Introduction
The immune system distinguishes between self from
non-self components, and induces a defensive response against an
invading pathogen; it has evolved to protect the host against
pathogenic microbes (1). Tissue
injury or infection causes a highly regulated defensive process
called inflammation (2), which is
a key process in the host defense system. However, excessive
inflammation can be a cause for the development of various human
diseases including inflammatory disorders, autoimmune disease,
cancer, and cardiovascular diseases (3).
Autoimmunity is the loss of immunological
self-tolerance in the host organism, which brings about an abnormal
immune response against its own self (4). Autoimmune diseases are pathological
conditions that lead to chronic inflammation and tissue and organ
damage. Among them, neuromyelitis optica spectrum disorder (NMOSD)
is caused by demyelination in the central nervous system due to the
binding of serum autoantibodies to astrocyte water channel
aquaporin-4 (5). Recent
metabolomic analyses revealed that lactate levels in cerebrospinal
fluid (CSF) from clinical patients with NMOSD are elevated
(6,7).
Lactate is the final product resulting from
nonoxidative glycolysis, and is considered a waste product of
cellular metabolism (8,9). However, abnormal lactate levels have
been detected in many diseases such as cancer and rheumatoid
arthritis synovitis (9,10). Additionally, lactate has begun to
be recognized as an active molecule that regulates the immune
response (9). Lactate
dehydrogenase is the enzyme that catalyzes the forward and backward
conversion of pyruvate to lactate. Lactate dehydrogenase A (LDHA)
converts pyruvate to lactate and produces NAD+. On the
contrary, lactate dehydrogenase B (LDHB) catabolizes lactate to
pyruvate. Previous studies showed that targeting LDHA is an
attractive strategy for the development of cancer therapeutics
(11,12). In this study, we investigated
whether the inhibition of LDHA has anti-inflammatory effects in
lipopolysaccharide (LPS)-induced RAW 264.7 macrophages.
Materials and methods
RAW 264.7 cell culture
Murine RAW 264.7 macrophages were obtained from the
American Type Culture Collection (ATCC; Manassas, VA, USA). Cells
were cultured in complete high glucose Dulbecco's modified Eagle's
Medium (DMEM; Welgene, Inc., Daejeon, Korea) containing 10% fetal
bovine serum (FBS), 100 U/ml penicillin, and 100 µg/ml streptomycin
at 37°C in a humidified incubator under conditions of 5%
CO2.
Cell viability assay
Cell viability was analyzed by a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. RAW 264.7 cells were seeded in a 96-well culture plate at a
cell density of 2.0×104 cells/well. Then, MTT
colorimetric solutions were added to each well, and the absorbance
was measured at 570 nm using a microplate spectrophotometer (BioTek
instruments, Winooski, VT, USA).
RNA preparation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using the easy-spin Total RNA
Extraction kit (iNtRON Biotechnology, Sungnam, Korea), according to
the manufacturer's procedure. cDNA was synthesized from RNA in a
PCR Thermal Cycler (Takara Bio, Inc., Otsu, Japan) using the
iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). RT-qPCR analysis was performed with the SYBR Green Master
Mix (Takara Bio, Inc.), using gene-specific primers, in a
StepOnelus Real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). PCR conditions were initially
maintained at 95°C for 5 min, followed by 40 cycles of 95°C for 15
sec, 60°C for 15 sec and 72°C for 45 sec. The level of mRNA
expression was normalized to the level of GAPDH expression. The
relative value of gene expression was analyzed using the
2−∆∆Cq method (13).
The primer sequences used in this study are presented in Table I.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Target gene | Primer sequences
(5′-3′) |
|---|
| GAPDH | F:
CAAGGTCATCCATGACAACTTTG |
|
| R:
GGCCATCCACAGTCTTCTGG |
| TNF-α | F:
CAGGCGGTGCCTATGTCTC |
|
| R:
CGATCACCCCGAAGTTCAGTAG |
| IL-1β | F:
GAAATGCCACCTTTTGACAGTG |
|
| R:
TGGATGCTCTCATCAGGACAG |
| IL-6 | F:
TCCAGTTGCCTTCTTGGGAC |
|
| R:
GTGTAATTAAGCCTCCGACTTG |
| iNOS | F:
GTTCTCAGCCCAACAATACAAGA |
|
| R:
GTGGACGGGTCGATGTCAC |
| COX-2 | F:
TGAGTACCGCAAACGCTTCTC |
|
| R:
TGGACGAGGTTTTTCCACCAG |
| HO-1 | F:
TGAAGGAGGCCACCAAGGAGG |
|
| R:
AGAGGTCACCCAGGTAGCGGG |
| LDHA | F:
TGTCTCCAGCAAAGACTACTGT |
|
| R:
GACTGTACTTGACAATGTTGGGA |
Measurement of lactate in culture
media
RAW 264.7 cells were seeded in a 6-well culture
plate in the presence and absence of FX11 or LPS. Culture medium
was collected and lactate levels were measured by UPLC-tandem mass
spectrometry (Triple Quad 5500; Sciex, Framingham, MA, USA), as
described previously, with modifications (14).
Western blot analyses
RAW 264.7 cells were seeded in a 6-well culture
plate. The next day, cells were treated with different
concentrations of FX11 or LDHA-specific small interfering RNA
(siRNA) (50 nM). The cells were harvested by lysis buffer (150 mM
Sodium chloride, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS,
50 mM Tris-HCl, pH 7.5 and 2 mM EDTA) containing protease
inhibitors (Roche Diagnostics, Indianapolis, IN, USA) to obtain
whole cell lysates. Protein concentrations were measured by using a
BCA protein assay kit (Thermo Fisher Scientific, Inc.). Equal
amounts of the proteins were subjected to SDS-PAGE, and then
transferred onto a PVDF membrane. The membrane was blocked with 5%
skim milk and incubated with specific primary antibodies against
p-JNK, JNK, p-ERK, ERK, p-p38, p38, inducible nitric oxide synthase
(iNOS), cyclooxygenase 2 (COX-2), HO-1, and LDHA (Cell Signaling
Technology, Inc., Danvers, MA, USA), and β-actin (EMD Millipore,
Billerica, MA, USA). The blots were then incubated with the
horseradish peroxidase (HRP)-linked Goat anti-Rabbit (or Mouse)
secondary IgG and IgM (EMD Millipore) at room temperature for 1 h.
The signals were detected using the SuperSignal West Pico
Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.). A
chemiluminescence image analyzer (Vilber Lourmat Corporation,
Torcy, France) was used for the detection of signals.
Quantification of nitrite
The concentrations of nitrite in the supernatant of
cell culture media were measured by Griess reagent (Molecular
Probes; Thermo Fisher Scientific, Inc.). The cell culture media and
Griess reagent were taken in a 96-well plate. This mixture was then
incubated for 30 min at room temperature. The nitrite
concentrations were measured by measuring the absorbance at 548 nm
using a microplate spectrophotometer (BioTek Instruments Inc.,
Winooski, VT, USA).
Determination of the levels of
pro-inflammatory cytokines secreted
The concentrations of pro-inflammatory cytokines
[tumor necrosis factor (TNF)-α, interleukin (IL)-1β, and IL-6] in
the supernatant of cell culture media were measured using a mouse
ELISA kit (Enzo Life Sciences, Ann Arbor, MI, USA), according to
the manufacturer's instructions.
Transfection of lactate dehydrogenase
A-specific siRNA
LDHA silencing in RAW 264.7 macrophages was
performed by transiently transfecting the cells with mouse
LDHA-specific siRNAs. LDHA-siRNA duplexes and scrambled siRNA were
purchased from OriGene Technologies (Rockville, MD, USA). RAW 264.7
cells were seeded in a 6-well culture plate at a cell density of
4×105 cells/well. The cells were transfected with
LDHA-specific siRNA for 48 h using the Lipofectamine 2000 reagent,
according to the manufacturer's instructions (Invitrogen; Thermo
Fisher Scientific, Inc.).
Affymetrix whole transcript expression
array methods
The Affymetrix whole transcript expression array
process was performed according to the manufacturer's protocol
(Genechip whole transcript plus reagent kit). cDNA was synthesized
using the genechip whole transcript amplification kit according to
the manufacturer's instructions. The sense cDNA was then fragmented
and biotin-labeled with TdT (terminal deoxynucleotidyl transferase)
using the Genechip WT terminal labeling kit. Approximately 5.5 µg
of the labeled DNA target was hybridized to the Affymetrix genechip
mouse 2.0 ST array at 45°C for 16 h. The hybridized arrays were
washed and stained on a geneChip fluidics station 450 and scanned
on a GCS3000 scanner (Affymetrix; Thermo Fisher Scientific, Inc.).
Signal values were determined by using the Affymetrix®
GeneChip™ command console software.
Raw data preparation and statistical
analysis
Raw data were extracted automatically in Affymetrix
data extraction protocol using the software provided by Affymetrix
genechip Command Console Software (AGCC). After importing the CEL
files, the data were summarized and normalized using the robust
multi-average (RMA) method implemented in the Affymetrix expression
console software (EC). We exported the result with gene level RMA
analysis and performed the differentially expressed gene (DEG)
analysis. For a DEG set, a hierarchical cluster analysis was
performed using complete linkage and Euclidean distance as a
measure of similarity. Gene-Enrichment and functional annotation
analysis for significant probe lists was performed using gene
ontology (www.geneontology.org/) and KEGG (www.genome.jp/kegg/). All data analysis and
visualization of DEGs were performed by using R 3.1.2 (www.r-project.org).
Isolation of human peripheral blood
mononuclear cells (PBMCs)
Experiments were performed using whole heparinized
blood obtained by venopuncture from healthy donors and from
patients with Behçet's disease (Healthy controls, n=4. Behçet's
disease, n=6). The present study was conducted in accordance with
the provisions of the Declaration of Helsinki for the participation
of human subjects in research and was approved by the Institutional
Review Board of Ajou University Medical Center (IRB no.
BMR-GEN-14-463); written informed consent was obtained from all
participants. PBMCs were isolated using histopaque-1077
density-centrifugation and were cultured in RPMI-1640 medium
supplemented with 10% FBS, 1% non-essential amino acids (100 mM),
and 1% L-glutamine (200 mM). The cultures were maintained at 37°C
in a 5% CO2 atmosphere with 95% humidity.
Data analysis and statistics
All the data are expressed as mean ± standard error
mean. Statistical differences among groups were calculated using
one-way analysis of variance with a Bonferroni post hoc test.
P<0.05 was considered to indicate a statistically significant
difference. Statistical analysis was performed with GraphPad prism
6 software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Inhibition of lactate dehydrogenase A
suppresses expression of cytokines
Lactate is implicated in excessive cell
proliferation and autoimmune diseases (6,11).
Therefore, we hypothesized that modulation of lactate regulates
inflammatory response induced by LPS. We investigated whether the
pharmacological inhibition of LDHA affects the transcriptional
expression of cytokines. Among various LDH inhibitors, we utilized
FX11
[3-dihydroxy-6-methyl-7-(phenylmethyl)-4-propylnaphthalene-1-carboxylic
acid], a gossypol derivative, which demonstrates a higher
selectivity for the forward reaction of LDHA, than for the reverse
reaction of LDHB (Ki=0.05 µM for LDHA, Ki=20 µM for LDHB) (15,16).
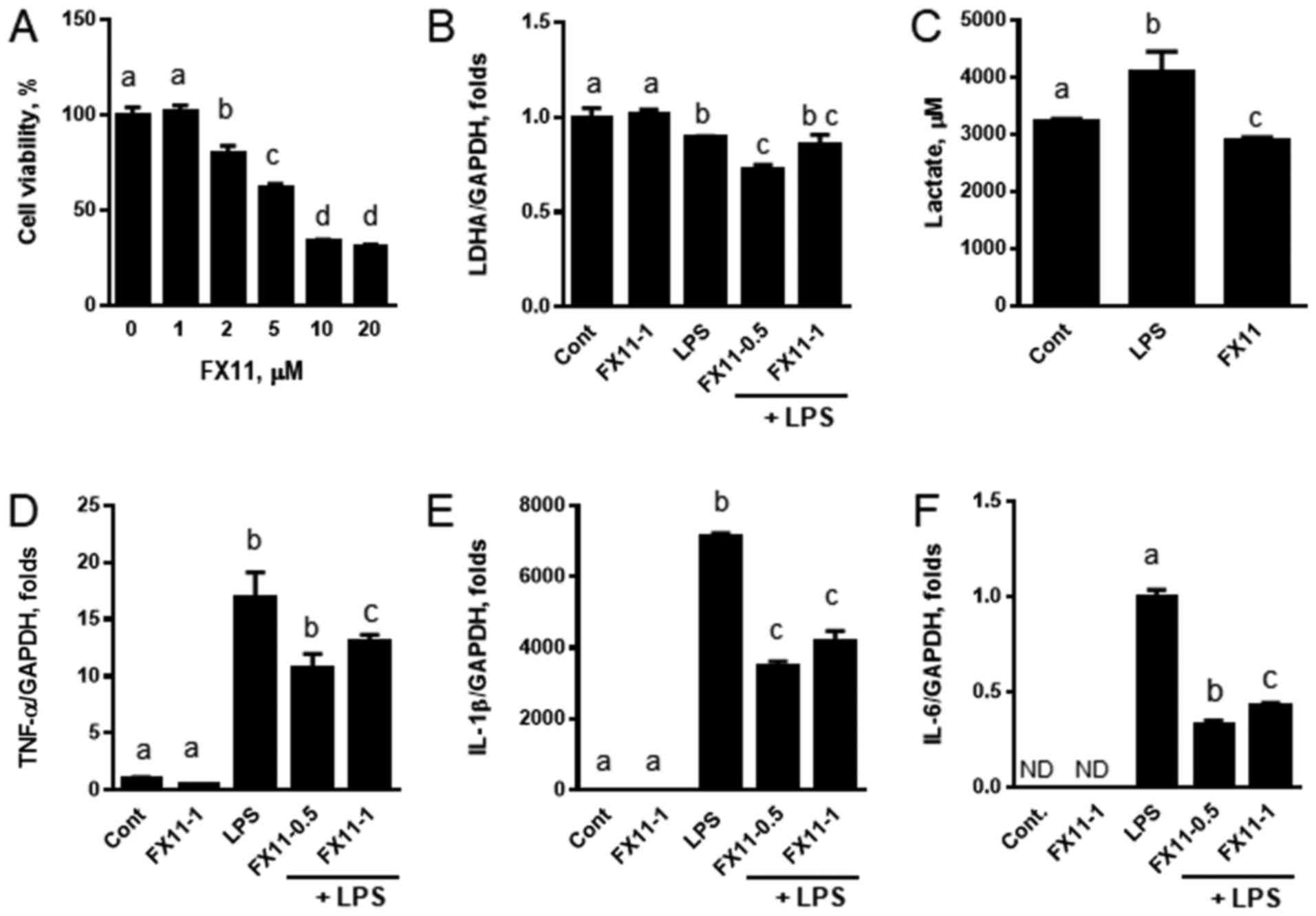
First, we measured the cell viability, to determine a tolerable
dose range of FX11 for the study. When RAW 264.7 cells were treated
with various concentrations of FX11 for 24 h, it was seen that the
cell survival was reduced above FX11 concentrations of 2 µM
(Fig. 1A); we thus decided to
treat the cells with FX11 concentrations below 2 µM. We treated RAW
264.7 cells with LPS to activate the inflammatory response in the
presence or absence of FX11. Inhibition of LDHA did not alter the
expression of LDHA. However, a slight decrease in LDHA expression
was found in samples treated with 0.5 µM FX11, regardless of LPS
treatment (Fig. 1B). When the
cells were treated with LPS, the lactate levels in the media
increased, and FX11 treatment reduced the lactate levels to values
lower than those observed in the untreated control (Fig. 1C). This suggests that FX11
pharmacologically inhibited LDHA in RAW 264.7 cells. The
transcriptional induction of cytokines including TNFα, IL-1β, and
IL-6 by LPS was suppressed when the cells were co-treated with FX11
(Fig. 1D-F). The most notable
effect was found in IL-6. These results suggest that reduced
lactate levels correlate with the suppression of the LPS-induced
expression of cytokines in macrophages.
Inhibition of lactate dehydrogenase A
downregulates inflammatory genes and inhibits the phosphorylation
of p38 MAP kinase
Inflammatory response induced by bacterial infection
activates cytokine production and inflammatory mediators such as
nitric oxide (NO) and eicosanoids (17,18).
Anti-inflammatory effects were indicated by the suppression of iNOS
and COX-2 expression, leading to decreased NO and PGE2 production,
respectively (19). In contrast,
heme oxygenase 1 (HO-1) downregulates the expression of cytokines
such as TNF-α, IL-1β, and IL-6, and reduces NO production. Thus,
HO-1 is suggested as an anti-inflammatory mediator (2). To determine the degree of
inflammation regulated by lactate, we examined the effects of FX11
on the transcriptional expression of iNOS and COX-2. Inhibition of
LDHA by FX11 downregulated the LPS-induced expression of iNOS and
COX-2 (Fig. 2A and B). However,
the expressions of iNOS, LDHA, and HO-1 proteins were not changed
(Fig. 2D), and the production of
nitrite (Fig. 2C) and PGE2 was not
altered (data not shown). Only COX-2 was downregulated after
treatment with 1 µM FX11.
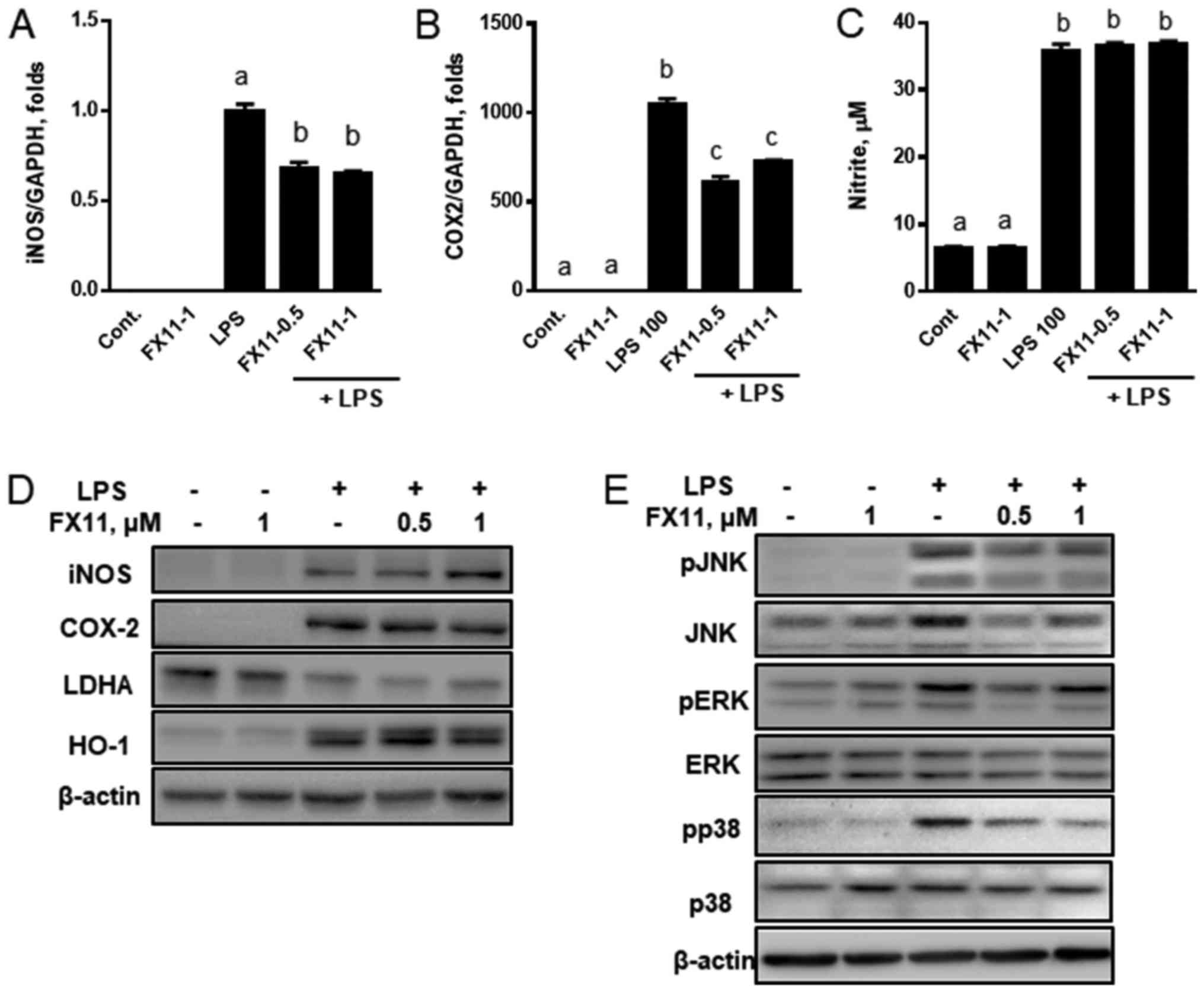 | Figure 2.LDHA inhibition by FX11 downregulates
the expression of inflammatory genes and inhibits MAP kinase
pathways in LPS-treated RAW 264.7 cells. RAW 264.7 cells were
treated with LPS in the presence or absence of various
concentrations of FX11 (0.5 and 1 µM) for 24 h. The mRNA
expressions of (A) iNOS and (B) COX-2 were then measured by reverse
transcription-quantitative polymerase chain reaction and the levels
of (C) nitrite were measured by Griess reagent assay. (D) Protein
levels of iNOS, COX-2, LDHA and HO-1 were measured by immunoblot
analyses. (E) The degrees of MAP kinase phosphorylation were
measured by immunoblot analyses. Data are presented as the mean ±
standard error of the mean (n=3). mRNA expression was normalized to
the level of GAPDH expression. Different letters indicate a
significant difference among treatments at P<0.05. LDHA, lactate
dehydrogenase; LPS, lipopolysaccharide; iNOS, nitric oxide
synthase; COX-2, cyclooxygenase 2; HO-1, heme oxygenase 1; MAP,
mitogen-activated protein; Cont., control; p-, phosphorylated; JNK,
c-Jun N terminal kinase; ERK, extracellular signal-regulated
kinase. |
Since FX11 transcriptionally altered the expression
of iNOS, COX-2, HO-1, and inflammatory cytokines, we investigated
whether the anti-inflammatory effects of FX11 inhibit the
phosphorylation of MAP kinases in LPS-induced inflammatory
response. We found that the phosphorylation of JNK, ERK, and p38
was reduced by FX11 treatment (Fig.
2E). These results suggest that FX11 downregulates the
transcriptional expression of iNOS and COX-2, and inhibits the
phosphorylation of MAP kinases.
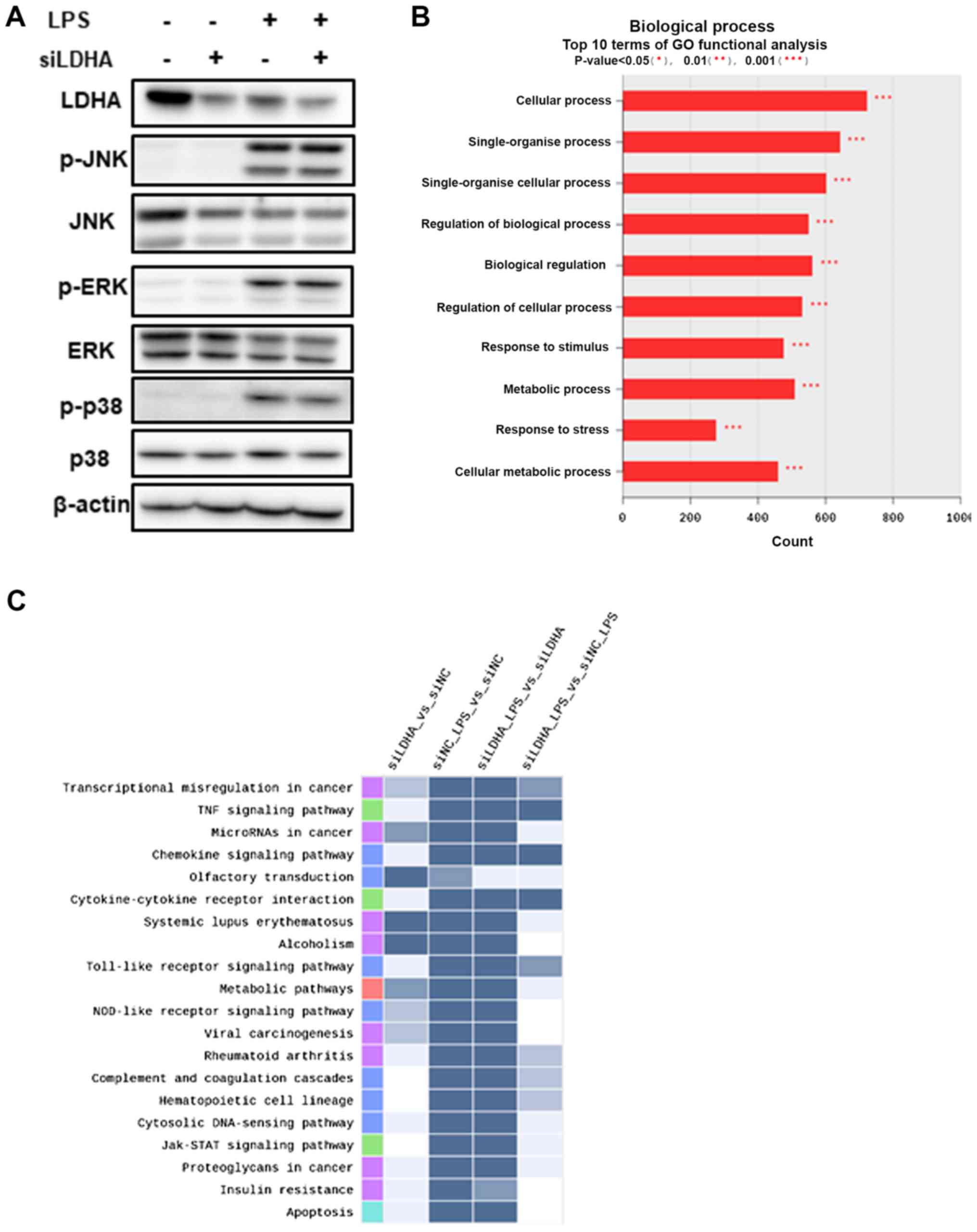
Suppression of LDHA exerts
anti-inflammatory effects via the reduced production of IL-6 and
nitrites
To support the role of lactate production in
anti-inflammatory response, we suppressed LDHA expression by siRNA
and examined its effects on the expression of inflammatory genes
and signal mediators. LDHA-specific siRNA (siLDHA) downregulated
LDHA expression and siLDHA also reduced the LPS-mediated protein
induction of iNOS and COX-2 (Fig.
3A). However, the protein levels of HO-1 were not altered. The
transcriptional upregulation of IL-6, iNOS, and COX-2 by LPS was
attenuated by siLDHA (Fig. 3B-D).
Consistent with these expression results, the production of IL-6
and nitrites was reduced when siLDHA was transiently transfected
into the cells (Fig. 3E and F). We
then examined the effects of genetic downregulation of LDHA on MAP
kinase signaling. Although we did not find any changes in the
phosphorylation of JNK and ERK, the phosphorylation of p38 was
slightly reduced by siLDHA (Fig.
4A). These results suggest that siLDHA-mediated genetic
suppression resulted in anti-inflammatory effects by the
downregulation of inflammatory gene expression and inhibition of
p38 phosphorylation.
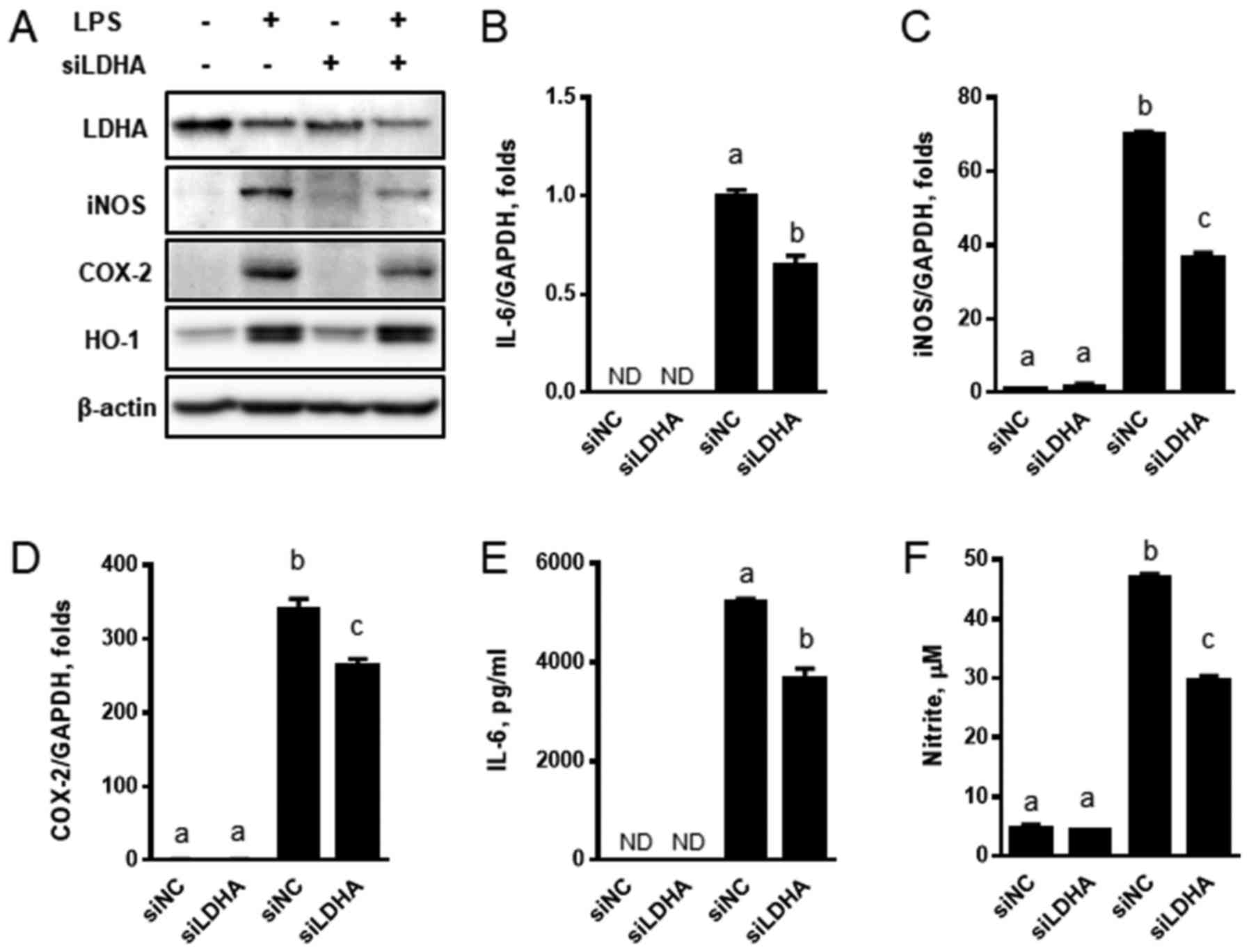 | Figure 3.Downregulation of LDHA by siRNA
suppresses the expression of inflammatory genes and cytokines. (A)
RAW 264.7 cells transfected with LDHA-specific siRNA were treated
with LPS in the presence or absence of FX11 for 24 h, and
immunoblot analyses were performed with LDHA, iNOS, COX-2, and
HO-1. The expressions of (B) IL-6, (C) iNOS and (D) COX-2 were
measured by reverse transcription-quantitative polymerase chain
reaction. The levels of (E) IL-6 and (F) nitrite were measured by
ELISA and Griess reagent respectively. Data are presented as the
mean ± standard error of the mean (n=3). mRNA expression was
normalized to the level of GAPDH expression. Different letters
indicate a significant difference among treatments at P<0.05.
LDHA, lactate dehydrogenase; LPS, lipopolysaccharide; siRNA, small
interfering RNA; iNOS, nitric oxide synthase; COX-2, cyclooxygenase
2; HO-1, heme oxygenase 1; ND, not detectable; siNC, scrambled
negative control. |
siRNA-mediated LDHA suppression alters
gene expression in transcriptional misregulation in
inflammation
Alteration of inflammatory gene expression was found
in LDHA-specific siRNA transfected RAW 264.7 cells. To determine
the general effects of LDHA suppression, we performed microarray
gene analysis using mRNA from both LPS-treated and untreated
siLDHA-transfected cells. Gene ontology functional analyses
(Fig. 4B) and pathway analysis
(Fig. 4C) revealed that LDHA
silencing by siRNA is implicated in diverse signaling pathways. The
downregulated genes were clearly enriched in ‘cytokine activity’
(GO: 0005125), including TNF signaling pathway, chemokine
signaling, cytokine-cytokine receptor interaction, and TLR
signaling pathway, when we compared cells treated with LPS alone
and those treated with LPS plus siLDHA. These results suggest that
LDHA knockdown is associated with suppression of cytokine-related
inflammatory responses.
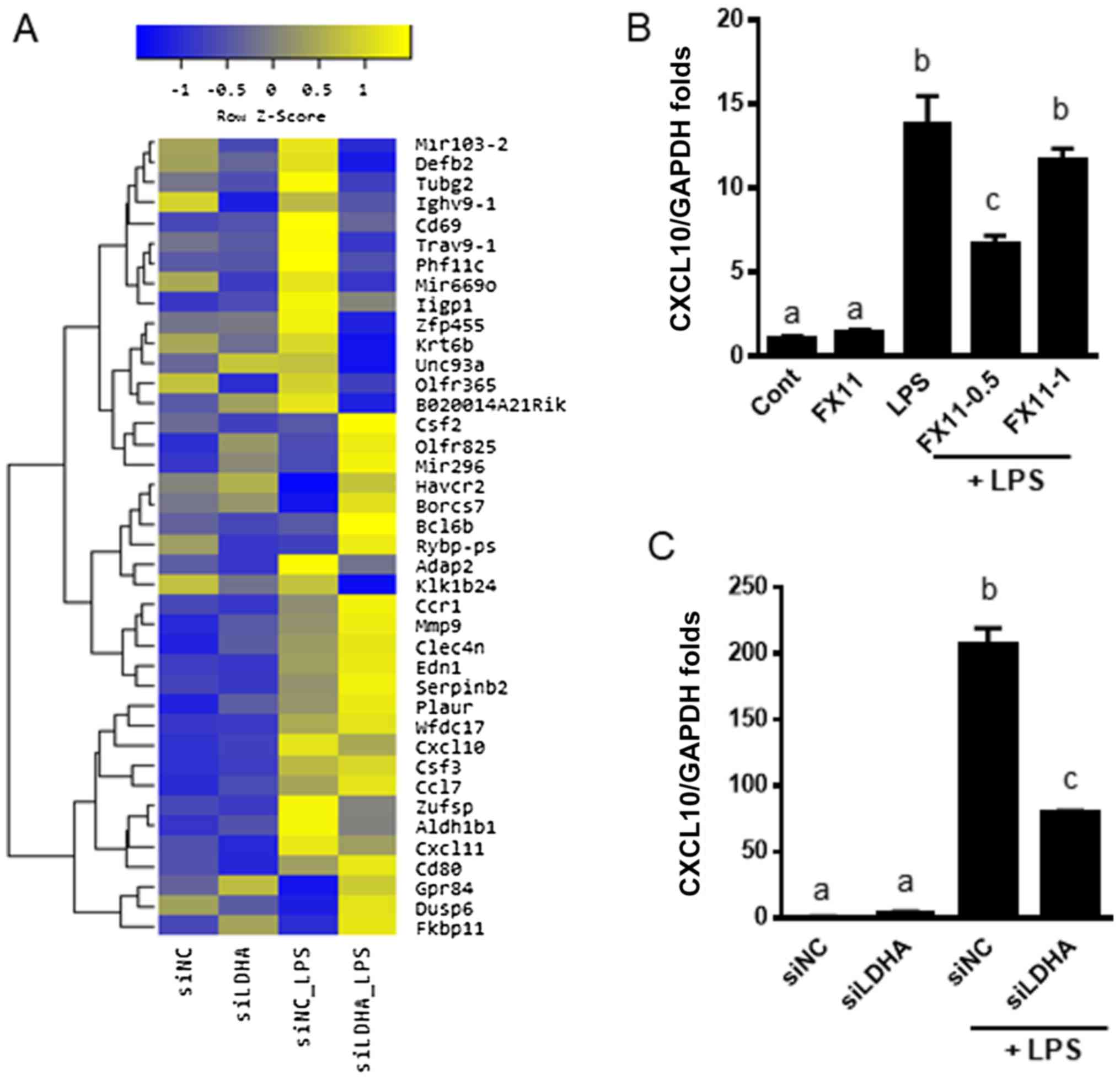
We found that the expression of inflammatory and
immune-related genes was mainly altered (Fig. 5A). To confirm the involvement of
LDHA in inflammation, we measured the expression of CXCL10, which
was downregulated as shown in the microarray analyses. We found
that pharmacologic or genetic inhibition of LDHA by FX11 treatment
or siLDHA transfection, respectively, downregulated CXCL10 in
LPS-treated cells (Fig. 5B and C).
These results were consistent with those of the microarray
analyses. These results suggest that lactate metabolism is
positively correlated with inflammatory and immune responses.
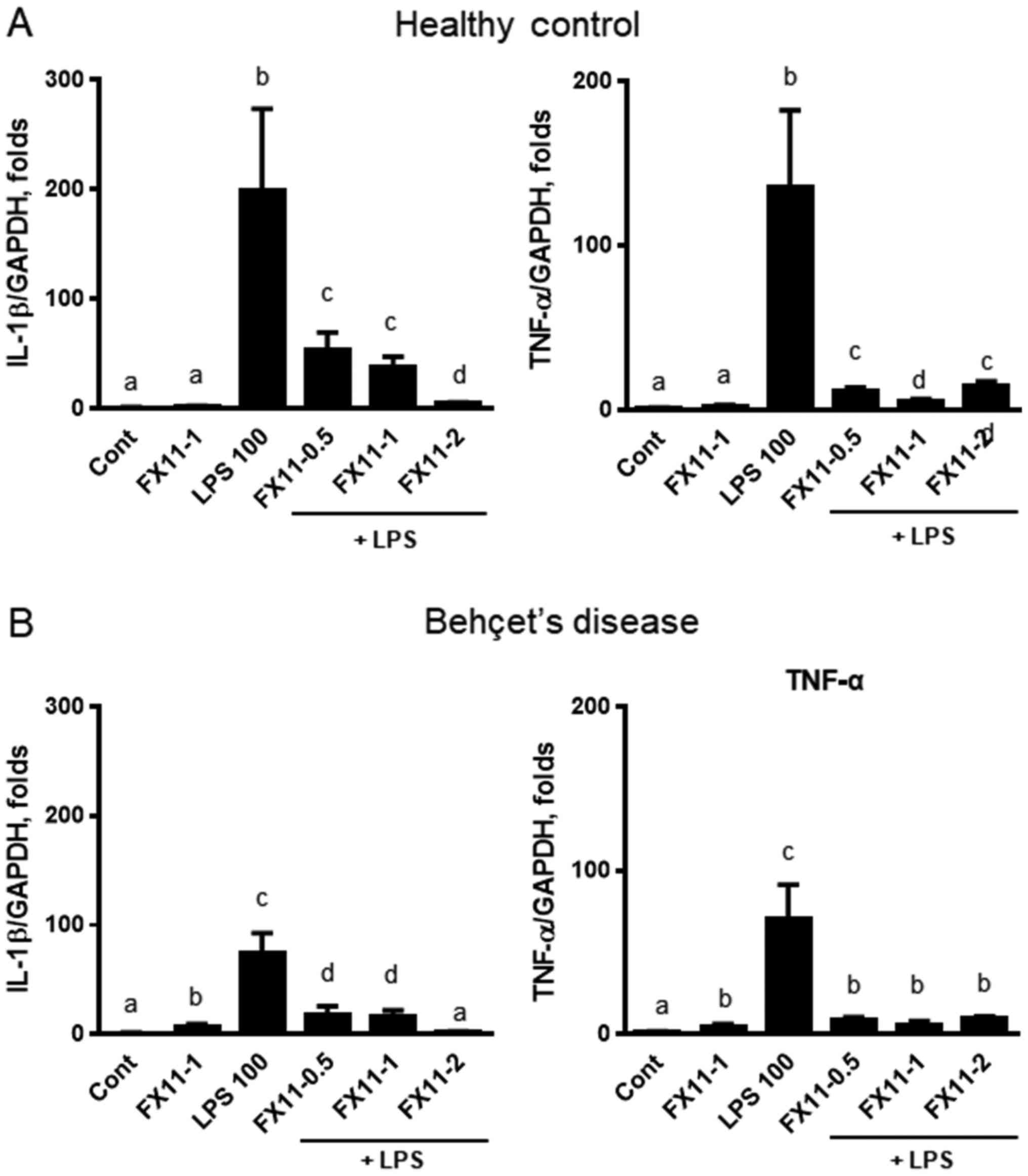
Clinical implication of LDHA
suppression in PBMCs
To determine the clinical relevance of LDHA
inhibition, we treated PBMCs isolated from healthy controls and
patients with Behçet's disease with FX-11 in the presence or
absence of LPS pre-treatment for 6 h. Behçet's disease is an
autoimmune disease characterized by ulceration of the mouth and
genitals, inflammation in the eye, and arthritis. IL-1β and TNFα
were significantly upregulated by LPS treatment (Fig. 6). When cells were treated with FX11
in the presence of LPS, the expression of IL-1β and TNFα was
suppressed in both PBMCs from healthy controls (Fig. 6A) and from patients with Behçet's
disease (Fig. 6B). These results
suggest that LDHA suppression downregulates the inflammatory
response in PBMCs isolated from clinical subjects.
Discussion
Lactate has long been considered a waste product of
cellular metabolism. Currently, lactate is known to serve as an
indicator of the status of cell metabolism (20). There have been reports about the
correlation of increased lactate levels and development of various
diseases including cancer and rheumatoid arthritis synovitis
(9,14). Recent reports have demonstrated
that the lactate levels in CSF from clinical patients with NMOSD
were elevated (6,7). We hypothesized that the suppression
of LDHA would downregulate LPS-induced inflammatory response in RAW
264.7 macrophages. To suppress LDHA activity, we administered a
pharmacological inhibitor, FX11, and a genetic suppressor,
LDHA-specific siRNA. In this study, we demonstrated that: i) The
LDHA inhibitor FX11 downregulated the production of inflammatory
cytokines, iNOS, and COX-2 via the inhibition of MAP kinase
phosphorylation; ii) siRNA-mediated suppression of LDHA
downregulated the gene expression of iNOS and COX-2, IL-6
production, and NO levels; iii) Suppression of LDHA by
LDHA-specific siRNA modulates gene expression of cancer and
inflammatory genes, as seen in the results of the microarray
analyses.
Lactate is the final product resulting from
glycolysis, and is accumulated at inflammatory sites (8). Lactate dehydrogenase catalyzes the
conversion of pyruvate to lactate and the reverse reaction. Reports
that LDHA inhibition prevents cancer development have drawn
attention for the development of cancer therapeutics, and suggest
an important role of lactate in the regulation of cell metabolism
and survival (11,21,22).
Additionally, metabolomics analyses of clinical CSF samples from
NMOSD patients suggest that lactate is a critical regulator in the
development of autoimmune diseases (6,7). In
contrast, the fact that siLDHA-mediated suppression reduced IL-6
and NO production suggests that LDHA inhibition clearly alleviates
LPS-mediated inflammatory response. Inconsistency of the results
from FX11 and siLDHA treatment could be due to the off-target
effects of FX-11. However, the exact reason for this is not clear
yet.
Another point is that the phosphorylation of MAP
kinases is inhibited by LDHA inhibition. In the immune system, MAP
kinases including ERK, JNK, and p38 play an important role in
cytokine production, which is a cellular response (23). Cytokines increase the levels of
leukocyte adhesion molecules that induce leukocyte extravasation
(24). Among the MAP kinase
family, ERK is mostly related to anabolic processes including cell
division, growth, and differentiation. On the other hand, JNK and
p38 are mainly involved in cellular responses in stress condition
(25). Some reports reveal that
p38 plays an important role in the production of various
inflammatory mediators such as TNF-α, IL-1β, IL-6, COX-2, and PGE2
(26–28). We expected that the
anti-inflammatory effects of LDHA downregulation were associated
with the modulation of MAP kinase signaling pathways. Indeed, our
studies demonstrated that immunosuppression by FX11-induced LDHA
inhibition was closely associated with general MAP kinase signaling
pathways. In contrast, LDHA suppression by siRNA only reduced p38
phosphorylation. These results implied that the anti-inflammatory
effect of FX11 on phosphorylation of MAP kinases is due to the
compound-specific effect. Lactate reduction specifically modulates
the p38 MAP kinase pathway, but further studies will be needed to
determine the outcome associated with p38 and immunosuppression by
LDHA suppression.
Our microarray results suggest that LDHA suppression
is associated with various signaling pathways involving ‘cytokine
activity’. These microarray results suggest that lactate metabolism
is positively correlated with inflammatory response. Detailed
analysis linking the altered signaling pathways and activation of
inflammatory response should be conducted.
The clinical status of autoimmune diseases is that
macrophages are already activated, and the secreted inflammatory
cytokines exacerbate the inflammatory state. Knowing whether
reduction in lactate levels is effective in preventing early
activation of macrophages can be critical for applying this
strategy in treating autoimmune diseases. The fact that
inflammatory cytokine expression was downregulated in PBMCs from
healthy control subjects and in patients with autoimmune Behçet's
disease upon treatment with LDHA inhibitor suggests that LDHA
inhibition is associated with suppression of inflammatory responses
in the clinical setting.
Collectively, we found that the suppression of LDHA
has anti-inflammatory effects due to the downregulation of several
inflammatory mediators including cytokines and NO. Consequently,
our findings suggest that LDHA can be a potential therapeutic
target for autoimmune diseases via its anti-inflammatory effects in
macrophages.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Bio and
Medical Technology Development Program through the National
Research Foundation of Korea, funded by the Korean government
(MSIP; grant no. NRF-2014M3A9B6069338).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YJS performed the experiments, and prepared the
figures and the manuscript. AK, GT and HYY performed the
experiments evaluating protein expression. ESL and MJP performed
the experiments on clinical samples from patients. YJK and SMS
performed virtual screening of the bioactive compounds. TSP
conceived and designed the present study, supervised the project
and assisted with data interpretation. All authors approved the
final manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the provisions of the Declaration of Helsinki for the participation
of human subjects in research and was approved by the Institutional
Review Board of Ajou University Medical Center (IRB no.
BMR-GEN-14-463); written informed consent was obtained from all
participants.
Patient consent for publication
Patient consent for publication was obtained from
all participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chaplin DD: Overview of the immune
response. J Allergy Clin Immunol. 125((2 Suppl 2)): S3–S23. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jeong YH, Oh YC, Cho WK, Yim NH and Ma JY:
Anti-inflammatory effect of rhapontici radix ethanol extract via
inhibition of NF-kB and MAPK and induction of HO-1 in macrophages.
Mediators Inflamm. 2016:72169122016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kaminska B: MAPK signalling pathways as
molecular targets for anti-inflammatory therapy-from molecular
mechanisms to therapeutic benefits. Biochim Biophys Acta.
1754:253–262. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hewagama A and Richardson B: The genetics
and epigenetics of autoimmune diseases. J Autoimmun. 33:3–11. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alberga D, Trisciuzzi D, Lattanzi G,
Bennett JL, Verkman AS, Mangiatordi GF and Nicolotti O: Comparative
molecular dynamics study of neuromyelitis optica-immunoglobulin G
binding to aquaporin-4 extracellular domains. Biochim Biophys Acta.
1859:1326–1334. 2017. View Article : Google Scholar :
|
|
6
|
Park SJ, Jeong IH, Kong BS, Lee JE, Kim
KH, Lee DY and Kim HJ: Disease type- and status-specific alteration
of CSF metabolome coordinated with clinical parameters in
inflammatory demyelinating diseases of CNS. PLoS One.
11:e01662772016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim HH, Jeong IH, Hyun JS, Kong BS, Kim HJ
and Park SJ: Metabolomic profiling of CSF in multiple sclerosis and
neuromyelitis optica spectrum disorder by nuclear magnetic
resonance. PLoS One. 12:e01817582017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Samuvel DJ, Sundararaj KP, Nareika A,
Lopes-Virella MF and Huang Y: Lactate boosts TLR4 signaling and
NF-kappaB pathway-mediated gene transcription in macrophages via
monocarboxylate transporters and MD-2 up-regulation. J Immunol.
182:2476–2484. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pucino V, Bombardieri M, Pitzalis C and
Mauro C: Lactate at the crossroads of metabolism, inflammation, and
autoimmunity. Eur J Immunol. 47:14–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hirschhaeuser F, Sattler UG and
Mueller-Klieser W: Lactate: A metabolic key player in cancer.
Cancer Res. 71:6921–6925. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Le A, Cooper CR, Gouw AM, Dinavahi R,
Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL and Dang
CV: Inhibition of lactate dehydrogenase A induces oxidative stress
and inhibits tumor progression. Proc Natl Acad Sci USA.
107:2037–2042. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miao P, Sheng S, Sun X, Liu J and Huang G:
Lactate dehydrogenase A in cancer: A promising target for diagnosis
and therapy. IUBMB Life. 65:904–910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scheijen JL, Hanssen NM, van de Waarenburg
MP, Jonkers DM, Stehouwer CD and Schalkwijk CG: L(+) and D(−)
lactate are increased in plasma and urine samples of type 2
diabetes as measured by a simultaneous quantification of L(+) and
D(−) lactate by reversed-phase liquid chromatography tandem mass
spectrometry. Exp Diabetes Res. 2012:2348122012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doherty JR and Cleveland JL: Targeting
lactate metabolism for cancer therapeutics. J Clin Invest.
123:3685–3692. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rellinger EJ, Craig BT, Alvarez AL, Dusek
HL, Kim KW, Qiao J and Chung DH: FX11 inhibits aerobic glycolysis
and growth of neuroblastoma cells. Surgery. 161:747–752. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park JY, Pillinger MH and Abramson SB:
Prostaglandin E2 synthesis and secretion: The role of PGE2
synthases. Clin Immunol. 119:229–240. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao Y, Vanhoutte PM and Leung SW:
Vascular nitric oxide: Beyond eNOS. J Pharmacol Sci. 129:83–94.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou HY, Shin EM, Guo LY, Youn UJ, Bae K,
Kang SS, Zou LB and Kim YS: Anti-inflammatory activity of
4-methoxyhonokiol is a function of the inhibition of iNOS and COX-2
expression in RAW 264.7 macrophages via NF-kappaB, JNK and p38 MAPK
inactivation. Eur J Pharmacol. 586:340–349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Andersen LW, Mackenhauer J, Roberts JC,
Berg KM, Cocchi MN and Donnino MW: Etiology and therapeutic
approach to elevated lactate levels. Mayo Clin Proc. 88:1127–1140.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fantin VR, St-Pierre J and Leder P:
Attenuation of LDH-A expression uncovers a link between glycolysis,
mitochondrial physiology, and tumor maintenance. Cancer Cell.
9:425–434. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie H, Valera VA, Merino MJ, Amato AM,
Signoretti S, Linehan WM, Sukhatme VP and Seth P: LDH-A inhibition,
a therapeutic strategy for treatment of hereditary leiomyomatosis
and renal cell cancer. Mol Cancer Ther. 8:626–635. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jeong DH, Kim KB, Kim MJ, Kang BK and Ahn
DH: Skipjack tuna (Katsuwonus pelamis) eyeball oil exerts an
anti-inflammatory effect by inhibiting NF-kB and MAPK activation in
LPS-induced RAW 264.7 cells and croton oil-treated mice. Int
Immunopharmacol. 40:50–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Newton K and Dixit VM: Signaling in innate
immunity and inflammation. Cold Spring Harb Perspect Biol. 4(pii):
a0060492012.PubMed/NCBI
|
|
25
|
Sun J, Ramnath RD, Zhi L, Tamizhselvi R
and Bhatia M: Substance P enhances NF-kappaB transactivation and
chemokine response in murine macrophages via ERK1/2 and p38 MAPK
signaling pathways. Am J Physiol Cell Physiol. 294:C1586–C1596.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang Y, Kim SC, Yu T, Yi YS, Rhee MH, Sung
GH, Yoo BC and Cho JY: Functional roles of p38 mitogen-activated
protein kinase in macrophage-mediated inflammatory responses.
Mediators Inflamm. 2014:3523712014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Herlaar E and Brown Z: p38 MAPK signalling
cascades in inflammatory disease. Mol Med Today. 5:439–447. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zarubin T and Han J: Activation and
signaling of the p38 MAP kinase pathway. Cell Res. 15:11–18. 2005.
View Article : Google Scholar : PubMed/NCBI
|