protocol.
Western blotting analysis
Total proteins were extracted with RIPA buffer
(Beyotime Institute of Biotechnology, Shanghai, China). The nuclear
and cytoplasmic proteins were separately extracted using Nuclear
and Cytoplasmic Protein Extraction kit (Beyotime Institute of
Biotechnology), according to the manufacturer's protocol, and the
protein concentration was measured by Enhanced BCA Protein Assay
kit (Beyotime Institute of Biotechnology), according to the
manufacturer's protocol. Next, 50 µg heat-denatured proteins were
separated by 8% or 10% SDS-PAGE and then transferred to
polyvinylidene difluoride membranes, followed by blocking with 1%
bovine serum albumin solution at room temperature for 1 h. The
membranes were then incubated at 4°C overnight with primary
antibodies, including anti-HMGB1 (cat. no. ab128129; 1:1,000;
Abcam), anti-NF-κB p65 (cat. no. ab193238; 1:1,000; Abcam),
anti-GAPDH (cat. no. TA336768; 1:1,000; Zhongshan Jinqiao
Biotechnology, Beijing, China) and anti-Lamin A (cat. no. ab26300;
1:1,000; Abcam), followed by incubation with horseradish peroxidase
conjugated goat anti-rabbit or goat anti-mouse immunoglobulin G
(cat. no. E030120-01; 1:4,000; EarthOx Life Sciences, Millbrae, CA,
USA) at room temperature for 30 min. Detection of protein bands was
performed using the enhanced chemiluminescence for western blotting
kit (Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. GAPDH or Lamin A was used as an internal
control. Relative protein expression was determined by densitometry
using Image J2× analysis software (National Institutes of Health,
Bethesda, MD, USA).
Statistical analysis
All experiments were repeated three times. Data are
presented as the mean ± standard error of the mean. Statistical
analysis was performed with SPSS version 17.0 software (SPSS, Inc.,
Chicago, IL, USA). Differences between groups were initially
evaluated using one-way analysis of variance, and multiple
comparison analysis was further performed in the case that
differences were statistically significant using Fisher's least
significant difference test. P<0.05 was considered to indicate a
statistically significant difference.
Results
GAS5 expression is induced by PA
treatment in H9c2 myocardial cells
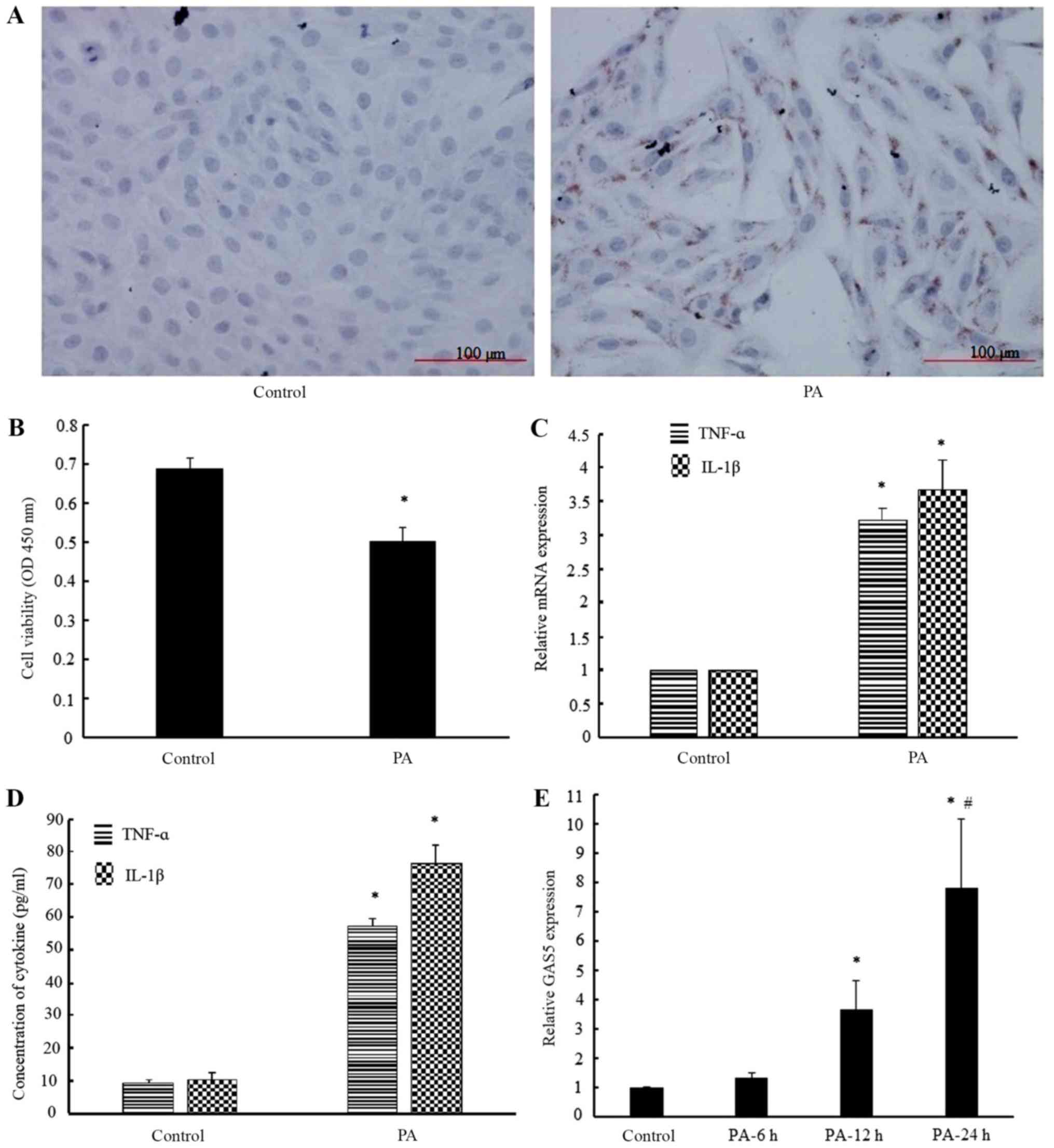
Oil Red O staining illustrated that lipids were
accumulated in H9c2 cells following treatment with 400 µM PA
(Fig. 1A), and the results of
CCK-8 assay demonstrated that cell viability was markedly decreased
by PA treatment (Fig. 1B), which
suggested that PA induced lipotoxic injury in cardiomyocytes. In
addition, the pro-inflammatory cytokine TNF-α and IL-1β mRNA and
protein levels were significantly increased by PA treatment in H9c2
cells, as compared with the control group (Fig. 1C and D). This further suggests that
PA exposure caused inflammation. Furthermore, the results of
RT-qPCR revealed that the expression of GAS5 in H9c2 cells was
significantly increased by 3.65- and 7.82-fold following treatment
with 400 µM PA for 12 and 24 h, respectively (Fig. 1E). Therefore, these results
demonstrated that GAS5 expression was induced by PA treatment in
H9c2 myocardial cells.
Knockdown of GAS5 expression
alleviates PA-induced myocardial injury
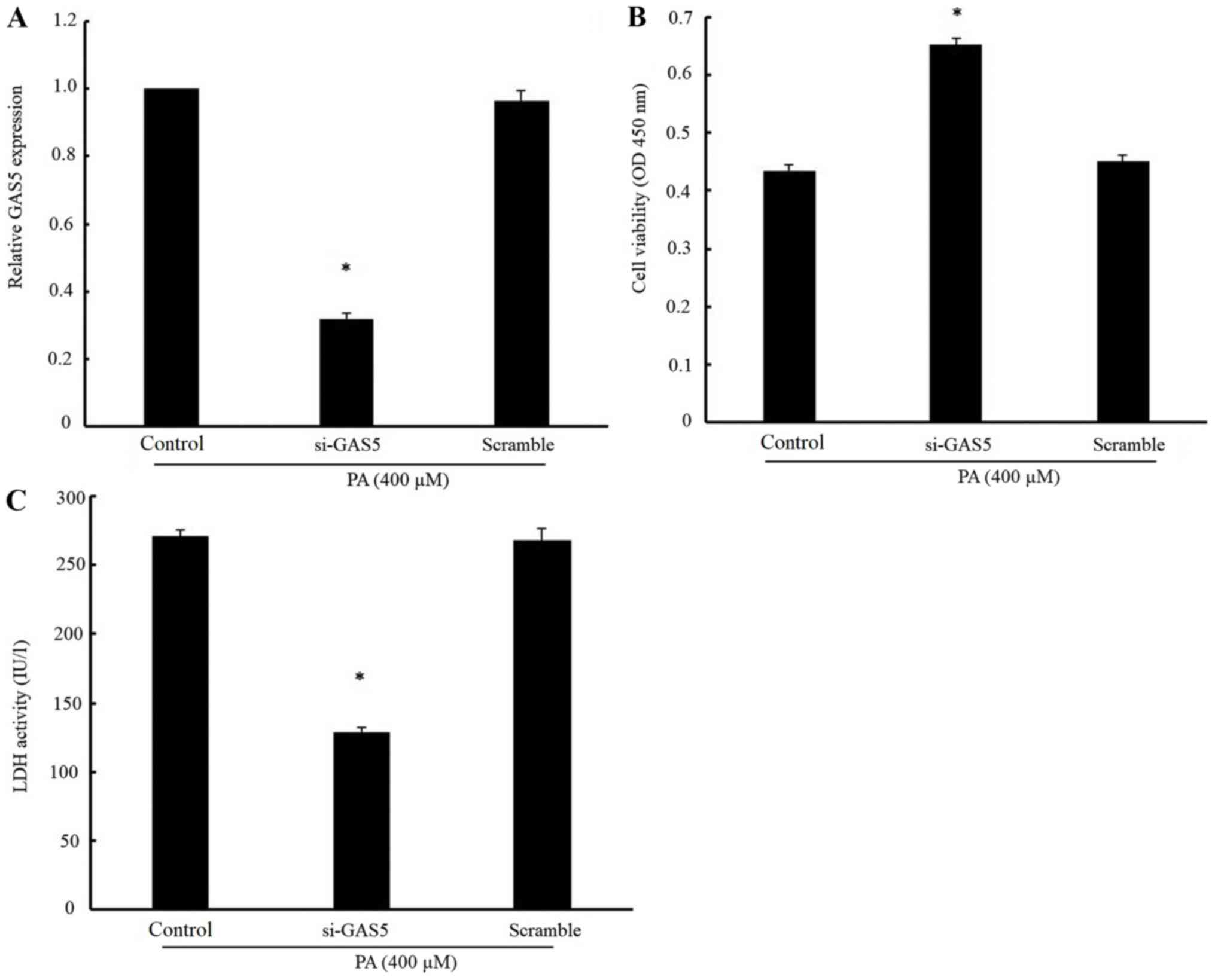
The expression of GAS5 in H9c2 cells was reduced by
nearly 70% following transfection with GAS5-specific siRNA
(Fig. 2A), suggesting that GAS5
siRNA effectively inhibited the endogenous expression of GAS5 in
cardiomyocytes. It was further observed that the cell viability was
markedly increased (Fig. 2B) and
the activity of LDH was significantly decreased by the
downregulation of GAS5 in PA-treated cells (Fig. 2C). Taken together, these results
demonstrated that the knockdown of GAS5 expression alleviated
PA-induced myocardial injury.
Knockdown of GAS5 expression inhibits
PA-induced inflammation
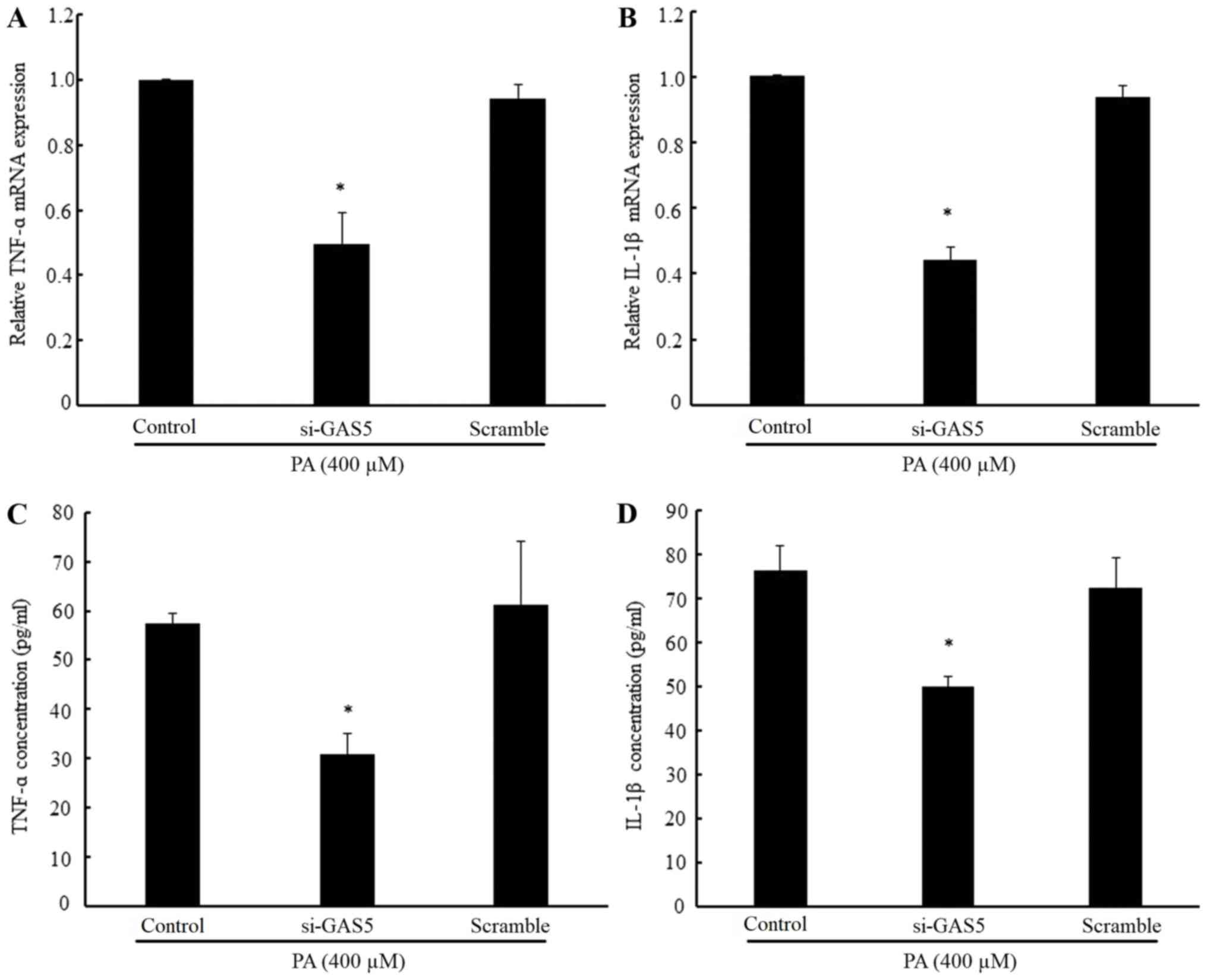
To evaluate whether GAS5 regulated PA-induced
inflammation, the present study detected the mRNA expression and
concentration levels of the pro-inflammatory cytokines TNF-α and
IL-1β. The results demonstrated that the mRNA expression levels of
TNF-α and IL-1β in PA-treated H9c2 cells were notably reduced by
the downregulation of GAS5 as compared with the non-transfected
PA-treated group (Fig. 3A and B).
In addition, the concentrations of TNF-α and IL-1β in PA-treated
H9c2 cells were significantly decreased by the downregulation of
GAS5 (Fig. 3C and D). Taken
together, these results demonstrated that the downregulation of
GAS5 inhibits PA-induced inflammation.
Interaction between GAS5 and
miR-26a
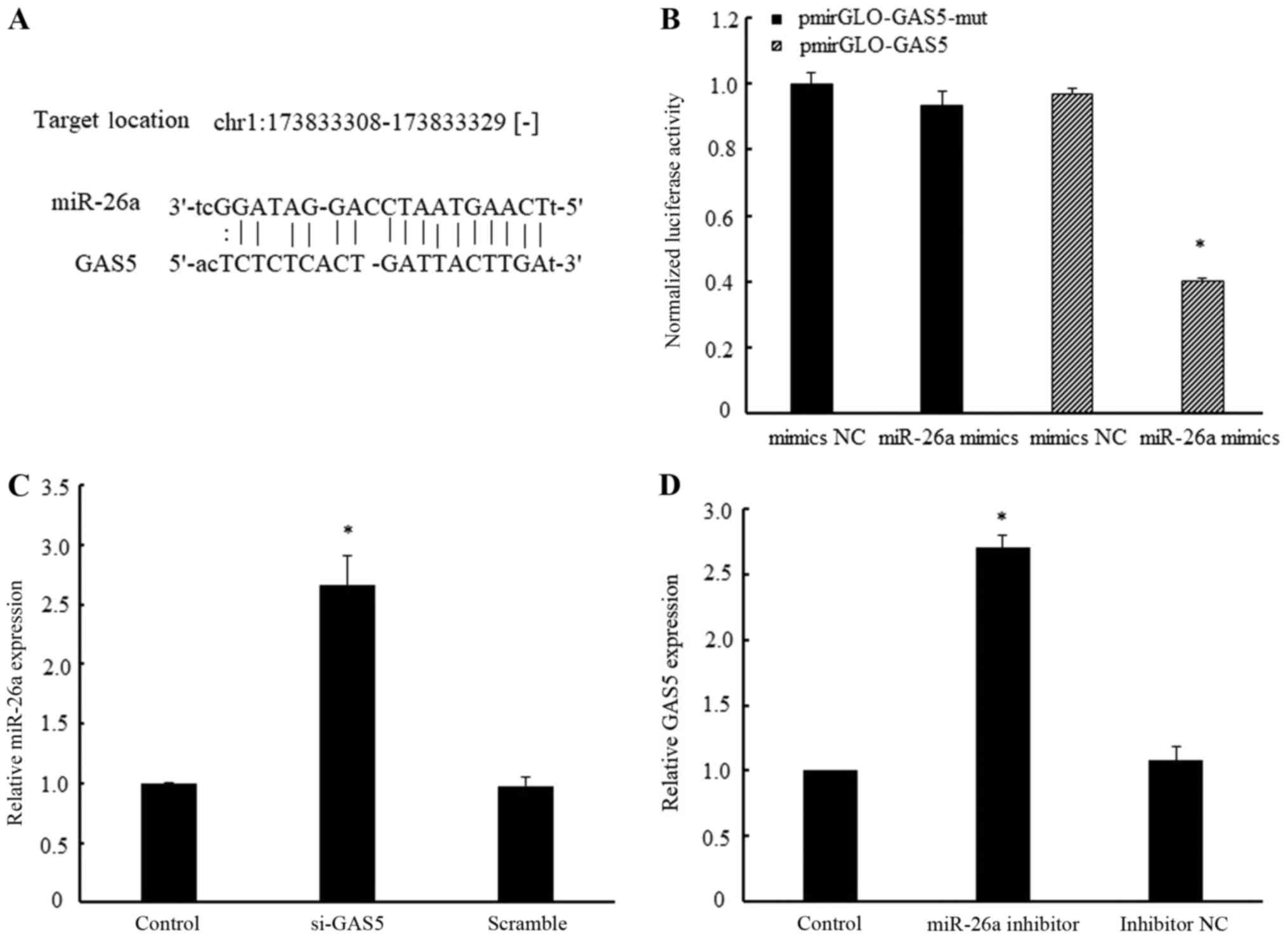
Bioinformatics analysis (starBase v2.0; http://starbase.sysu.edu.cn/index.php)
revealed that the GAS5 transcript sequence contained a miR-26a
binding region (Fig. 4A).
Subsequently, the results of the luciferase activity assay
demonstrated that luciferase activity in the pmirGLO-GAS5 group was
reduced by 61% in the group transfected with miR-26a mimics
compared with mimics NC (Fig. 4B).
In the pmirGLO-GAS5-mut group, miR-26a mimics did not have a
significant inhibitory effect on luciferase activity when compared
with the mimics NC (Fig. 4B).
These results demonstrated that GAS5 specifically binds with
miR-26a.
Furthermore, the knockdown of GAS5 was demonstrated
to markedly upregulate miR-26a expression, while treatment with
miR-26a inhibitor significantly increased GAS5 expression (Fig. 4C and D). Therefore, these results
further demonstrated that a reciprocal negative regulation exists
between miR-26a and GAS5.
Knockdown of GAS5 expression inhibits
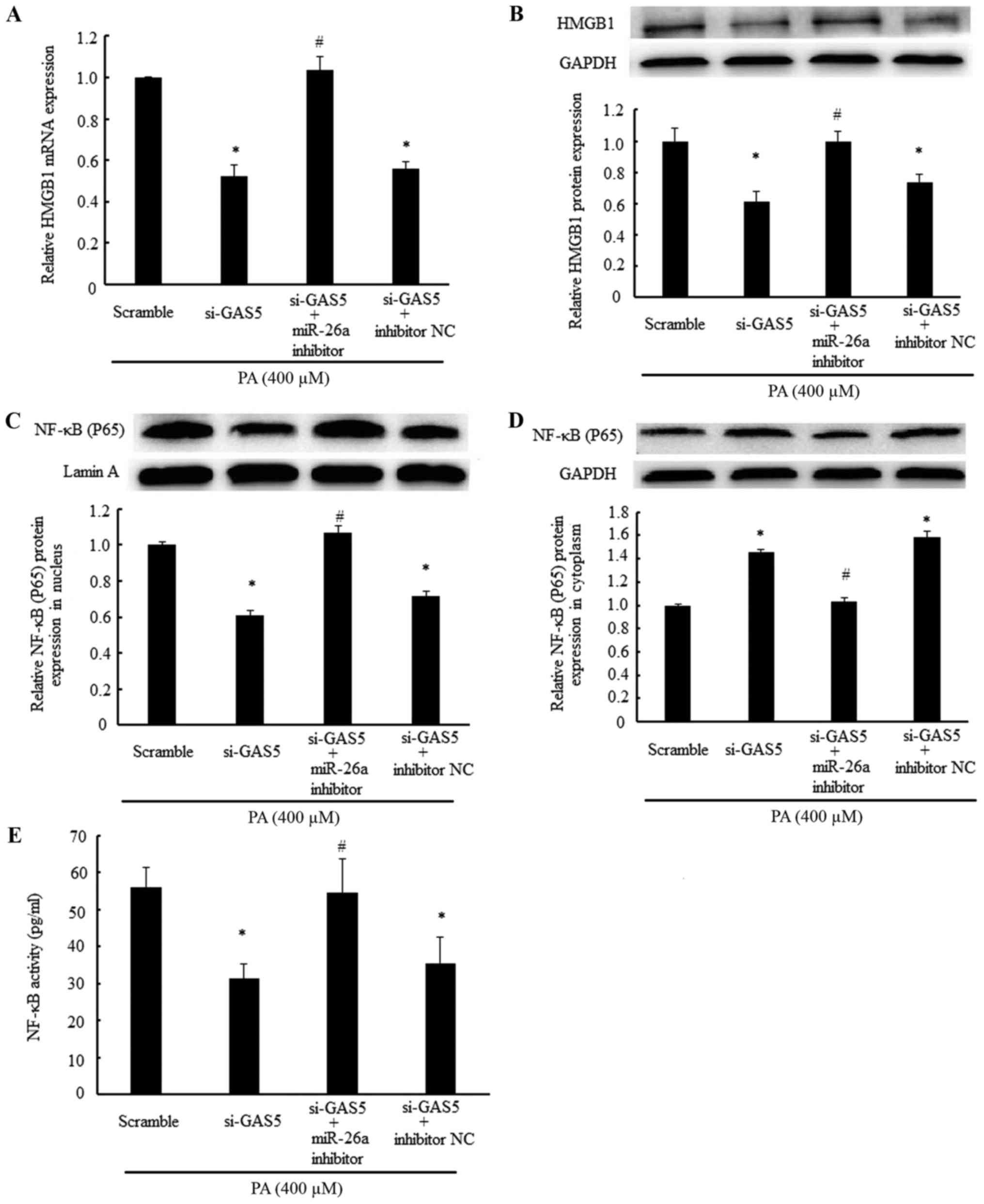
HMGB1/NF-κB signaling mediated by miR-26a
As HMGB1 is a known target of miR-26a (24), the present study investigated
whether GAS5, acting as a ceRNA, regulated the expression of HMGB1,
and examined whether the regulation of GAS5 on the expression of
HMGB1 was mediated by miR-26a. The results demonstrated that the
mRNA and protein expression levels of HMGB1 were repressed by
transfection with the GAS5-specific siRNA alone in PA-treated
cells. However, co-transfection with the GAS5-specific siRNA and
miR-26a inhibitor prevented the inhibition of the mRNA and protein
expression levels of HMGB1 (Fig. 5A
and B). Similarly, transfection with the GAS5-specific siRNA
alone decreased the protein expression of NF-κB in the nucleus,
increased the cytoplasmic protein expression of NF-κB and inhibited
the activity of NF-κB in cardiomyocytes. However, these effects of
GAS5 knockdown on NF-κB were abolished by co-transfection with the
miR-26a inhibitor (Fig. 5C and D).
Taken together, these results demonstrated that the knockdown of
GAS5 expression inhibited the HMGB1/NF-κB signaling pathway, and
this effect was mediated by miR-26a.
Discussion
In the present study, the expression and role of
GAS5 on PA-induced myocardial injury were examined. Previous
studies have revealed that lncRNAs serve a regulatory role in cell
and tissue damage induced by PA irritation (21,22,25,26).
It has also been demonstrated that lncRNA hypoxia-inducible factor
1α-antisense RNA 1 (ΗIF1A-AS1) expression is upregulated in
vascular smooth muscle cells and human vascular endothelial cells
exposed to PA, and that downregulation of HIF1A-AS1 expression
reduced PA-induced apoptosis in these cells (25,26).
However, the regulatory role of lncRNAs in PA-induced myocardial
injury remains unknown. To the best of our knowledge, the present
study is the first to report the role of lncRNAs in PA-induced
myocardial injury.
GAS5 is typically induced under conditions of
cellular stress, such as ischemia (17) and serum starvation (27). PA is an inducer of cellular
oxidative stress and endoplasmic reticulum stress (22). As hypothesized, when rat myocardial
H9c2 cells were exposed to PA, the expression levels of GAS5 were
increased in the present study. Furthermore, it was demonstrated
that the downregulation of GAS5 alleviated the extent of PA-induced
myocardial injury. Taken together, the results of the present study
suggest that GAS5 may be a novel putative therapeutic target for
PA-induced myocardial injury.
Recent studies have provided growing evidence that
PA-induced myocardial injury is associated with the activation of
inflammatory and innate immune responses (5,6). PA
may promote the development of myocardial injury through a
mechanism of direct interactions with myeloid differentiation
protein 2 and a Toll-like receptor 4 (TLR4) accessory protein,
resulting in the activation of the TLR4/NF-κB signaling pathway for
the regulation of pro-inflammatory molecules (5). GAS5 is a repressor of the
glucocorticoid receptor (GR) through binding to its DNA-binding
domain by acting as a molecular decoy (27). Furthermore, decreasing GAS5 levels
can enhance the suppressive effect of glucocorticoid on
inflammatory responses (28). As
hypothesized, the current study revealed that the downregulation of
GAS5 decreased the mRNA expression levels and activities of TNF-α
and IL-1β in PA-treated myocardial cells. Thus, these results
strengthen the evidence that GAS5 is a modulator of inflammatory
response.
The regulatory type of lncRNA for gene function
typically depends on its cellular location. The majority of lncRNAs
in the cytoplasm regulate gene expression by sponging with miRNAs,
functioning as ceRNAs (29).
Several studies have demonstrated that GAS5 is abundantly expressed
in the cytoplasm and regulates gene expression in a ceRNA
regulatory mechanism (15,18). For instance, GAS5 has been reported
to increase p27 expression as a ceRNA for miR-222, thereby
inhibiting liver fibrosis progression (18). Thus, it is hypothesized that GAS5
may form a regulatory network of ceRNAs to regulate PA-induced
myocardial injury. The present study demonstrated that GAS5 binds
to miR-26a, and that a reciprocal negative regulation exists
between miR-26a and GAS5. These results provide novel evidence for
the interaction between miR-26a and GAS5.
HMGB1, a well-established miR-26a target gene, can
activate NF-κB and increase the expression of pro-inflammatory
cytokines (24). Therefore, in the
present study, it was speculated that GAS5 functions as a ceRNA to
regulate HMGB1 expression by binding with miR-26a. The results
demonstrated that the downregulation of GAS5 inhibited the
activation of the HMGB1/NF-κB signaling pathway, and this
repressive effect was mediated by miR-26a. Taken together, the
results of the present study demonstrated that GAS5 regulates HMGB1
expression through miR-26a binding, thereby inhibiting the
activation of NF-κB.
However, there are limitations to the present
research. The expression and role of GAS5 on PA-induced myocardial
injury was only confirmed in vitro. Therefore a myocardial
lipotoxic injury in vivo model induced by PA or a high-fat
diet should be used to further confirm the role of GAS5 in
myocardial lipotoxic injury.
In conclusion, the present study demonstrated that
GAS5 is upregulated in cardiomyocytes exposed to PA, while
knockdown of GAS5 expression alleviates PA-induced myocardial
inflammatory injury through the miR-26a/HMGB1/NF-κB axis. These
findings provide a novel insight into the underlying mechanism of
myocardial lipotoxic injury.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81670320, 81800232,
81401413 and 81871373) and the Natural Science Foundation of
Liaoning Province (grant no. 201602826).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
QY, DJ and CM conceived and designed the
experiments. QY and CZ conducted the experiments. YW, LZ, QZ and GL
participated in the completion of the experiments. QY, LZ, QZ and
GL analyzed the data. QY and NW wrote the paper. NW acquired and
interpreted the data, and drafted and revised the manuscript. All
the authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
NCD Risk Factor Collaboration (NCD-RisC):
Worldwide trends in body-mass index, underweight, overweight, and
obesity from 1975 to 2016: A pooled analysis of 2416
population-based measurement studies in 128·9 million children,
adolescents, and adults. Lancet. 390:2627–2642. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boden G: Obesity and free fatty acids.
Endocrinol Metab Clin North Am. 37(635–646): viii–ix. 2008.
|
|
3
|
Djoussé L, Benkeser D, Arnold A, Kizer JR,
Zieman SJ, Lemaitre RN, Tracy RP, Gottdiener JS, Mozaffarian D,
Siscovick DS, et al: Plasma free fatty acids and risk of heart
failure: The Cardiovascular Health Study. Circ Heart Fail.
6:964–969. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zou L, Li X, Wu N, Jia P, Liu C and Jia D:
Palmitate induces myocardial lipotoxic injury via the endoplasmic
reticulum stress-mediated apoptosis pathway. Mol Med Rep.
16:6934–6939. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Qian Y, Fang Q, Zhong P, Li W,
Wang L, Fu W, Zhang Y, Xu Z, Li X and Liang G: Saturated palmitic
acid induces myocardial inflammatory injuries through direct
binding to TLR4 accessory protein MD2. Nat Commun. 8:139972017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeng C, Zhong P, Zhao Y, Kanchana K, Zhang
Y, Khan ZA, Chakrabarti S, Wu L, Wang J and Liang G: Curcumin
protects hearts from FFA-induced injury by activating Nrf2 and
inactivating NF-κB both in vitro and in vivo. J Mol Cell Cardiol.
79:1–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun W, Yang Y, Xu C and Guo J: Regulatory
mechanisms of long noncoding RNAs on gene expression in cancers.
Cancer Genet. 216–217:105–110. 2017. View Article : Google Scholar
|
|
8
|
Ratajczak MZ, Shin DM, Schneider G,
Ratajczak J and Kucia M: Parental imprinting regulates insulin-like
growth factor signaling: A Rosetta Stone for understanding the
biology of pluripotent stem cells, aging and cancerogenesis.
Leukemia. 27:773–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee JT: The X as model for RNA's niche in
epigenomic regulation. Cold Spring Harb Perspect Biol.
2:a0037492010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sen R, Ghosal S, Das S, Balti S and
Chakrabarti J: Competing endogenous RNA: The key to
posttranscriptional regulation. ScientificWorldJournal.
2014:8962062014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Zhang Y, Yang T, Zhao W, Wang N,
Li P, Zeng X and Zhang W: Long non-coding RNA MALAT1 for promoting
metastasis and proliferation by acting as a ceRNA of miR-144-3p in
osteosarcoma cells. Oncotarget. 8:59417–59434. 2017.PubMed/NCBI
|
|
14
|
Coccia EM, Cicala C, Charlesworth A,
Ciccarelli C, Rossi GB, Philipson L and Sorrentino V: Regulation
and expression of a growth arrest-specific gene (gas5) during
growth, differentiation, and development. Mol Cell Biol.
12:3514–3521. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C,
Xu M, Wu F and Mo YY: Negative regulation of lncRNA GAS5 by miR-21.
Cell Death Differ. 20:1558–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y and
Song Y: The growth arrest-specific transcript 5 (GAS5): A pivotal
tumor suppressor long noncoding RNA in human cancers. Tumour Biol.
37:1437–1444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen F, Zhang L, Wang E, Zhang C and Li X:
LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA
for miR-137 to regulate the Notch1 signaling pathway. Biochem
Biophys Res Commun. 496:184–190. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G,
Guo C, Liu Z and Fan X: Long Non-coding RNA growth arrest-specific
transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism
of competing endogenous RNA. J Biol Chem. 290:28286–28298. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tao H, Zhang JG, Qin RH, Dai C, Shi P,
Yang JJ, Deng ZY and Shi KH: LncRNA GAS5 controls cardiac
fibroblast activation and fibrosis by targeting miR-21 via
PTEN/MMP-2 signaling pathway. Toxicology. 386:11–18. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song J, Ahn C, Chun CH and Jin EJ: A long
non-coding RNA, GAS5, plays a critical role in the regulation of
miR-21 during osteoarthritis. J Orthop Res. 32:1628–1635. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan C, Chen J and Chen N: Long noncoding
RNA MALAT1 promotes hepatic steatosis and insulin resistance by
increasing nuclear SREBP-1c protein stability. Sci Rep.
6:226402016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brookheart RT, Michel CI, Listenberger LL,
Ory DS and Schaffer JE: The non-coding RNA gadd7 is a regulator of
lipid-induced oxidative and endoplasmic reticulum stress. J Biol
Chem. 284:7446–7454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yao L, Lv X and Wang X: MicroRNA 26a
inhibits HMGB1 expression and attenuates cardiac
ischemia-reperfusion injury. J Pharmacol Sci. 131:6–12. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He Q, Tan J, Yu B, Shi W and Liang K: Long
noncoding RNA HIF1A-AS1A reduces apoptosis of vascular smooth
muscle cells: Implications for the pathogenesis of thoracoabdominal
aorta aneurysm. Pharmazie. 70:310–315. 2015.PubMed/NCBI
|
|
26
|
Wang J, Chen L, Li H, Yang J, Gong Z, Wang
B and Zhao X: Clopidogrel reduces apoptosis and promotes
proliferation of human vascular endothelial cells induced by
palmitic acid via suppression of the long non-coding RNA HIF1A-AS1
in vitro. Mol Cell Biochem. 404:203–210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Keenan CR, Schuliga MJ and Stewart AG:
Pro-inflammatory mediators increase levels of the noncoding RNA
GAS5 in airway smooth muscle and epithelial cells. Can J Physiol
Pharmacol. 93:203–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rashid F, Shah A and Shan G: Long
non-coding RNAs in the cytoplasm. Genomics Proteomics
Bioinformatics. 14:73–80. 2016. View Article : Google Scholar : PubMed/NCBI
|