Introduction
Hepatitis B virus (HBV) infection represents a major
risk factor for the development of hepatocellular carcinoma (HCC),
ranking fifth in global cancer incidence and representing the third
leading cause of cancer-associated mortality (1). Following the infection of
hepatoctyes, HBV relaxed-circle DNA (rcDNA) is transferred to the
nucleus, where it forms covalently closed circular DNA (cccDNA).
Within infected cells, the pregenomic RNA is then transcribed from
the cccDNA and transported to the cytoplasm, where the mature
capsids of the rcDNA are reverse transcribed and either secreted
from the cells or returned to the nucleus to form the cccDNA pool.
During this process, some HBV DNA genes integrate into the host
chromosomal DNA. HBV integration is suspected to be one of the most
important etiological events in HBV-induced HCC (2).
Previous isolation of HBV integration sites using
polymerase chain reaction (PCR)-based methods, including Alu-PCR or
ligation-mediated PCR, has suggested that the HBV insertional sites
occur randomly throughout the genome, leading to the suggestion
that there are no preferential integration sites with little
oncogenic annotation (3–5). Through the application of whole
genome sequencing (WGS), a large cohort of integration sites have
been identified and, among them, a number of hotspots have been
found, including TERT, MLL4, KMT2B, CCNE1, and FN1. This suggests
that HBV may have preferential integration sites associated with
distinct biological consequences, for example, altering the
function of endogenous genes, causing genetic damage and
chromosomal instability, which may lead to tumorigenesis in
HBV-infected patients (6–9). In the present study, according to WGS
detection in patients with HBV infection, several types of
viral-host junction were we found in HBV integration breakpoints.
Certain expression characteristics of viral-human chimeric
transcripts may assist in further understanding the molecular
mechanisms of HBV integration and its role in
hepatocarcinogenesis.
Materials and methods
Patients and samples
Liver biopsy specimens were collected from a total
of 18 patients from Renmin Hospital of Wuhan University (Wuhan,
China) between March and December 2016, comprising 16 men and two
women, aged between 21 and 43 (32.16±7.11) years old. The patients
included three patients with acute hepatitis B (AHB) achieving
virological seroclearance (VR) spontaneously (group 1), two
inactive HBsAg carriers (group 2), 13 patients with chronic
hepatitis B (CHB) receiving nucleos(t)ide analogs (adefovir, 10
mg/day or lamivudine, 100 mg/day) either as monotherapy or with
pegylated-interferon (IFN)α (100 µg/week). Among the patients with
CHB, six patients had primary treatment failure (group 3), five
patients had achieved VR (group 4), and two patients had achieved
both VR and HBsAg seroclearance and had not relapsed for >6
months (group 5) (10). Every
enrolled patient signed an informed consent form approved by the
Ethics Committee of Renmin Hospital of Wuhan University. The biopsy
specimens were frozen in liquid nitrogen and then stored at −80°C
until further experimental analysis.
HBV integration detection
Liver DNA was extracted from biopsy specimens using
the QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany). A
‘short-read’ WGS for HBV integration was performed by the Beijing
Genomics Institute (Shenzhen, China), as previously reported
(7). A cluster of multiple read
pairs was considered to be a candidate HBV integration breakpoint
when it was identified with close mapping positions, linking an end
of the human genome and an end of the HBV genome.
PCR and Sanger sequencing
validation
In order to confirm the newly identified events and
detect the unknown HBV sequences within the chimeric fragments,
conventional PCR and Sanger sequencing were used to verify the HBV
integration breakpoints with reads N≥2 in the partial HBV
integration breakpoints detected by WGS, the strategy of which is
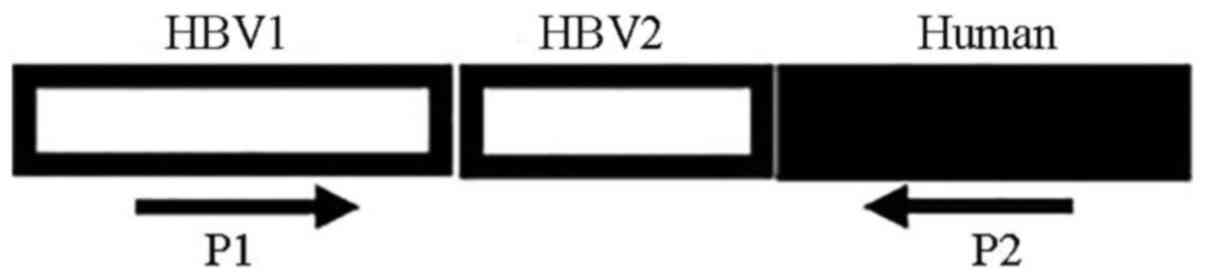
shown in Fig. 1. The PCR primers
were designed based on WGS-assembled fragments, in which one primer
was located in the human genome and the other in the HBV genome.
The PCR mix was prepared as follows: 1 µl of DNA; 2 µl of 10X Taq
buffer; 11.5 µl of H2O; 2.5 µl of dNTPs; 1 µl of forward
and reverse primers (10 µM, respectively) and 1 µl of Taq™ enzyme
(Takara Bio, Inc., Otsu, Japan). PCR was subjected to the following
cycling conditions: Initial denaturation for 10 min at 95°C; 40
cycles of denaturation for 10 sec at 95°C, annealing for 10 sec and
extension at 72°C, final extension for 7 min at 72°C. The forward
and reverse primer sequences the denaturation temperature and the
duration of extension were determined by preliminary tests. The PCR
products were electrophoresed on a 1% agarose gel and were then
extracted and sequenced by Sanger sequencing (Shanghai Sangon
Biology Engineering Technology & Service Co., Ltd., Shanghai,
China). Finally, the results of sequencing were compared with the
HBV and human genomes using the Basic local Alignment Search Tool
(BLAST; blast.ncbi.nlm.nih.gov/Blast.cgi).
Viral-host chimeric transcripts
Total RNA was extracted as previously described
(11). Reverse transcription-PCR
(RT-PCR) was used to synthesize cDNA according to the
manufacturer's protocol (PrimeScript RT™ Reagent kit with gDNA
Eraser, Takara Bio, Inc.). The expression of viral-human chimeric
transcripts was surveyed by conventional PCR, the conditions of
which were designed by the preliminary tests above, and Sanger
sequencing. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
gene was used as a control. The primer sequence for GAPDH was
GAPDH-A1, sense 5′-ACCACAGTCCATGCCATCAC-3′ and antisense
5′-TCCACCACCCTGTTGCTGTA-3′. The PCR amplification conditions of
GAPDH consisted of initial denaturation at 95°C for 10 min,
followed by 95°C for 10 sec, 60°C for 10 sec, and 72°C for 20 sec
for 30 cycles. The PCR product was 452 bp.
Intrahepatic (IH) HBV covalently
closed circular (cccDNA) quantification
The IH HBV cccDNA levels were measured using
quantitative PCR analysis as described previously (12). The cccDNA copy number for the
extracted liver samples is calculated by dividing the copies/µl. In
order to illuminate the influence of variation in the amount of
liver tissue among samples, β-globin, the housekeeping gene
(LightCycler Control Kit DNA, Roche Diagnostics) used to calculate
the number of cells based on one copy of β-globin per genome, was
used to allow for the standardization of the extracted DNA and
expression of HBV cccDNA as copies per cell (copies/cell) (12).
Statistical analysis
The statistical analysis was performed using SPSS
13.0 statistical software (SPSS, Inc., Chicago, IL, USA).
Continuous variables are expressed as the mean ± standard
deviation. Correlations were evaluated using Pearson's correlation
test. P<0.05 (two-sided) was considered to indicate a
statistically significant difference.
Results
IH cccDNA quantification
With a lower limit of 0.00024 copies/cell, IH cccDNA
levels were detected in two patients with AHB, six patients with
CHB with primary treatment failure, five patients achieving VR, and
one patient achieving both VR and HBsAg seroclearance (38.41±106.18
copies/cell).
Identification of global HBV
integrations in HBV-infected patients
The average sequencing depth coverage of WGS for HBV
integration was 4,879×. HBV integration breakpoints were identified
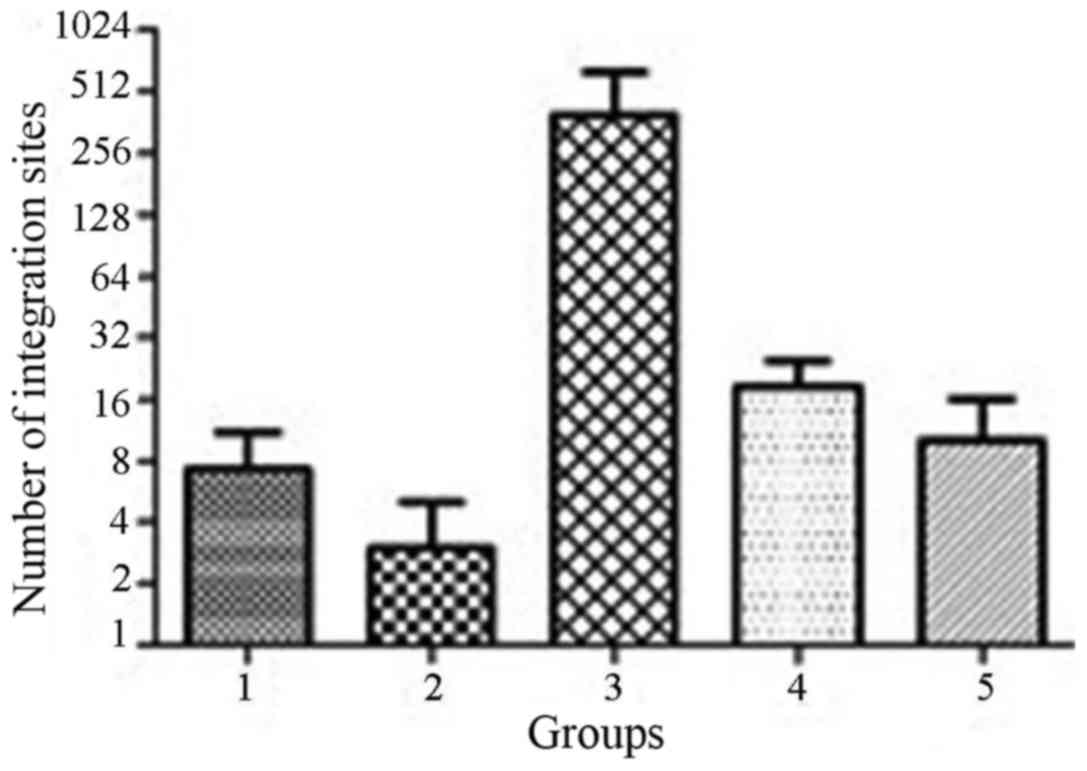
within all 18 patients with a total of 2,083 and with an average of
138.2±379.9 breakpoints per sample (1–1,596 breakpoints per
sample), whereas the higher number was 248.5±57.3 in group 3 and
18.6±13.7 in group 4, respectively (Fig. 2). The number of HBV integration
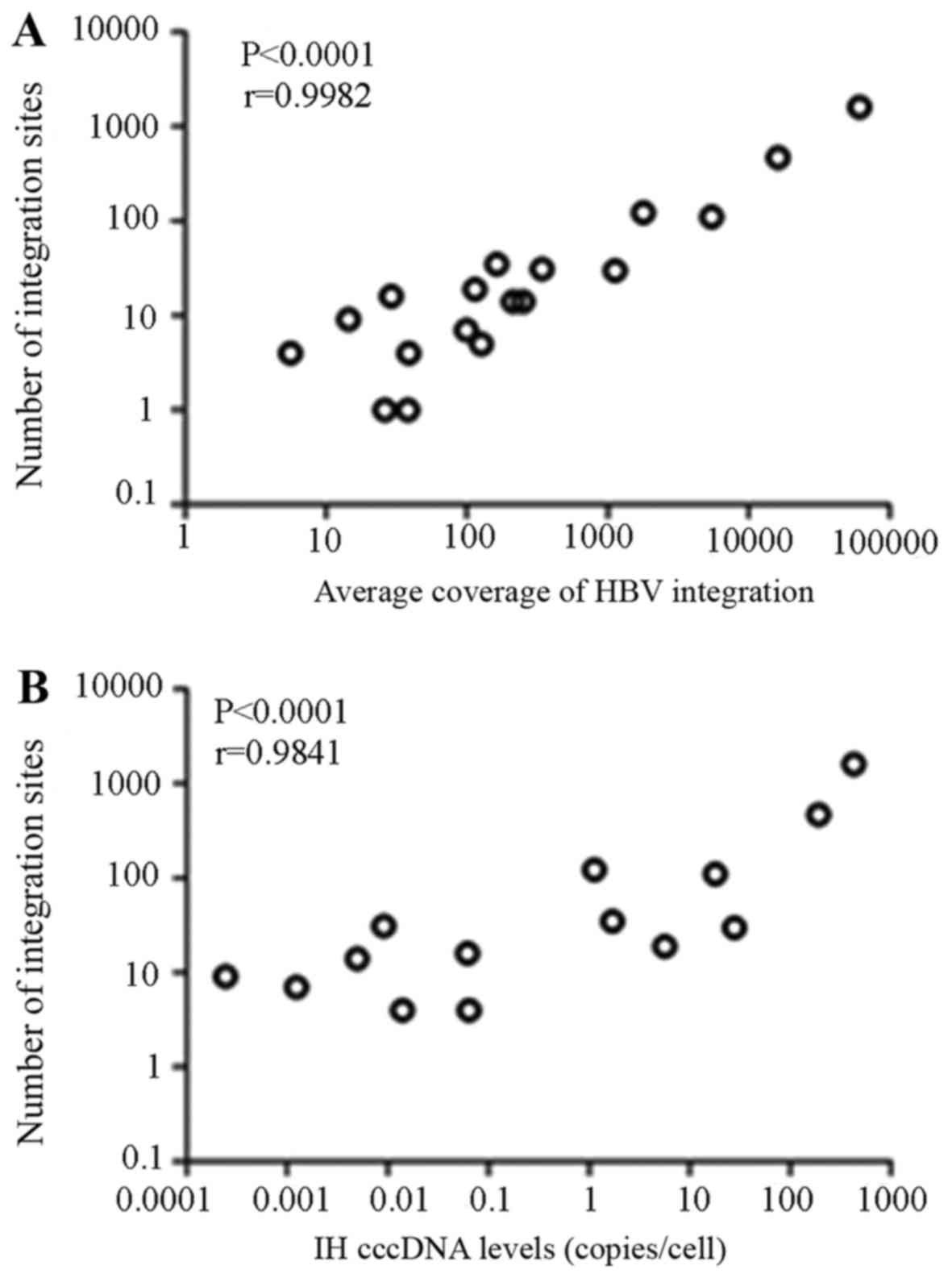
breakpoints was positively associated with the sequencing depth
coverage and the IH cccDNA levels, respectively (P<0.0001 and
P<0.0001; Fig. 3).
Characteristics of HBV integration
breakpoints
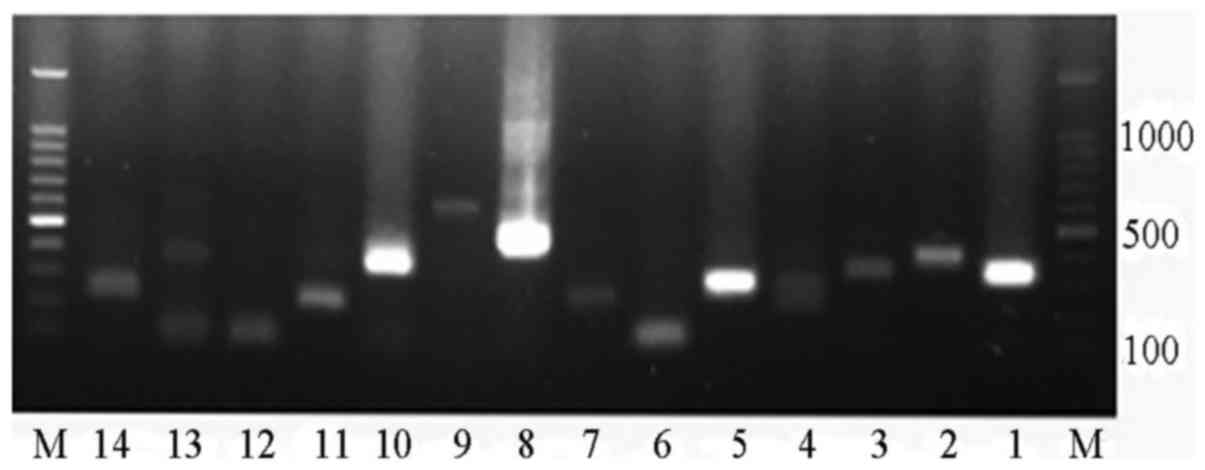
A total of 14 putative insertions (Table I) with at least two supporting
paired-end reads were selected for PCR analysis and successful
validation of 100% of these integration sites was achieved
(Fig. 4); the unknown HBV
sequences (HBV2) were detected within the chimeric fragments of
numbers 1, 2, 3, 4, 7, 8, 9, 10, 11, 13 and 14 (Fig. 5). The designed primer sequences of
conventional PCR for these breakpoints and their detected
characteristics are shown in Table
II. There were four types of viral-host junction: Forward
simple junction, found in the breakpoints of numbers 2, 3, 8 and
12; reverse simple junction, found in those of numbers 4, 5, 6, 9,
10, 11, 13 and 14; and forward and reverse complicated junctions,
found in those of numbers 1 and 7, respectively (Fig. 5). Microhomology was found in
several viral-cellular junctions. For example, 3 bp (GCT) of
microhomology were found in breakpoint of number 1, 5 bp (AAAAG) of
microhomology were found in number 2, 2 bp (GC) of microhomology
were found in number 8, and 7 bp (GACCTTC) of microhomology were
found in number 11. A detailed analysis of the inserted viral
fragments revealed that, with the exception of the breakpoints of
number 4 and number 7 mapped in the S gene and that of number six
mapped in the Precore gene, the others were localized within the
HBx gene. The 3′-end of HBx in several of these breakpoints was
found to be deleted.
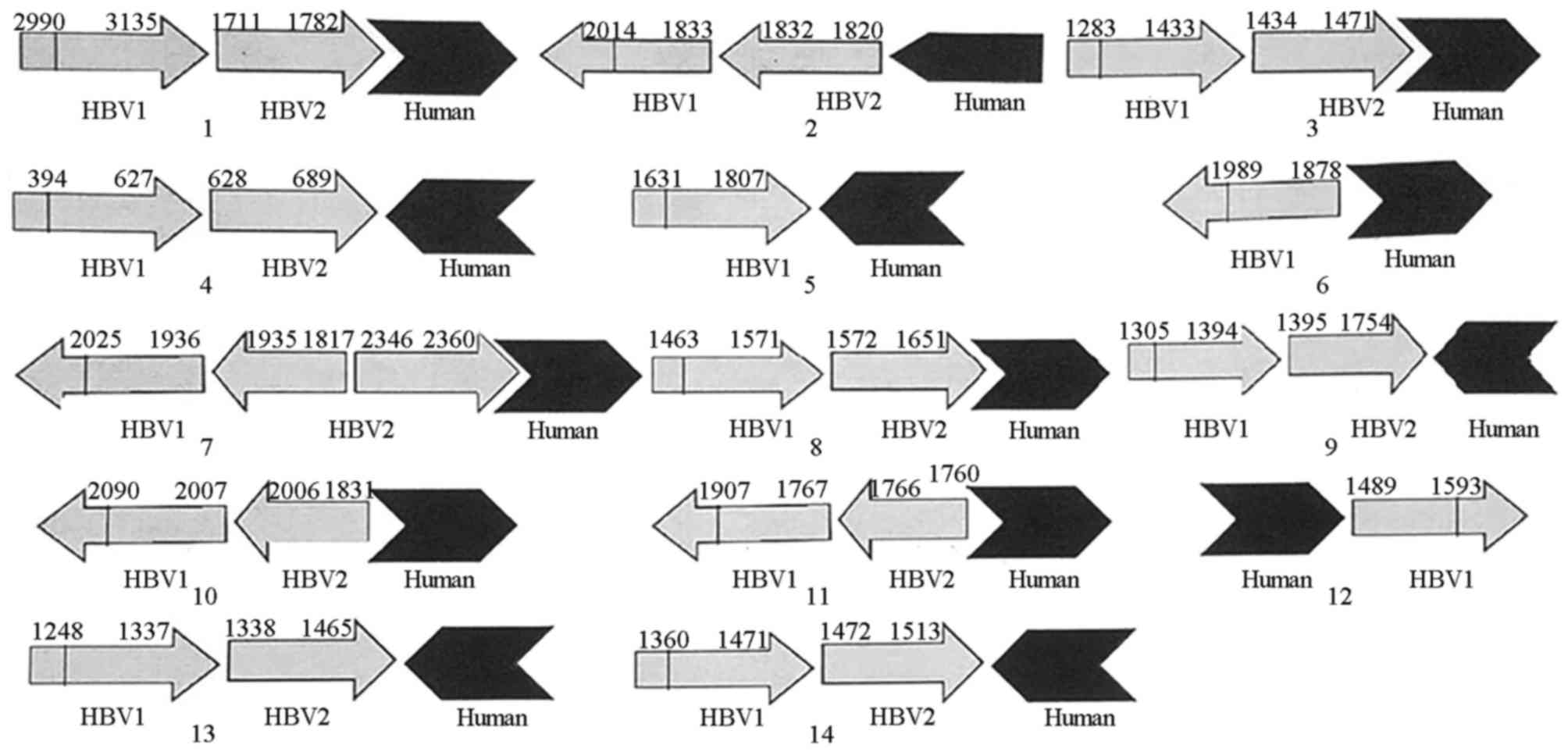 | Figure 5.Sequences of viral-host junctions.
Numbers 1–14 represent HBV sequences of 14 viral-host junctions.
HBV1, HBV sequences detected by WGS. HBV2, HBV sequences detected
by conventional PCR. Human, host gene sequences detected by WGS.
There were four types of viral-host junction: Forward simple
junction (2, 3, 8 and 12); reverse simple junction (4, 5, 6, 9, 10,
11, 13 and 14); forward complicated junction (1); and reverse complicated junction
(7). Viral sequences are shown in
gray, human sequences are shown in black. The open arrows represent
the orientation of the genes (increasing bases). HBV, hepatitis B
virus; WGS, whole genome sequencing. |
 | Table I.Viral-host junctions of 14 HBV
integration breakpoints with at least two supporting paired-end
reads. |
Table I.
Viral-host junctions of 14 HBV
integration breakpoints with at least two supporting paired-end
reads.
| Number | Code | HBV (nt) | Gene | Host | Location (nt) | Gene |
|---|
| 1 | 92096 | 1,782 | X | chr9 | 100,940,295 | CORO2A|intron |
| 2 | 92096 | 1,820 | PreC/X | chr9 | 101,030,026 | Intergenic |
| 3 | 73958 | 1,471 | P/X | chr3 |
2,002,381 | Intergenic |
| 4 | 24454 | 689 | S/P | chr2 | 216,281,034 | Intergenic |
| 5 | 32827 | 1,807 | X | chr16 |
51,320,015 | Intergenic |
| 6 | 32827 | 1,878 | PreC | chr16 |
51,320,070 | Intergenic |
| 7 | 68892 | 2,360 | P/S | chr10 |
34,836,578 | PARD3|intron |
| 8 | 68892 | 1,651 | X | chr14 |
19,828,316 | Intergenic |
| 9 | 68892 | 1,754 | X | chr14 |
57,417,625 | Intergenic |
| 10 | 68892 | 1,831 | X | chr14 |
57,417,636 | Intergenic |
| 11 | 68892 | 1,760 | X | chr16 |
34,990,970 | Intergenic |
| 12 | 94220 | 1,546 | X/P | chr9 |
98,928,137 | Intergenic |
| 13 | 95658 | 1,465 | X/P | chrX |
36,919,408 | Intergenic |
| 14 | 43985 | 1,513 | X/P | chr5 |
42,009,760 | Intergenic |
 | Table II.Sequences of primers and reaction
conditions of 14 HBV integration breakpoints with at least two
supporting paired-end reads. |
Table II.
Sequences of primers and reaction
conditions of 14 HBV integration breakpoints with at least two
supporting paired-end reads.
| Number | Primer
sequence | Denaturation
temperature (°C) | Extension
(sec) | Amplification
fragment length (bp) |
|---|
| 1 |
5′-ACTTCGCTTCACCTCTGC-3′ | 64 | 14 | 321 |
|
|
5′-TATGATGGCACCACTGCA-3′ |
|
|
|
| 2 |
5′-GAGGCGGTGTCTAGGAGA-3′ | 54 | 17 | 415 |
|
|
5′-GGTCAAATTGTTTGGATAAA-3′ |
|
|
|
| 3 |
5′-GCAAAACTCATCGGGACT-3′ | 56 | 16 | 360 |
|
|
5′-TTGTGATGACTTGCTGGA-3′ |
|
|
|
| 4 |
5′-AAGACCTGCACGATTC-3′ | 50 | 14 | 310 |
|
|
5′-TATGCTCACTTCCACA-3′ |
|
|
|
| 5 |
5′-GTCTTGCCCAAGGTCTTA-3′ | 56 | 14 | 305 |
|
|
5′-CAGATGGCGCACTAACAA-3′ |
|
|
|
| 6 |
5′-TGAGTAACTCCACAGAAGC-3′ | 56 | 10 | 136 |
|
|
5′-CAAGAAATAGCCCCAACT-3′ |
|
|
|
| 7 |
5′-CCCGATACAGAGCAGAGG-3′ | 62 | 12 | 270 |
|
|
5′-GGTCATGGCATGGGAAGA-3′ |
|
|
|
| 8 |
5′-CGCTTCTCCGCCTATTGT-3′ | 58 | 18 | 435 |
|
|
5′-ACCCTGGCATCCCTGGTTC-3′ |
|
|
|
| 9 |
5′-ATGGCTGCTAGGCTGTGC-3′ | 58 | 25 | 622 |
|
|
5′-CCATTCCCAACTTGAAGATTTA-3′ |
|
|
|
| 10 |
5′-AGTATAGCTTGCCTGAGT-3′ | 56 | 16 | 354 |
|
|
5′-CTGCTCTGAGGCAATTAA-3′ |
|
|
|
| 11 |
5′-GCTTGGAGGCTTGAACAG-3′ | 56 | 12 | 253 |
|
|
5′-AGAGCCCCTTGGAAAATA-3′ |
|
|
|
| 12 |
5′-GAGGTGGCAATGAGGTGAG-3′ | 56 | 10 | 103 |
|
|
5′-CAGAGGTGAAGCGAAGTG-3′ |
|
|
|
| 13 |
5′-AAAACTCATCGGGACTGA-3′ | 48 | 15 | 319 |
|
|
5′-CATTTTGTTGACCTGGAA-3′ |
|
|
|
| 14 |
5′-CTGTGCTGCCAACTGGAT-3′ | 60 | 12 | 254 |
|
|
5′-GCTAAGTGGAGCTTATTTCA-3′ |
|
|
|
Expression of viral-human chimeric
transcripts
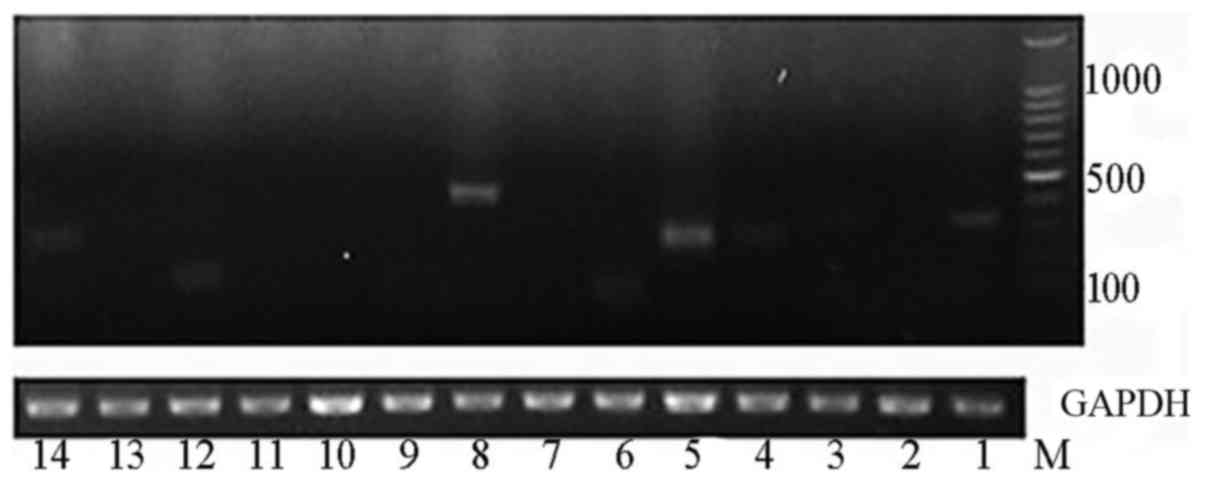
Among the 14 viral-host junctions with at least two
supporting paired-end reads, chimeric transcripts were observed in
breakpoints of numbers 1, 3, 4, 5, 6, 8, 12 and 14 (Fig. 6).
Discussion
HBV, a notorious DNA virus using cccDNA as its
stable transcriptional template for HBV mRNAs, can insert its
genome into the human genome and induce multiple
hepatocarcinogenesis events. Current antiviral drugs, including
INF-α and tenofovir, cannot completely eliminate intrahepatic
cccDNA, which has significant involvement in HBV infection relapse
and the pathogenesis of HCC and cirrhosis (13). However, HBV integration, the
process by which underlying cancer-driving genetic events,
including somatic mutation, structural rearrangement and clonal
expansion, and its in-depth mechanisms and timing remain to be
fully elucidated.
HBV DNA integration events occur with low frequency
(0.01–0.1% of infected hepatocytes) in transient infections
(14). Compared with traditional
PCR-based methods, which do not have the required sensitivity for
the detection of virus-cell junctions with such a low frequency of
occurrence, WGS may provide insights into HBV integration detection
by yielding greater specificity and sensitivity (6–9). In
the present study, using ‘short reads’ WGS with an average
sequencing depth of 4,879×, it was found that the rates of HBV
integration were all 100% in patients with AHB, carriers of
inactive HBsAg, and patients with CHB achieving different prognoses
following antiviral treatment. The average number of integration
breakpoints was 138.2±379.9 per sample. There may be several
reasons for detecting a greater number of integration breakpoints
per sample in the present study compared with other studies
(15). Firstly, a previous study
indicated that the ability to identify HBV insertion events
depended on the HBV insertion allele frequency and the sequencing
depth and coverage (16). This was
supported by the positive correlation between the number of HBV
integration sites and the sequencing depth coverage in the present
study. Secondly, it was not possible to acquire significant
differences in the number of integration sites among different
groups due to a small number of patients in each group. The
positive association between the number of HBV integration sites
and the IH cccDNA level, which is associated with the overall
survival rate of patients (17),
suggested that there exists a close association between the
frequency of HBV integration and the prognosis of HBV-infected
patients, which requires validation in further investigations with
larger sample sizes.
During HBV infection, host chromosomal DNA double
stranded breaks (DSBs), provoking cellular responses with the most
deleterious effects, may be caused by oxidative DNA damage
(18,19). Under this condition and in order to
maintain DNA integrity, a mixture of DNA repair mechanisms for host
chromosomes may be involved in HBV DNA integration, for example,
homologous recombination repair (HRR), classical non-homologous end
joining (c-NHEJ), alternative end joining [including
microhomology-mediated end joining (MMEJ)] and single strand
annealing (20). HRR is an
evolutionarily conserved, error-free repair mechanism, using an
undamaged sister chromatid as a template to accurately repair the
damage. By contrast, c-NHEJ is an error-prone repair pathway,
repairing DSBs by joining two non-homologous DNA segments together,
which may lead to the potential risks of gene deletion, insertion,
indirect or direct repeats, and the phenomenon that HBV DNA
molecules integrate into the host chromosomal DNA. Enriched
microhomology (MH) exists between human and HBV genome sequences,
and MMEJ may be another important mechanism mediating virus
integration processes (21).
In the present study, several forms of junction were
found between viral and host genes: Forward and reverse simple
junctions, and forward and reverse complicated junctions,
suggesting that they may be formed by NHEJ, as NHEJ is typically
associated with deletions and sometimes insertions of different
sequences at the termini (22).
Secondly, the finding of MH in viral-host junctions in the present
study, for example, 3 bp (GCT) in number 1, 5 bp (AAAAG) in number
2, and 7 bp (GACCTTC) in number 11, indicates that MMEJ may be key
in their formation (23). This
finding is similar to the result of a previous study, where
significant enrichment of MH sizes of 2 and 5 bp was found in the
integration junction (24).
Thirdly, several HBV integration breakpoints were observed within
the HBx genome in the present study, confirming that the HBx gene
had more integration opportunities than other regions of HBV, due
to the existence of a large number of viral transcriptional
regulators in the HBx gene (8).
In the viral-human chimeric transcription analysis,
it was found that several transcribed viral-human sequences were
located within the X gene, demonstrating that chimeric transcripts
were observed only when the site of integration was at 3′-end of
HBx and often when its deletion occurred (7). Several independent lines of evidence
have demonstrated that HBsAg is not only expressed from the
episomal cccDNA minichromosome, but also from transcripts arising
from HBV DNA integrated into the host genome (25). In the present study, this was
demonstrated by the expression of chimera transcription of number
4, where the breakpoint of the inserted viral fragment was mapped
in the S gene. This finding was also in accordance with a previous
report, in which serum HBsAg and IH cccDNA levels were not
correlated in patients with HBeAg-negative CHB (26), whereas patients with HBeAg-negative
CHB usually have a longer, mostly perinatal HBV infection history
and are expected to have more extensive HBV DNA integration than
HBeAg-positive cases (27).
There were several limitations in the present study.
Firstly, the full HBV integration sequences of these sites were not
detected, the left end of which may not exactly match the
double-stranded linear DNA ends, but may instead include terminal
truncations of ~100 bp (14,28).
Secondly, in order to further validate the hypothesis that HBV
integration sites may have another origin for HBsAg production,
chimeric protein expression requires detection from the integrated
S gene. Finally, further experiments with larger sample sizes are
required to further elucidate the molecular mechanisms and
tumorigenesis of HBV integration.
In conclusion, the present study showed that HBV
integration was detected in 100% of HBV-infected patients with a
high number of integration sites. A close association existed
between HBV integration and the prognoses of patients. HBx
integration may be indispensable for viral-host chimeric
transcripts and the expression of the HBs-host chimeric transcript
suggests that HBsAg may be produced from integrated DNA.
Acknowledgements
No applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XFD and JS conducted the experiments. CPH conducted
statistical analysis. CH and RZ designed all the primers and
modified the English language of the manuscript. ZC and LY
collected the patient data. PR wrote the manuscript and made
substantial contributions to the design of the present study. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Liver tissue in the present study was collected from
patients between March and December 2016 and written informed
consent was provided. The present study was approved by the Ethics
Committee of Renmin Hospital of Wuhan University in March 2016.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lok AS and McMahon BJ: Chronic Hepatitis
B. Hepatology. 45:507–539. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Murakami Y, Saigo K, Takashima H, Minami
M, Okanoue T, Bréchot C and Paterlini-Bréchot P: Large scaled
analysis of hepatitis B virus (HBV) DNA integration in HBV related
hepatocellular carcinomas. Gut. 54:1162–1168. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saigo K, Yoshida K, Ikeda R, Sakamoto Y,
Murakami Y, Urashima T, Asano T, Kenmochi T and Inoue I:
Integration of hepatitis B virus DNA into the myeloid/lymphoid or
mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in
human hepatocellular carcinoma. Hum Mutat. 29:703–708. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tamori A, Yamanishi Y, Kawashima S,
Kanehisa M, Enomoto M, Tanaka H, Kubo S, Shiomi S and Nishiguchi S:
Alteration of gene expression in human hepatocellular carcinoma
with integrated hepatitis B virus DNA. Clin Cancer Res.
11:5821–5826. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang Z, Jhunjhunwala S, Liu J, Haverty
PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S,
et al: The effects of hepatitis B virus integration into the
genomes of hepatocellular carcinoma patients. Genome Res.
22:593–601. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sung WK, Zheng H, Li S, Chen R, Liu X, Li
Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, et al: Genome-wide
survey of recurrent HBV integration in hepatocellular carcinoma.
Nat Genet. 44:765–769. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Toh ST, Jin Y, Liu L, Wang J, Babrzadeh F,
Gharizadeh B, Ronaghi M, Toh HC, Chow PK, Chung AY, et al: Deep
sequencing of the hepatitis B virus in hepatocellular carcinoma
patients reveals enriched integration events, structural
alterations and sequence variations. Carcinogenesis. 34:787–798.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao LH, Liu X, Yan HX, Li WY, Zeng X,
Yang Y, Zhao J, Liu SP, Zhuang XH, Lin C, et al: Genomic and
oncogenic preference of HBV integration in hepatocellular
carcinoma. Nat Commun. 7:129922016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chinese Society of Hepatology and Chinese
Society of Infectious Diseases. Chinese Medical Association, The
guideline of prevention and treatment for chronic hepatitis B (2010
version). Zhonghua Gan Zang Bing Za Zhi (Chinese J Hepatol).
19:13–24. 2011.
|
|
11
|
Belloni L, Allweiss L, Guerrieri F,
Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M
and Levrero M: IFN-α inhibits HBV transcription and replication in
cell culture and in humanized mice by targeting the epigenetic
regulation of the nuclear cccDNA minichromosome. J Clin Invest.
122:529–537. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Werle-Lapostolle B, Bowden S, Locarnini S,
Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z,
Delaney WE 4th, et al: Persistence of cccDNA during the natural
history of chronic hepatitis B and decline during adefovir
dipivoxil therapy. Gastroenterology. 126:1750–1758. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng Q, Bai L, Zheng S, Liu M, Zhang J,
Wang T, Xu Z, Chen Y, Li J and Duan Z: Efficient inhibition of duck
hepatitis B virus DNA by the CRISPR/Cas9 system. Mol Med Rep.
16:7199–7204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tu T, Budzinska MA, Vondran FWR, Shackel
NA and Urban S: Hepatitis B virus DNA integration occurs early in
the viral life cycle in an in vitro infection model via
NTCP-dependent uptake of enveloped virus particles. J Virol. Feb
7–2018.(Epub ahead of print). View Article : Google Scholar
|
|
15
|
Murakami Y, Minami M, Daimon Y and Okanoue
T: Hepatitis B virus DNA in liver, serum, and peripheral blood
mononuclear cells after the clearance of serum hepatitis B virus
surface antigen. J Med Virol. 72:203–214. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sims D, Sudbery I, Ilott NE, Heger A and
Ponting CP: Sequencing depth and coverage: Key considerations in
genomic analyses. Nat Rev Genet. 15:121–132. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hosaka T, Suzuki F, Kobayashi M, Hirakawa
M, Kawamura Y, Yatsuji H, Sezaki H, Akuta N, Suzuki Y, Saitoh S, et
al: HBcrAg is a predictor of post-treatment recurrence of
hepatocellular carcinoma during antiviral therapy. Liver Int.
30:1461–1470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dandri M, Burda MR, Bürkle A, Zuckerman
DM, Will H, Rogler CE, Greten H and Petersen J: Increase in de novo
HBV DNA integrations in response to oxidative DNA damage or
inhibition of poly(ADP-ribosyl)ation. Hepatology. 35:217–223. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bill CA and Summers J: Genomic DNA
double-strand breaks are targets for hepadnaviral DNA integration.
Proc Natl Acad Sci USA. 101:11135–11140. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X,
Ding W, Yu L, Wang X and Wang L: Genome-wide profiling of HPV
integration in cervical cancer identifies clustered genomic hot
spots and a potential microhomology-mediated integration mechanism.
Nat Genet. 47:158–163. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mladenov E, Magin S, Soni A and Iliakis G:
DNA double-strand-break repair in higher eukaryotes and its role in
genomic instability and cancer: Cell cycle and
proliferation-dependent regulation. Semin. Cancer Biol. 37–38.
51–64. 2016.
|
|
22
|
Hu X, Lin J, Xie Q, Ren J, Chang Y, Wu W
and Xia Y: DNA double-strand breaks, potential targets for HBV
integration. J Huazhong Univ Sci Technolog Med Sci. 30:265–270.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hastings PJ, Ira G and Lupski JR: A
microhomology-mediated break-induced replication model for the
origin of human copy number variation. PLoS Genet. 5:e10003272009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoo S, Wang W, Wang Q, Fiel MI, Lee E,
Hiotis SP and Zhu J: A pilot systematic genomic comparison of
recurrence risks of hepatitis B virus-associated hepatocellular
carcinoma with low- and high-degree liver fibrosis. BMC Med.
150:2142017. View Article : Google Scholar
|
|
25
|
Wooddell CI, Yuen MF, Chan HL, Gish RG,
Locarnini SA, Chavez D, Ferrari C, Given BD, Hamilton J, Kanner SB,
et al: RNAi-based treatment of chronically infected patients and
chimpanzees reveals that integrated hepatitis B virus DNA is a
source of HBsAg. Sci Transl Med. 9:eaan02412017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ruan P, Zhou B, Dai X, Sun Z, Guo X, Huang
J and Gong Z: Predictive value of intrahepatic hepatitis B virus
covalently closed circular DNA and total DNA in patients with acute
hepatitis B and patients with chronic hepatitis B receiving
anti-viral treatment. Mol Med Rep. 9:1135–1141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thompson AJ, Nguyen T, Iser D, Ayres A,
Jackson K, Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W, et
al: Serum hepatitis B surface antigen and hepatitis B e antigen
titers: Disease phase influences correlation with viral load and
intrahepatic hepatitis B virus markers. Hepatology. 51:1933–1944.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mason WS, Low HC, Xu C, Aldrich CE,
Scougall CA, Grosse A, Clouston A, Chavez D, Litwin S, Peri S, et
al: Detection of clonally expanded hepatocytes in chimpanzees with
chronic hepatitis B virus infection. J Virol. 83:8396–8408. 2009.
View Article : Google Scholar : PubMed/NCBI
|