Introduction
Spindle and kinetochore-associated (SKA)2 is located
on chromosome 17 of the human genome and has been identified as a
conserved protein involved in the kinetochore complex (1,2).
SKA2, together with its cofactors SKA1 and SKA3, constitute the SKA
complex, which maintains the metaphase plate and/or spindle
checkpoint silencing (3–5). Checkpoint-dependent delays in a
metaphase-like state are prolonged by RNA interference-mediated
SKA2 depletion (6,7). In addition, SKA2 has been reported to
serve a role in tumorigenesis. Aberrant patterns of SKA2 expression
have been observed in several types of cancer, including lung
cancer (8–10), kidney cancer (11), pancreatic cancer (12), gastric cancer (13), glioma (14) and osteosarcoma (15,16).
However, to the best of our knowledge, the roles of SKA2 in breast
cancer migration remain to be elucidated.
Breast cancer is common in women worldwide. The
first highest incidence rates and second highest mortality rates
have been demonstrated in the developed world (17,18).
Furthermore, breast cancer is prone to metastasis and results in
poor prognosis (19,20). Although marked progress has been
achieved in breast cancer therapy due to modern technology, unknown
molecular mechanisms of metastasis remain and require further
investigation. As a fundamental process of migration,
epithelial-mesenchymal transition (EMT) serves a crucial role in
breast cancer progression (21–24).
However, the specific regulatory mechanism mediating EMT and SKA2
in breast cancer progression remains to be investigated. In the
present study, it was demonstrated that SKA2 was upregulated in
human breast cancer cell lines and tissues by reverse transcription
quantitative polymerase chain reaction (RT-qPCR) and western
blotting. Small interfering (si)SKA2 was used to knockdown the
expression of SKA2 in MCF-7 and T47D cells, the results
demonstrated that decreasing the level of SKA2 inhibited the
metastasis of the breast cancer cells by wound healing assay and
cell invasion assays. However, although the results are promising,
further studies are needed to confirm this. Therefore, the present
study further investigated whether SKA2 regulates metastasis via
EMT-associated proteins and matrix metalloproteinase (MMP)-2/9. The
current study aimed to determine the potential role of SKA2 in
breast cancer invasion, and its molecular mechanism.
Patients and methods
Patients
Tumor specimens and adjacent tissues were collected
from patients who were diagnosed with breast cancer (stages I–IV)
and underwent surgery at Ningbo No. 2 Hospital (Ningbo, China)
between March 2015 and August 2017. None of these patients
underwent local or systemic therapy prior to surgery. The clinical
characteristics of these 160 patients are described in Table I. The present study was approved by
the Research Ethics Committee of Ningbo No. 2 Hospital, and all
patients provided written informed consent.
 | Table I.SKA2 expression and
clinicopathological characteristics of patients. |
Table I.
SKA2 expression and
clinicopathological characteristics of patients.
|
|
| SKA2 expression
levels |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Number of
patients | Low (%) | High (%) | P-value |
|---|
| Age (years) |
|
|
| 0.839 |
|
≤50 | 72 | 24 (33.3) | 48 (66.7) |
|
|
>50 | 88 | 28 (31.8) | 60 (68.2) |
|
| Tumor (cm) |
|
|
| 0.077 |
|
<2 | 78 | 32 (41) | 46 (59) |
|
|
2-5 | 55 | 14 (22.5) | 41 (74.5) |
|
|
>5 | 27 | 6 (22.2) | 21 (77.8) |
|
| Tumor TNM
staging |
|
|
| 0.001 |
| I | 48 | 25 (52.1) | 23 (47.9) |
|
| II | 40 | 14 (35) | 26 (65) |
|
|
III | 64 | 11 (17.2) | 53 (82.8) |
|
| IV | 8 | 2 (25) | 6 (75) |
|
| Tumor histological
grade |
|
|
| 0.485 |
| 1 | 9 | 4 (44.4) | 5 (55.6) |
|
| 2 | 96 | 28 (29.2) | 68 (70.8) |
|
| 3 | 55 | 20 (36.4) | 35 (63.6) |
|
| Pathological
type |
|
|
| 0.052 |
|
Infiltrative | 128 | 37 (28.9) | 91 (71.1) |
|
| Not
infiltrative | 32 | 15 (46.9) | 17 (53.1) |
|
| Molecular type |
|
|
| 0.198 |
| Luminal
A |
|
|
| 0.001 |
|
Lymphatic
metastasis | 27 | 5 (18.5) | 22 (81.5) |
|
|
No lymphatic
metastasis | 18 | 12 (66.7) | 6 (33.3) |
|
| Luminal
B |
|
|
| 0.002 |
|
Lymphatic
metastasis | 50 | 10 (20) | 40 (80) |
|
|
No lymphatic
metastasis | 21 | 12 (57.1) | 9 (42.9) |
|
|
HER2 |
|
|
| 0.021 |
|
Lymphatic
metastasis | 16 | 2 (12.5) | 14 (87.5) |
|
|
No lymphatic
metastasis | 9 | 5 (55.6) | 4 (44.4) |
|
|
Basal |
|
|
| 0.025 |
|
Lymphatic
metastasis | 13 | 2 (15.4) | 11 (84.6) |
|
|
No lymphatic
metastasis | 6 | 4 (66.7) | 2 (33.3) |
|
| Lymphatic
metastasis |
|
|
| <0.001 |
|
Yes | 106 | 19 (17.9) | 87 (82.1) |
|
| No | 54 | 33 (61.1) | 21 (38.9) |
|
Cell lines and transfection
Human breast cancer cell lines MCF-7, T47D and
MDA-MB-231, and normal MCF-10A cells were purchased from the
American Type Culture Collection (Manassas, VA, USA). All cells
were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone;
GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (FBS; ExCell Bio, Shanghai, China) at 37°C in an
incubator containing 5% CO2.
Cells (MCF-7 and T47D) were transfected with small
interfering RNA (si)SKA2 (sense, 5′-GGCUGGAAUAUGAAAUCAATT-3′ and
antisense, 5′-UUGAUUUCAUAUUCCAGCCTT-3′), si-negative control (NC;
sense, 5′-UCCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′) (both Shanghai GenePharma Co., Ltd.,
Shanghai, China), SKA2cDNA plasmid (Ref Seq: M_001100595.1, Atgg
cctcggaggt ggggcacaat ttggagtcgc cggaaactcc gcggcgga ggctggacca
gagtcgagtt ctcctcct gcaccaaagg gagccgccac tctggtgt ctaaaccgcc
tcggttccag gctgagt ctgatctgga ttacattcaa caggctgg aatatgaaat
caagactaat tcctgatt cagcaagtga gctgtcacca ctga) or NC plasmid (both
Shanghai Genechem Co., Ltd., Shanghai, China) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. The cells were seeded in 6-well plates
(siRNA transfection: 100 pmol siRNA and 5 µl Lipofectamine 2000,
plasmid DNA transfection: 4 µg DNA and 10 µl Lipofectamine 2000, at
room temperature) and were grown to 50% confluence prior to
transfection. RNA and protein were extracted 24 h
post-transfection. The MOCK was not transfected with any agent.
Extraction of cytoplasmic and nuclear
proteins
A total of 24 h post-transfection, MCF-7 cells were
harvested and washed three times with cold PBS. Cytoplasmic and
nuclear protein fractions were extracted using a Mammalian Nuclear
and Cytoplasmic Protein Extraction kit (Beijing Transgen Biotech,
Co., Ltd., Beijing, China), according to the manufacturer's
protocol; these proteins were then used for western blotting.
Western blot analysis
Total protein extracted from the cells by 1XSDS
(Sigma-Aldrich; Merck KGaA) was quantified by bicinchoninic acid
analysis (Beyotime Institute of Biotechnology, Shanghai, China).
Cellular proteins (40 µg) were separated by 12% SDS-PAGE and were
transferred onto polyvinylidende difluoride membranes (EMD
Millipore, Billerica, MA, USA) for immunoblotting. After blocking
with 5% bovine serum albumin (BSA; Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China) for 2 h at room temperature,
the membranes were incubated with specific primary antibodies
diluted in 1X Tris-buffered saline-0.1% Tween-20 overnight at 4°C.
The following antibodies were used: SKA2 (1:1,500; cat. no.
ab91551), MMP2 (1:1,500; cat. no. ab7033), MMP9 (1:1,500; cat. no.
ab137651), vimentin (1:2,000; cat. no. ab8978), fibronectin
(1:2,000; cat. no. ab2413; all Abcam, Cambridge, UK), N-cadherin
(1:2,000; cat. no. 14215), E-cadherin (1:2,000; cat. no. 3195),
β-actin (1:1,500; cat. no. 8457), Histone H3 (1:1,500; cat. no.
9728) and GAPDH (1:1,500; cat. no. 5174; all Cell Signaling
Technology, Inc., Danvers, MA, USA). Subsequently, membranes were
incubated for 1 h at room temperature with goat anti-rabbit
immunoglobulin (Ig)G-horseradish peroxidase (HRP) or goat
anti-mouse IgG-HRP (1:5,000; cat. nos. BA1054/BA1050; Wuhan Boster
Biological Technology, Ltd., Wuhan, China). The protein bands on
the membrane were visualized using a chemiluminescence imaging
system (LI-COR Biosciences, Lincoln, CA, USA) and detected by
chemiluminescence. Densitometric analysis was performed by Tanon
GIS version 4.1.2 software (Tanon Science and Technology Co., Ltd.,
Shanghai, China).
RT-qPCR
The mRNA expression levels of SKA2 were detected
using RT-qPCR. Briefly, total RNA was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and 1 µg total RNA was subjected to first-strand cDNA
synthesis for 15 min at 37°C and 5 sec at 85°C using a reverse
transcription kit (Thermo Fisher Scientific, Inc.). cDNA was
amplified by RT-qPCR using SYBR-Green PCR Master Mix (Roche Applied
Science, Madison, WI, USA) on a LightCycler® 480 system
(95°C/10 min/1 cycle, 95°C/10 sec/45 cycles, 60°C/60 sec/45 cycles;
Roche Applied Science) and fold-changes were calculated by relative
quantification (2−∆∆Cq) (25). The primers used were as follows:
SKA2 forward, 5′-CTGAAACTATGCTAAGTGGGGGAG-3′, reverse,
5′-TTCCAAACATCCTGACACTCAAAAG-3′; and GAPDH forward,
5′-AAGCCTGCCGGTGACTAAC-3′ and reverse,
5′-GCATCACCCGGAGGAGAAAT-3′.
Wound healing assay
Cells were seeded in 6-well plates at a density of
1×105 cells/well. Once cellular density reached ~70%,
the cells were scraped in a straight line with a 1-ml blue
micro-pipette tip. After rinsing the dislodged cells with PBS, the
cell culture medium was replaced with fresh serum-free medium. The
widths at 0 and 24 h were compared to assess the distance of
migration using fluorescence microscopy.
Cell invasion assays
Cell invasion assays were conducted using a
Matrigel-coated invasion chamber (24-well plates, 8-µm pore size;
Corning Inc., Corning, NY, USA), in accordance with the
manufacturer's protocol. A total of 5×104 cells were
seeded in the upper chambers of the wells in 100 µl FBS-free
medium, and the lower chambers were filled with DMEM supplemented
with 20% FBS, in order to stimulate cell invasion. After 24 h
incubation at 37°C in an incubator containing 5% CO2,
the cells on the filter surface were stained with 0.1% crystal
violet for 30 min and images were captured with a fluorescence
microscope (Nikon Corporation, Tokyo, Japan). Absorbance was
measured at 590 nm.
Immunohistochemical staining
Immunohistochemical staining was performed on
formalin-fixed paraffin-embedded tissue sections (4-µm; formalin
fixation was performed at room temperature with no time limit set
for fixation). All sections were blocked with 5% BSA for 30 min at
room temperature and incubated with anti-SKA2 (1:500; cat. no.
ab91551; Abcam) at 4°C overnight. The sections were then incubated
with a horseradish peroxidase-goat anti-rabbit IgG secondary
antibody for 40 min at room temperature (1:100; cat. no. TA140003;
OriGene Technologies Inc., Beijing, China). Finally, Myer's
hematoxylin was used for background staining for 3 min at room
temperature and photographs were taken using fluorescence
microscopy. The appropriate positive (tumor tissues at different
stages) and negative controls (normal tissue) were included. The
positive cells and staining intensity of tumor tissues were scored
respectively. According to the percentage of positive cells, <5%
was 0, 5% ~<25% was 1, 25% ~<50% was 2, 50% ~<75% was 3,
and <75% was 4. According to the coloring strength score, it
could be divided into 4 grades: No coloring was 0, pale brown was
1, brown was 2, dark brown was 3. The result of the two products: 0
was negative, 1 was positive, in which <6 was low expression and
≥6 is high expression (26).
Fluorescent immunocytochemistry
MCF-7 cells were fixed in 4% paraformaldehyde for 30
min at room temperature and then permeabilized with 0.1% Triton
X-100 in PBS at room temperature for 15 min. The cells were blocked
with 1% BSA overnight and incubated overnight at 4°C with Alexa
Fluor® 488-conjugated β-tubulin rabbit monoclonal
antibody to (1:500; cat. no. 2116) or E-cadherin antibody (1:1,000;
cat. no. 3195; both Cell Signaling Technology, Inc.). Then all
cells were counterstained with 4′,6-diamidino-2-phenylindole
(5:100; cat. no. C0060; Beijing Solarbio Science & Technology
Co., Ltd., Beijing, China) for 15 min at room temperature and
E-cadherin stained cells were counterstained with CY3-labeled goat
anti-rabbit IgG (1:50; cat. no. BA1032; Wuhan Boster Biological
Technology, Ltd.) for 1 h at room temperature. Finally, the
coverslips were washed twice with PBS and images were captured
using confocal scanning microscopy.
Statistical analysis
All experiments were repeated three times and data
are expressed as the means ± standard deviation. The values between
the two groups in Fig. 1A was
compared with independent t-test. One-way analysis of variance and
followed by Fisher's least significant difference tests was used to
calculate P-values between the cell groups in the invasion and
wound healing assays, and to compare measurements of the protein
and mRNA expression levels in breast cancer cell lines. Data in
Table I were analyzed using the
χ2 test of four by four table. Statistical analyses were
performed using SPSS 15.0 software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
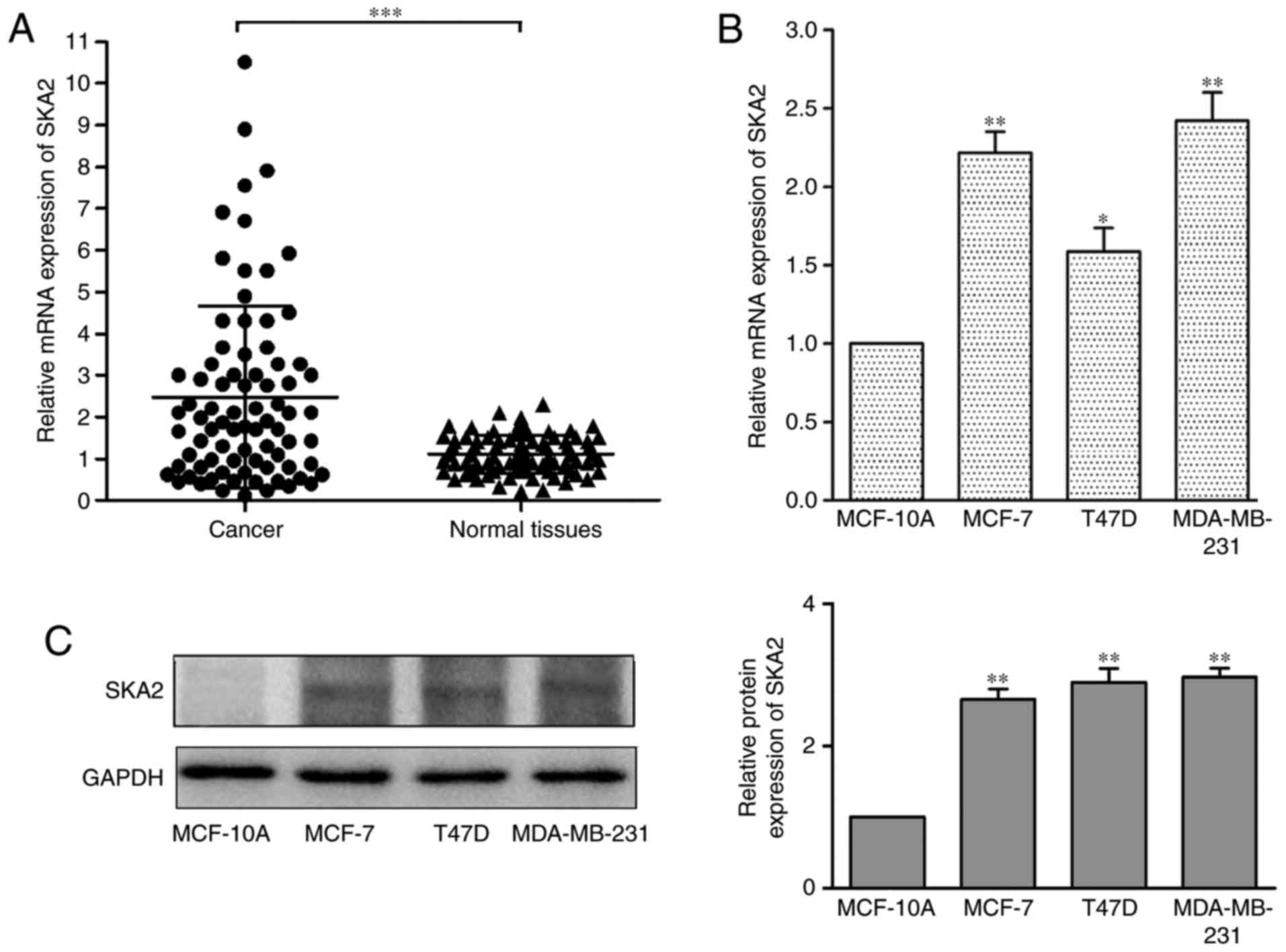
Results
SKA2 levels are upregulated in breast
cancer and are associated with breast cancer metastasis
To investigate the role of SKA2 in breast cancer,
SKA2 expression levels were measured in 80 patient samples by
RT-qPCR and the levels were normalized to GAPDH. The results
demonstrated that SKA2 mRNA was significantly upregulated in breast
cancer tissues compared with in adjacent normal tissues
(P<0.001; Fig. 1A). In
addition, the mRNA (Fig. 1B) and
protein (Fig. 1C) expression
levels of SKA2 in three human breast cancer cell lines (MCF-7, T47D
and MDA-MB-231) were evaluated and compared with the MCF-10A normal
human mammary epithelial cell line. SKA2 was significantly
increased in all three cell lines compared with in MCF-10A cells
(P<0.05).
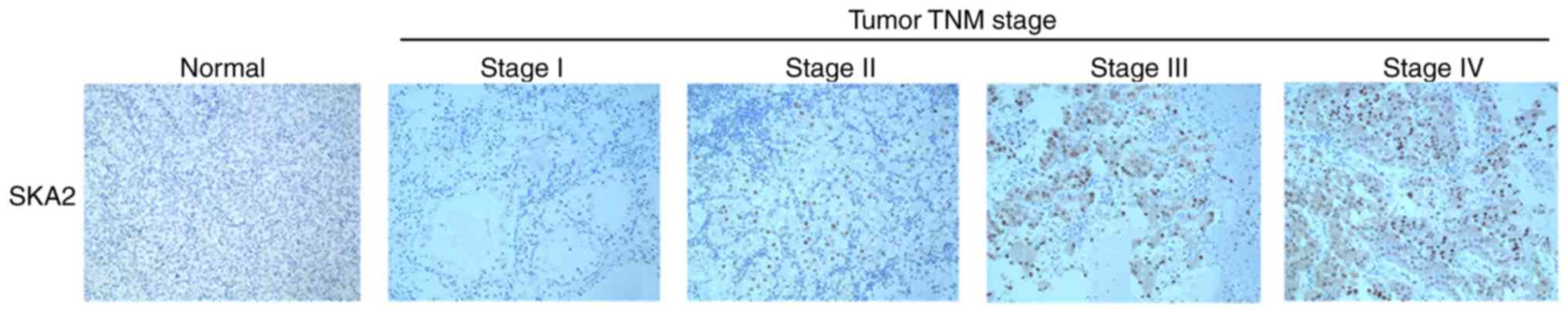
To investigate the clinical significance of SKA2 in
patients with breast cancer, SKA2 immunohistochemical staining in a
cohort of 160 specimens was analyzed. As demonstrated in Table I, high intratumoral SKA2 levels
were significantly associated with clinical stage (P=0.001) and all
lymph node metastasis groups (Luminal A, B HER2 and basal;
P<0.001) but not with patient age, tumor size, histological
grade, pathological type or molecular typing (Table I). In addition, intratumoral SKA2
levels increased gradually with disease progression from TNM stage
I to IV (Fig. 2).
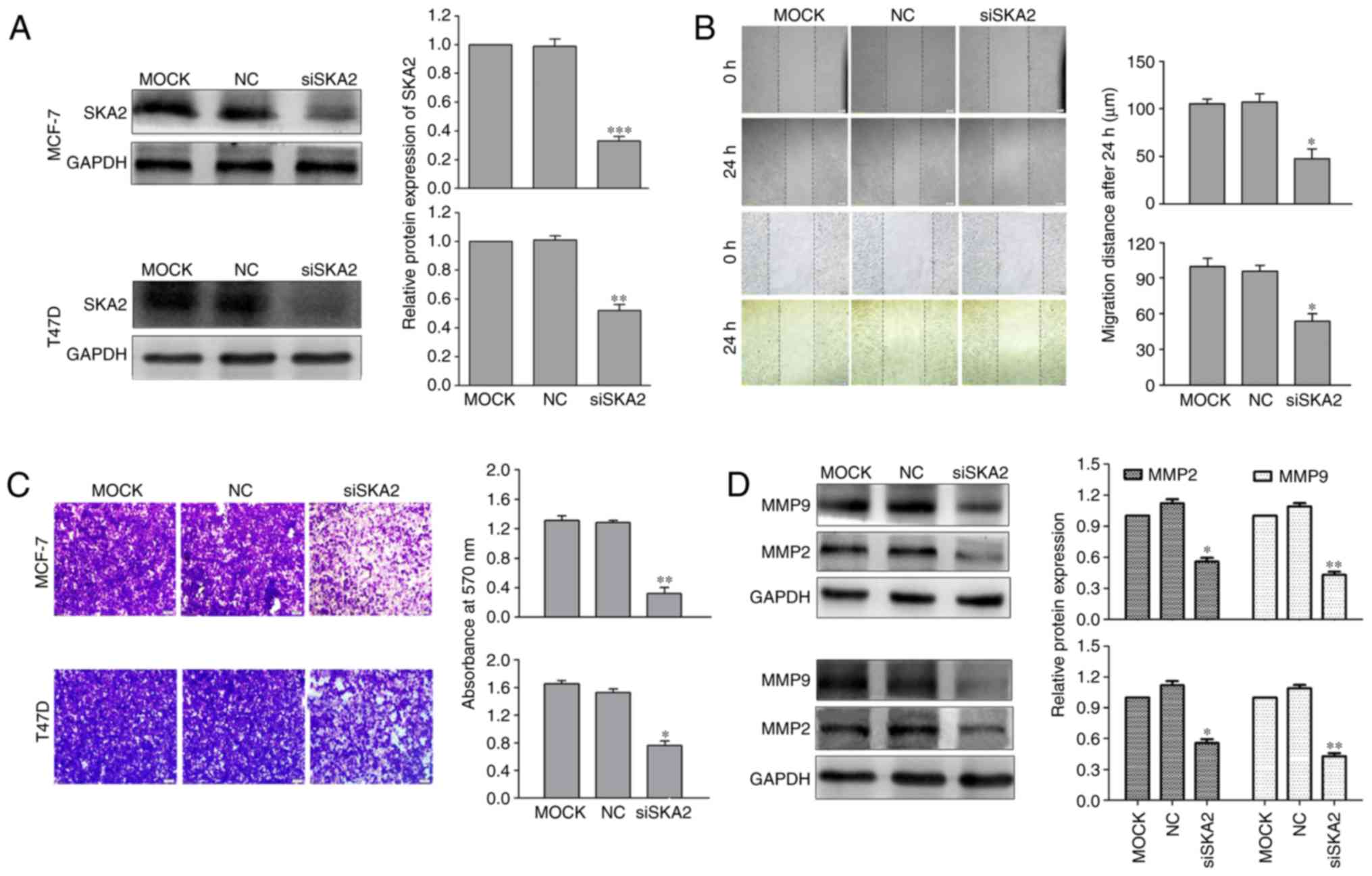
SKA2 knockdown inhibits the migration
and invasion of breast cancer cells
To investigate the function of SKA2 in breast cancer
cell migration, siSKA2 specifically targeting SKA2 was used.
Transfection with siSKA2 significantly decreased the protein
expression levels of SKA2 in breast cancer cells (MCF-7 and T47D
which were chosen as a result of their differential expression
levels of SKA2; P<0.01; Fig.
3A). Following knockdown of SKA2 expression, its effects on
cell migration and invasion in vitro were investigated using
Transwell and scratch wound healing assays. Compared with the
control cells, breast cancer cell migration and invasion were
significantly inhibited in siSKA2-transfected cells (P<0.05;
Fig. 3B and C).
It has previously been demonstrated that the
extracellular matrix (ECM) serves a critical role in tumor invasion
and metastasis (27). Since MMPs,
particularly MMP2 and MMP9, destroy ECM processing, they are
associated with tumor invasion and metastasis (28). The results of the present study
demonstrated that siSKA2 transfection significantly decreased the
expression levels of MMP2 and MMP9 in breast cancer cells
(P<0.05; Fig. 3D).
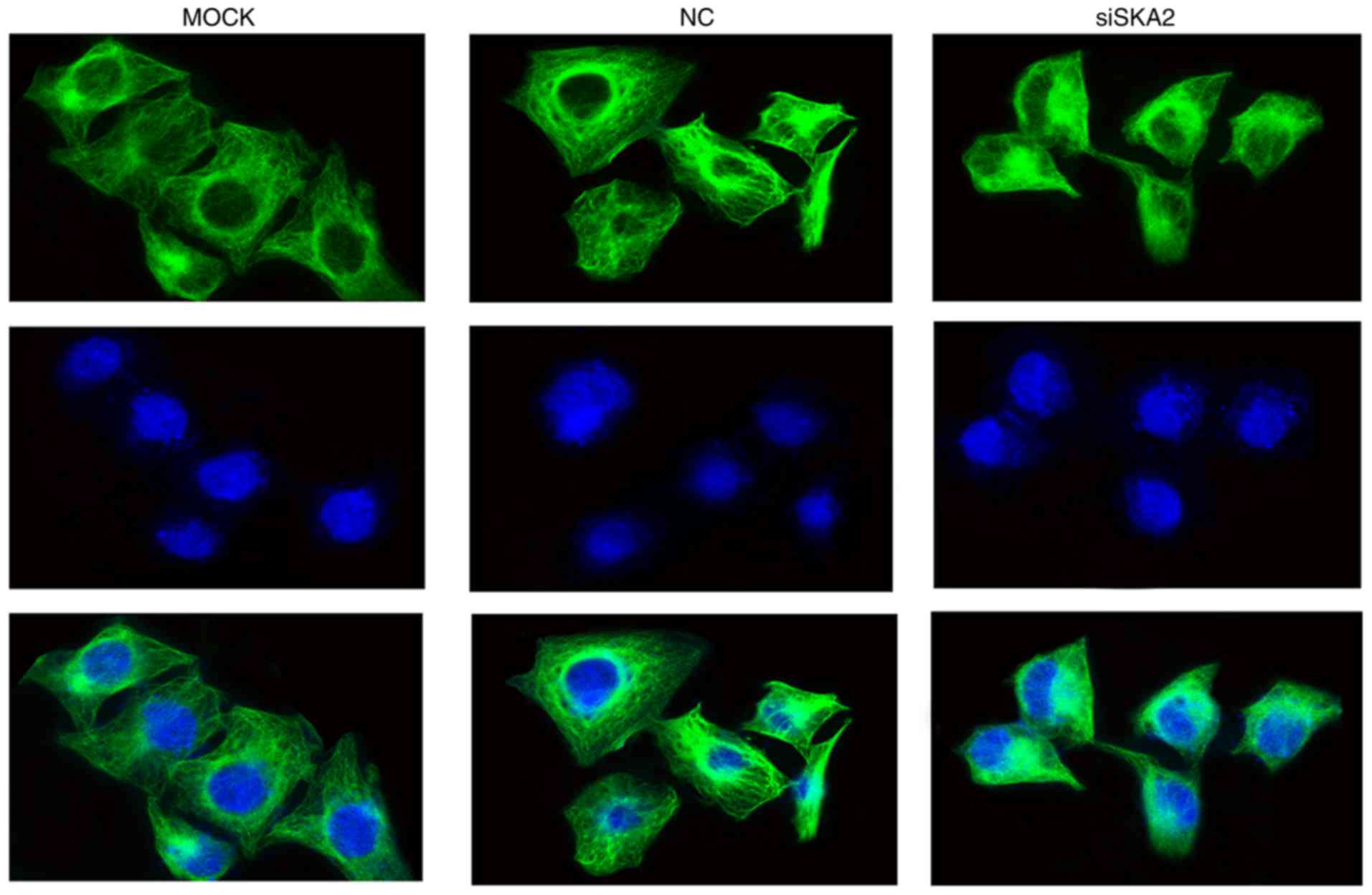
SKA2 regulates microtubule
organization in MCF-7 cells
MCF-7 cells were immunostained with anti-β-tubulin
antibodies to determine the presence of microtubules.
NC-transfected cells exhibited clear and intact microtubules with
well-formed dendrites. In siSKA2-transfected cells, the
microtubules were polymerized and the structure was spindle-shaped,
which may lead to inhibited cell migration (Fig. 4).
SKA2 promotes migration and invasion
via EMT
EMT is a process in which epithelial cells lose
epithelial characteristics and gain mesenchymal features, including
motility and invasiveness (29).
To identify the mechanisms underlying the decreased migration and
invasion of siSKA2-transfected breast cancer cells (MCF-7 and
T47D), the expression levels of E-cadherin, which is an epithelial
marker that is downregulated during EMT, were measured. In
addition, vimentin, fibronectin and N-cadherin, which are
mesenchymal markers that are upregulated during EMT, were also
measured (30). Notably, knockdown
of SKA2 significantly decreased the expression levels of vimentin,
fibronectin and N-cadherin (P<0.05); however, no significant
effects were detected on the expression levels of E-cadherin
(Fig. 5A). Subsequently, SKA2
expression was upregulated with SKA2cDNA, and the expression levels
of E-cadherin were significantly decreased (P<0.05), whereas
N-cadherin, fibronectin and vimentin levels were significantly
increased (P<0.05; Fig. 5B and
C). Therefore, SKA2 may mediate invasion and metastasis in
human breast cancer via EMT.
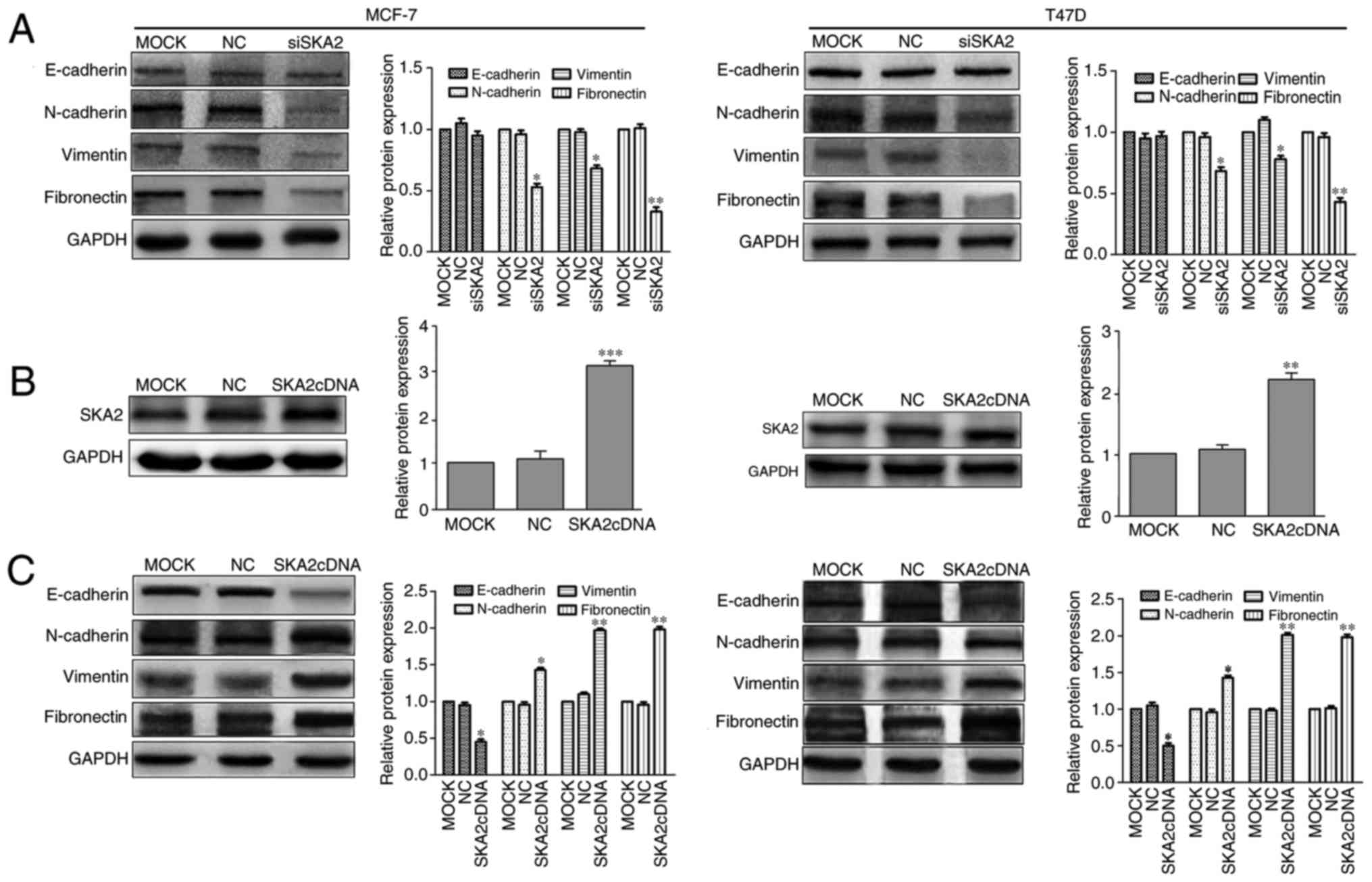 | Figure 5.SKA2 mediates breast cancer
metastasis via EMT. (A) After cells were transfected with siSKA2,
the expression levels of proteins associated with EMT were
determined by western blotting. Vimentin, fibronectin and
N-cadherin were decreased in cells transfected with siSKA2 compared
with in the MOCK group. However, E-cadherin levels were unaltered
in siSKA2-transfected breast cancer cells. (B) SKA2 expression was
upregulated with SKA2cDNA. (C) In SKA2cDNA-transfectd cells, the
expression levels of E-cadherin were decreased, whereas N-cadherin,
fibronectin and vimentin were increased. Data are presented as the
means ± standard deviation from three independent experiments.
*P<0.05, **P<0.01, ***P<0.001, vs. the MOCK group. EMT,
epithelial-mesenchymal transition; si, small interfering RNA; SKA2,
spindle and kinetochore-associated protein 2. |
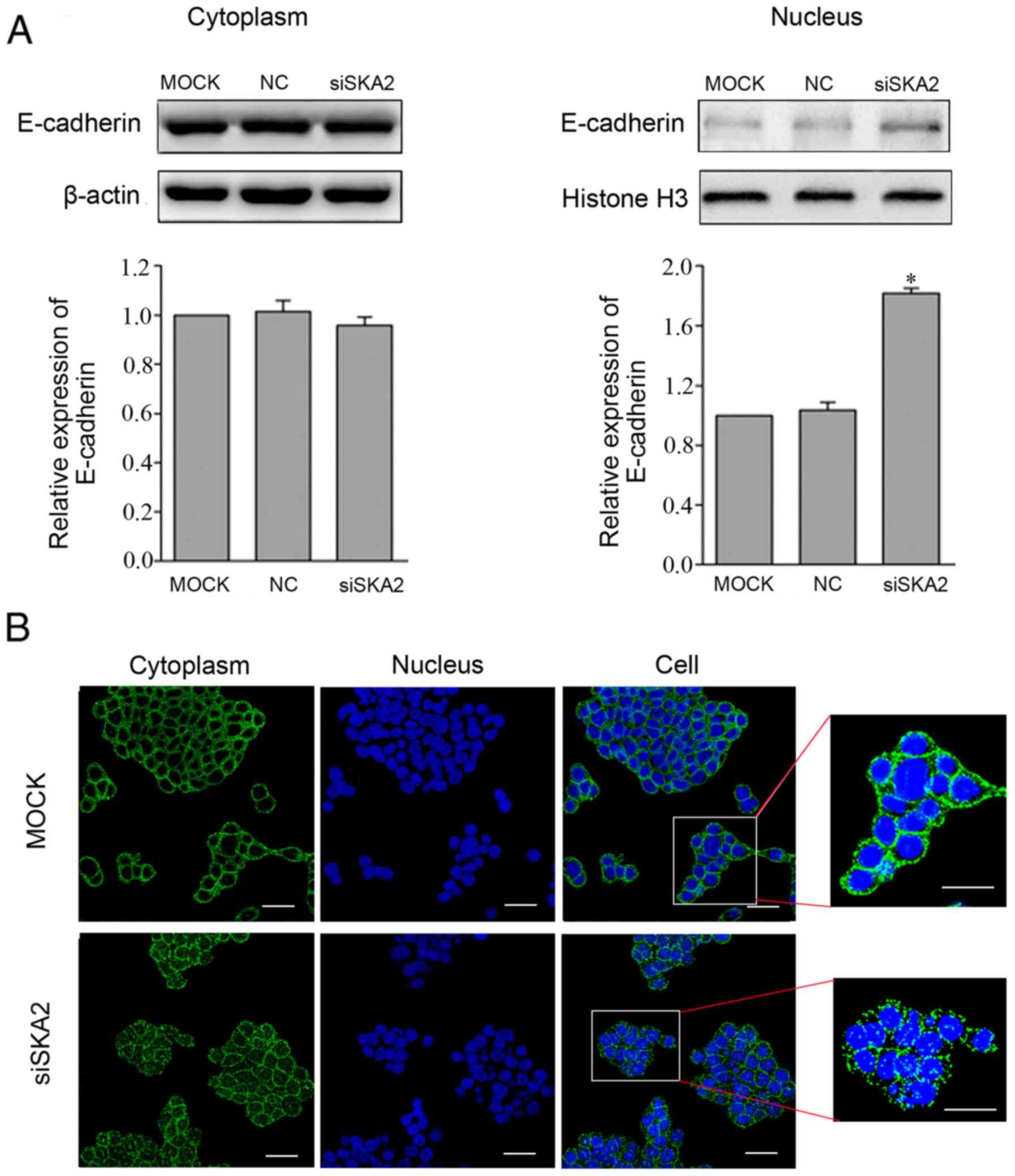
SKA2 knockdown induces translocation
of E-cadherin from the cytoplasm to the nucleus
To investigate the effects of SKA2 on the
translocation of E-cadherin, cytoplasmic and nuclear proteins were
isolated and extracted from MCF-7 cells (as SKA2 is highly
expressed in this cell line and MCF-7 is frequently used in breast
cancer studies this cell line was selected), and western blotting
was performed. The results demonstrated that there were no obvious
alterations in E-cadherin in the cytoplasm compared with in the
MOCK group; however, E-cadherin levels were significantly increased
in the nucleus following transfection with siSKA2 (P<0.05;
Fig. 6A). Similar results were
obtained by immunofluorescence (Fig.
6B). These findings indicated that the suppression of SKA2
facilitated the translocation of E-cadherin from the cytoplasm to
the nucleus.
Discussion
The development and progression of malignant tumors
constitutes a complex process that is affected by numerous factors,
including the biological characteristics of the tumor cells
themselves (31,32). The poor prognosis of breast cancer,
which is one of the most common and lethal malignant diseases, is
due to its metastasis (33,34).
The mechanism that induces and stimulates metastasis is complex and
remains unknown. In recent years, although specific evidence
(10) has confirmed that SKA2 may
contribute to cancer metastases, the mechanism is far from clear.
In the present study, the role of SKA2, a gene associated with
various types of human cancer, was investigated. The results
demonstrated that SKA2 participated in metastasis in breast cancer
tissues; this finding may aid in achieving a breakthrough in breast
cancer treatment.
To further evaluate the role of SKA2 in breast
cancer metastasis, experiments were designed in which the
expression levels of SKA2 were altered, in order to investigate
migration and invasion of breast cancer cells. The results
demonstrated that SKA2 expression was significantly upregulated in
breast cancer cells compared with in normal cells. Knockdown of
SKA2 significantly inhibited the ability of breast cancer cells to
migrate and invade, and decreased the expression levels of MMP2 and
MMP9. These results indicated that SKA2 may serve a crucial role in
the progression of breast cancer metastasis and may be considered a
novel biomarker for breast cancer prognosis.
Cytoskeletal function is associated with contraction
and migration; therefore, the cytoskeleton serves a vital role in
tumor metastasis. Microtubules are the backbone of the
cytoskeleton, which control cell movement. It has been reported
that anticancer drugs can promote the polymerization of
microtubules, destroy the stable structure and normal functions of
microtubules, and thus prevent cell division, movement and
migration (35). Therefore
microtubules serve an important role in cell migration. In cells
depleted of SKA2, microtubules were polymerized and exhibited a
spindle-shaped structure, which may lead to inhibition of cell
migration.
To investigate the molecular mechanism underlying
the effects of SKA2 on invasion and migration, the potential target
of SKA2 was investigated in breast cancer cells. EMT, which refers
to the biological phenomenon by which epithelial cells lose their
epithelial characteristics and obtain a mesenchymal phenotype, is
associated with embryonic development, tissue repair, fibrosis,
tumor invasion and migration (29,30).
During EMT, expression of the epithelial marker E-cadherin is lost,
whereas the expression levels of mesenchymal markers, including
vimentin, fibronectin and N-cadherin, are acquired. Furthermore,
EMT has been demonstrated to be associated with tumor invasion and
migration (36). Notably, siSKA2
decreased the protein expression levels of vimentin, fibronectin
and N-cadherin, but had no effect on E-cadherin, in breast cancer
cells. Nevertheless, in response to upregulation of SKA2, the
expression levels of E-cadherin were decreased, whereas N-cadherin,
fibronectin and vimentin were increased. Therefore, SKA2 may
mediate invasion and metastasis in human breast cancer via EMT.
To further analyze the regulation of E-cadherin by
SKA2, cytoplasmic and nuclear proteins were isolated, extracted and
analyzed by western blotting. Notably, the results demonstrated no
obvious alterations in E-cadherin in the cytoplasm; however, its
expression was upregulated in the nucleus when cells were
transfected with siSKA2. Therefore, it was hypothesized that
E-cadherin exists primarily in the cytoplasm and rarely in the
nucleus. When induced by siSKA2, translocation of E-cadherin to the
nucleus may be promoted, thus resulting in an increase in
E-cadherin levels in the nucleus. However, only a small amount of
E-cadherin in the cytoplasm moved to the nucleus; therefore, no
obvious alteration in E-cadherin levels in the cytoplasm was
detected. Similar results were obtained by immunofluorescence.
These results were similar to those reported previously (37,38).
Nevertheless, the specific mechanism through which SKA2 acts upon
E-cadherin remains unclear and requires further study.
In conclusion, the present study provided evidence
to suggest that SKA2 serves a promoting role in breast cancer cell
metastasis by regulating the expression levels of
mesenchymal-associated proteins. These findings have laid the
theoretical basis for further understanding the pathogenesis of
breast cancer, and developing novel diagnostic and therapeutic
strategies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372209), the Hua
Mei Research Foundation (grant no. 2016HMKY35) and the Ningbo
Natural Science Foundation of China (grant nos. 2013A610224 and
2017A610193).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZR and TY collected and analyzed the data, wrote and
revised the manuscript. PW conceived and designed the experiments.
PZ, KL and WL conducted the experiments. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Ningbo No. 2 Hospital and all patients provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jeyaprakash AA, Santamaria A, Jayachandran
U, Chan YW, Benda C, Nigg EA and Conti E: Structural and functional
organization of the ska complex, a key component of the
kinetochore-microtubule interface. Mol Cell. 46:274–286. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanisch A, Silljé HH and Nigg EA: Timely
anaphase onset requires a novel spindle and kinetochore complex
comprising Ska1 and Ska2. EMBO J. 25:5504–5515. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang QH, Qi ST, Wang ZB, Yang CR, Wei YC,
Chen L, Ouyang YC, Hou Y, Schatten H and Sun QY: Localization and
function of the Ska complex during mouse oocyte meiotic maturation.
Cell Cycle. 11:909–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gaitanos TN, Santamaria A, Jeyaprakash AA,
Wang B, Conti E and Nigg EA: Stable kinetochore-microtubule
interactions depend onthe Ska complex and its new component
Ska3/C13Orf3. EMBO J. 28:1442–1452. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boeszoermenyi A, Schmidt JC, Cheeseman IM,
Oberer M, Wagner G and Arthanari H: Resonance assignments of the
microtubule-binding domain of the C. elegans spindle and
kinetochore-associated protein 1. Biomol NMR Assign. 8:275–278.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guimaraes GJ and Deluca JG: Connecting
with Ska, a key complex at the kinetochore-microtubule interface.
EMBO J. 28:1375–1377. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Daum JR, Wren JD, Daniel JJ, Sivakumar S,
McAvoy JN, Potapova TA and Gorbsky GJ: Ska3 is required for spindle
checkpoint silencing and the maintenance of chromosome cohesion in
mitosis. Curr Biol. 19:1467–1472. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rice L, Waters CE, Eccles J, Garside H,
Sommer P, Kay P, Blackhall FH, Zeef L, Telfer B, Stratford I, et
al: Identification and functional analysis of SKA2 interaction with
the glucocorticoid receptor. J Endocrinol. 198:499–509. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia
W, Li J, Li S, Chen L, Ding H, et al: Intronic miR-301 feedback
regulates its host gene, ska2, in A549 cells by targeting MEOX2 to
affect ERK/CREB pathways. Biochem Biophys Res Commun. 396:978–982.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Zhang Y, Zhang C, Weng H, Li Y,
Cai W, Xie M, Long Y, Ai Q, Liu Z, et al: The gene pair PRR11 and
SKA2 shares a NF-Y-regulated bidirectional promoter and contributes
to lung cancer development. Biochim Biophys Acta. 1849:1133–1144.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhuang H, Meng X, Li Y, Wang X, Huang S,
Liu K, Hehir M, Fang R, Jiang L, Zhou JX, et al: Cyclic AMP
responsive element-binding protein promotes renal cell carcinoma
proliferation probably via the expression of spindle and
kinetochore-associated protein 2. Oncotarget. 7:16325–16337. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu Z and Li Y, Takwi A, Li B, Zhang J,
Conklin DJ, Young KH, Martin R and Li Y: miR-301a as an NF-κB
activator in pancreatic cancer cells. EMBO J. 30:57–67. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu
Y, Gu Q, Zhu Z and Liu B: Overexpressed miR-301a promotes cell
proliferation and invasion by targeting RUNX3 in gastric cancer. J
Gastroenterol. 48:1023–1033. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bian EB, Ma CC, He XJ, Wang C, Zong G,
Wang HL and Zhao B: Epigenetic modification of miR-141 regulates
SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget.
7:30610–30625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang IC, Chiang TI, Lo C, Lai YH, Yue CH,
Liu JY, Hsu LS and Lee CJ: Anemone altaica induces apoptosis in
human osteosarcoma cells. Am J Chin Med. 43:1031–1042. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin CC, Chao PY, Shen CY, Shu JJ, Yen SK,
Huang CY and Liu JY: Novel target genes responsive to apoptotic
activity by Ocimum gratissimum in human osteosarcoma cells. Am J
Chin Med. 42:743–767. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Libson S and Lippman M: A review of
clinical aspects of breast cancer. Int Rev Psychiatry. 26:4–15.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng Z, Chen X, Zhu D, Luo Z and Yang M:
Low expression of circulating MicroRNA-34c is associated with poor
prognosis in triple-negative breast cancer. Yonsei Med J.
58:697–702. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Wei B, Sonmez C, Li Z and Peng L:
High tumor budding count is associated with adverse
clinicopathologic features and poor prognosis in breast carcinoma.
Hum Pathol. 66:222–229. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma F, Li W, Liu C, Li W, Yu H, Lei B, Ren
Y, Li Z, Pang D and Qian C: MiR-23a promotes TGF-β1-induced EMT and
tumor metastasis in breast cancer cells by directly targeting CDH1
and activating Wnt/β-catenin signaling. Oncotarget. 41:69538–69550.
2017.
|
|
22
|
Tang X, Ding CK, Wu J, Sjol J, Wardell S,
Spasojevic I, George D, McDonnell DP, Hsu DS, Chang JT and Chi JT:
Cystine addiction of triple-negative breast cancer associated with
EMT augmented death signaling. Oncogene. 30:4235–4242. 2017.
View Article : Google Scholar
|
|
23
|
Neelakantan D, Zhou H, Oliphant MUJ, Zhang
X, Simon LM, Henke DM, Shaw CA, Wu MF, Hilsenbeck SG, White LD, et
al: EMT cells increase breast cancer metastasis via paracrine GLI
activation in neighbouring tumour cells. Nat Commun. 8:157732017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lakhtakia R, Aljarrah A, Furrukh M and
Ganguly SS: Epithelial mesenchymal transition (EMT) in metastatic
breast cancer in omani women. Cancer Microenviron. 10:25–37. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshida R, Nagira M, Kitaura M, Imagawa N,
Imai T and Yoshie O: Secondary lymphoid-tissue chemokine is a
functional ligand for the CC chemokine receptor CCR7. J Biol Chem.
273:7118–22. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoon SO, Park SJ, Yun CH and Chung AS:
Roles of matrix metalloproteinases in tumor metastasis and
angiogenesis. J Biochem Mol Biol. 36:128–37. 2003.PubMed/NCBI
|
|
28
|
John A and Tuszynski G: The role of matrix
metalloproteinases in tumor angiogenesis and tumor metastasis.
Pathol Oncol Res. 7:14–23. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zong H, Yin B, Zhou H, Cai D, Ma B and
Xiang Y: Inhibition of mTOR pathway attenuates migration and
invasion of gallbladder cancer via EMT inhibition. Mol Biol Rep.
41:4507–4512. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Czubaty A and Piekiełko-Witkowska A:
Protein kinases that phosphorylate splicing factors: Roles in
cancer development, progression and possible therapeutic options.
Int J Biochem Cell Biol. 10:102–115. 2017. View Article : Google Scholar
|
|
32
|
Okita Y, Kimura M, Xie R, Chen C, Shen LT,
Kojima Y, Suzuki H, Muratani M, Saitoh M, Semba K, et al: The
transcription factor MAFK induces EMT and malignant progression of
triple-negative breast cancer cells through its target GPNMB. Sci
Signal. 10:eaak93972017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shi Y, Zhao Y, Shao N, Ye R, Lin Y, Zhang
N, Li W, Zhang Y and Wang S: Overexpression of microRNA-96-5p
inhibits autophagy and apoptosis and enhances the proliferation,
migration and invasiveness of human breast cancer cells. Oncol
Lett. 13:4402–4412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma F, Li W, Liu C, Li W, Yu H, Lei B, Ren
Y, Li Z, Pang D and Qian C: MiR-23a promotes TGF-β1-induced EMT and
tumor metastasis in breast cancer cells by directly targeting CDH1
and activating Wnt/β-catenin signaling. Oncotarget. 41:69538–69550.
2017.
|
|
35
|
Hidaka M, Koga T, Kiyota H, Horiguchi T,
Shi QW, Hirose K and Uchida T: Relationship between the structures
of taxane derivatives and their microtubule polymerizationactivity.
Biosci Biotechnol Biochem. 76:349–352. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Micalizzi DS, Farabaugh SM and Ford HL:
Epithelial-mesenchymal transition in cancer: Parallels between
normal development and tumor progression. J Mammary Gland Biol
Neoplasia. 15:117–134. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ferber EC, Kajita M, Wadlow A, Tobiansky
L, Niessen C, Ariga H, Daniel J and Fujita Y: A role for the
cleaved cytoplasmic domain of E-cadherin in the nucleus. J Biol
Chem. 283:12691–12700. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Du W, Liu X, Fan G, Zhao X, Sun Y, Wang T,
Zhao R, Wang G, Zhao C, Zhu Y, et al: From cell membrane to the
nucleus: An emerging role of E-cadherin in gene transcriptional
regulation. J Cell Mol Med. 18:1712–1719. 2014. View Article : Google Scholar : PubMed/NCBI
|