Introduction
Breast cancer (BC) is the most common cancer in
women and a leading cause of cancer-associated mortalities in the
world (1). During BC development,
the epithelial-to-mesenchymal transition (EMT) serves a very
important role (2). EMT is
associated with wound healing, fibrosis and cancer progression; in
particular, the metastasis and invasive ability of cancer cells is
significantly enhanced (3).
Therefore, in the present study, the mechanism of the role of the
EMT in one BC cell line was investigated. Previous studies
identified that transforming growth factor β1 (TGF-β1) serves a
very important role in the EMT process (4,5).
TGF-β1 facilitates many responses by binding specifically and
activating cell surface receptor serine/threonine kinase complexes,
including TGFβ receptor (TβRI and TβRII) (4,6).
Activated TGF-β receptors can stimulate the receptor-regulated
phosphorylation of Smad family members, an important signal
transduction and modulator, by forming complexes (7). Additionally, phosphorylated Smad
family members 2 and 3 form a stable complex with Smad family
member 4 and move to the nucleus, where the transcription of the
target gene is regulated (8,9).
Non-Smad signaling pathways, including guanosine triphosphatase,
phosphoinositide 3 kinase and mitogen-activated protein kinase
signaling pathways, can also be activated by TGF-β (7,10);
however, the Smad-dependent signaling pathway is unique but is the
most critical to TGF-β-induced EMT (11,12).
During this process, cells lose epithelial markers, including
E-cadherin, and upregulate mesenchymal markers, including vimentin
(13,14).
There are several lines of evidence suggesting that
TGF-β receptors are distributed in lipid rafts/caveolae and
non-raft membrane microdomains (15). On the cell surface, TGF-β1
receptors are distributed between different microdomains of the
cellular membrane and may be internalized via clathrin-and
caveolae-mediated endocytic mechanisms (16,17).
Lipid rafts, enriched with cholesterol and sphingolipids, are
ordered microdomains within plasma membranes. Sphingomyelin (SM) is
an essential component of sphingolipids (18). SM is additionally associated with
the formation of other membrane microdomains, including
clathrin-coated pits and caveolae, thus serving an important role
in the regulation of trans-membrane signaling (19). Sphingomyelin synthase (SMS), with
two isoforms, SMS1 and SMS2, is a key enzyme involved in the
generation and development of SM. SMS can participate in
inflammation, atherosclerosis, proliferation, apoptosis,
differentiation and other functions (20).
Furthermore, bioinformatics retrieval from Gene
Expression Omnibus (GEO) datasets suggested that SMS1 and TβRI are
linked in the mammary gland and breast cancer cells (GSE54491 and
GSE89205; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89205).
The results showed that the expression of SMS1 was decreased, while
TβRI was increased, when breast cells were treated with TGF-β1.
Therefore, the accumulation of SMS1 inside cells may decrease
cellular TβRI expression levels. To test this hypothesis,
MDA-MB-231 cells were treated with or without TGF-β1 following
transfection with a SMS1 overexpression plasmid. Protein expression
levels associated with the development of the EMT were investigated
by western blotting and immunofluorescence, and migration and
invasion were investigated using a wound healing and a Matrigel
invasion assay, respectively.
Materials and methods
Microarray data
All available data on TGF-β1-treated breast cancer
cells or normal mammary gland cells from the Gene Expression
Omnibus (GEO) database were investigated and an mRNA microarray
GSE54491 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54491)
was identified. GSE54491 employed an Affymetrix mouse gene 1.0 ST
array [transcript (gene) version] to identify mRNAs that were
differentially expressed between TGF-β1-treated and
TGF-β1-untreated normal murine mammary gland (NMumG) cells.
MDA-MB-231 cells were selected for treatment with 10 ng/ml TGF-β1
to assess protein expression, according to previous studies
(14,21). After 72 h, the expression of SMS1
and SMS2 was examined by western blot analysis.
Expression of SMS1 in breast cancer
cells
MDA-MB-231, MCF-7 and BT549 cells were purchased
from The Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China). These cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare
Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine
serum (FBS; Tianjin Haoyang Biological Products Technology Co.,
Ltd., Tianjin, China) and incubated at 37°C in a humidified
atmosphere (90% relative humidity) containing 5% CO2. To
choose a suitable breast cancer cell line and determine the optimal
concentration of TGF-β1, western blotting was performed in order to
detect the expression levels of SMS1 in the three breast cancer
cells (MDA-MB-231, MCF-7 and BT549). Finally, MDA-MB-231 cells were
selected and treated with different concentrations (0, 5, 10, 15
and 20 ng/ml) of TGF-β1 (cat. no. 10804-HNAC; Sino Biological,
Inc., Beijing, China). After 72 h, the expression levels of SMS1,
E-cadherin and vimentin (EMT markers) were investigated by western
blotting.
Transfection and grouping
To construct an SMS1 overexpression cell model,
SMS1-overexpressing plasmids [pcDNA3.1(+)], which were constructed
by Magus Technology (Shanghai, China), were transfected into
MDA-MB-231 cells. Transfection was conducted according to the
manufacturer's protocol. First, 12 h prior to transfection,
1×106 cells were seeded into wells of a 6-well plate
(Beaver Nano-Technologies Co., Ltd., Suzhou, China) that contained
antibiotic-free medium. At the time of transfection, the cell
confluency was 60–70% (22). The
SMS1-overexpressing plasmid (4 µg; SMS1 group) or a negative
plasmid (4 µg; Control group) was diluted with 50 µl DMEM (FBS-free
and antibiotic-free medium) or 5 µl Entranster™-D-4000
(Engreen Biosystem New Zealand Ltd., Auckland, New Zealand) and 50
µl DMEM. After 5 min, the dilutions were mixed together and
incubated at 37°C for 20 min and subsequently dispensed into each
well. DMEM was replaced with DMEM containing 10% FBS after 6 h
(23). The cells were cultured for
24 h, and the Control and the SMS1 groups were treated with 10
ng/ml TGF-β1 at 37°C, as at that concentration, vimentin and
E-cadherin expression had significantly altered. Therefore, the
following four groups were created: Control, SMS1, TGF-β1 and
SMS1+TGF-β1. After 72 h, all cells were harvested for subsequent
experiments.
Wound healing assay
A total of 2×105 MDA-MB-231 cells were
seeded in 12-well tissue culture plates. After transfection and 10
ng/ml TGF-β1 treatment (36 h), the cells were maintained in
serum-free medium at 37°C for 8 h. Using a sterile 200-µl pipette
tip to gently swipe along the midline of the cell well, the cells
were scraped from the well and were washed with PBS three times.
Subsequently, the MDA-MB-231 cells were treated with TGF-β1 at the
above concentration for 24 h. Finally, the wound closure was
measured with a phase contrast inverted microscope (Olympus IX71;
Olympus Corporation, Tokyo, Japan; magnification, ×4).
Matrigel invasion analysis
The cells were treated as above. Briefly, cells were
starved for 8 h. Single-cell suspension (1×105 cells)
was added to serum-free medium with or without 10 ng/ml TGF-β1 on a
top well of a 24-well Transwell plate (cat. no. 3413; Corning
Incorporated, Corning, NY, USA) precoated with Matrigel (cat. no.
356234). After 36 h, the cells invading the lower chamber
containing medium was supplemented with 10% FBS were stained with
0.1% crystal violet solution for 5 min at room temperature
(CAS:548-62-9; Shanghai Macklin Biochemical Co., Ltd., Shanghai,
China) and counted using a phase contrast inverted light microscope
(Olympus IX71; Olympus Corporation, Tokyo, Japan; magnification,
×10).
Western blot analysis
Proteins were extracted using
radioimmunoprecipitation assay buffer (cat. no. ROO20; Beijing
Solarbio Bioscience & Technology Co., Ltd., Beijing, China),
and the protein concentration was measured using a bicinchoninic
acid assay (cat. no. CW0014; Beijing Kangwei Century Biotechnology
Co., Ltd., Beijing, China). Equal amounts of cleared lysates (~50
µg protein) were separated by 10–12% SDS-PAGE and subsequently
transferred onto polyvinylidene fluoride membranes (Immobilon-P;
EMD Millipore, Billerica, MA, USA). Equal transfer was validated by
staining with Ponceau red (cat. no. CW0057S; Beijing Kangwei
Century Biotechnology Co., Ltd.) for 30 min at room temperature.
The membranes were blocked with 10% skimmed milk or 10% bovine
serum albumin (BSA; cat. no. A8020; Beijing Solarbio Science &
Technology Co., Ltd.) in TBS for 1 h at room temperature and
subsequently incubated with primary antibodies in TBS containing
0.05% Tween-20, 2% BSA and 0.05% sodium azide overnight at 4°C
(24). The following antibodies
were used at the indicated dilutions: SMS1 (cat. no. sc-133135;
1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), vimentin
(cat. no. 10366-1-AP; 1:2,000; ProteinTech Group, Inc., Chicago,
IL, USA), E-cadherin (cat. no. 20874-1-AP; 1:2,000; ProteinTech
Group, Inc.), phospho (p)-Smad2 (cat. no. AF8314; 1:2,000; Affinity
Biosciences, Shanghai, China), Smad2 (cat. no. WL03369; 1:1,500;
Wanleibio Co., Ltd., Shanghai, China) and TβR1 (cat. no. AF5347;
1:2,000; Affinity Biosciences), and a GAPDH antibody (cat. no.
HRP-60004; 1:12,000; ProteinTech Group, Inc.). Subsequently, the
membranes were incubated at room temperature for 1.5 h with
secondary horseradish peroxidase-conjugated anti-rabbit antibodies
(cat. no. SA0000I-2; 1:8,000; ProteinTech Group, Inc.) or
anti-mouse antibodies (cat. no. SA0000I-I; 1:8,000; ProteinTech
Group, Inc.) in TBS containing 0.05% Tween 20. Signals were
determined using an enhanced chemiluminescence reagent (cat. no.
CW0049M; Beijing Kangwei Century Biotechnology Co., Ltd.) and an
autoradiography system (Image Lab; version 5.1; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) (14). Each assay was repeated at least
three times.
Immunofluorescence and confocal
microscopy imaging
A total of 2×104 cells were seeded on
coverslips in a 24-well plate. Followign the treatment, the cells
were gently washed with PBS. Subsequently, they were fixed in 4%
paraformaldehyde for 20 min at room temperature, followed by
permeabilization using 0.5% Triton X-100 (cat. no. T8200; Beijing
Solarbio Science & Technology Co., Ltd.) in PBS for 20 min at
room temperature. The coverslips were subsequently blocked in 5%
BSA in PBS for 60 min at room temperature. Subsequently, the cells
were incubated with anti-TGF-βRI (cat. no. AF5347; 1:100; Affinity
Biosciences), anti-vimentin (cat. no. 10366-1-AP; 1:200;
ProteinTech Group, Inc.) and anti E-cadherin (cat. no. 20874-1-AP;
1:50; ProteinTech Group, Inc.) antibodies overnight at 4°C and
subsequently washed in PBS. The cells were exposed to secondary
antibody conjugated with fluorescein isothiocyanate (cat. no.
BA1105; 1:50; Wuhan Boster Biological Technology, Ltd., Wuhan,
China) or TRITC (cat. no. E032420-01; 1:250; Earthox Life Sciences
Millbrae, CA, USA) for 60 min at room temperature. DAPI was used to
stain the nuclei (cat. no. AR1177; Wuhan Boster Biological
Technology, Ltd.) for 5 min at room temperature, and the cells were
washed with PBS. Finally, an inverted fluorescence (Olympus IX71;
Olympus Corporation; magnification, ×20) was used to acquire the
data of vimentin and E-cadherin and a confocal microscope (Leica
SP8; Leica Microsystems, Ltd., Milton Keynes, UK; magnification,
×40) was used to acquire the data of TGF-βRI. Images were analyzed
by Image J software (Ver. 2.1; National Institutes of Health,
Bethesda, MD, USA).
Statistical analysis
All data are presented as the mean ± standard
deviation. Unpaired t-test was used for single comparisons. For
multiple comparisons, one-way analysis of variance followed by
Tukey's or Games-Howell post-hoc test was used. All experiments
were repeated at least three times. P<0.05 was considered to
indicate a statistically significant difference.
Results
Bioinformatics analysis of the
possible trends of SMS1 and TGF-β type I receptors in the
development of EMT
The GSE54491 dataset was downloaded from National
Center for Biotechnology Information GEO DataSets. For the GSE54491
dataset, normal murine mammary gland (NMumG) cells transformed by
overexpression of endothelial growth factor receptor (NME) cells
were cultured in the presence of TGF-β1 (5 ng/ml) for 4 weeks, at
which point TGF-β1 supplementation was discontinued and the cells
were allowed to recover for an additional 4 weeks (Post-TGF-Rec).
Total RNA was prepared from unstimulated cells (Pre-TGF) at similar
passage numbers and compared by microarray analysis. In the present
study, the two groups were analyzed in triplicate, including three
Pre-TGF and three Post-TGF-Rec samples. The results showed that the
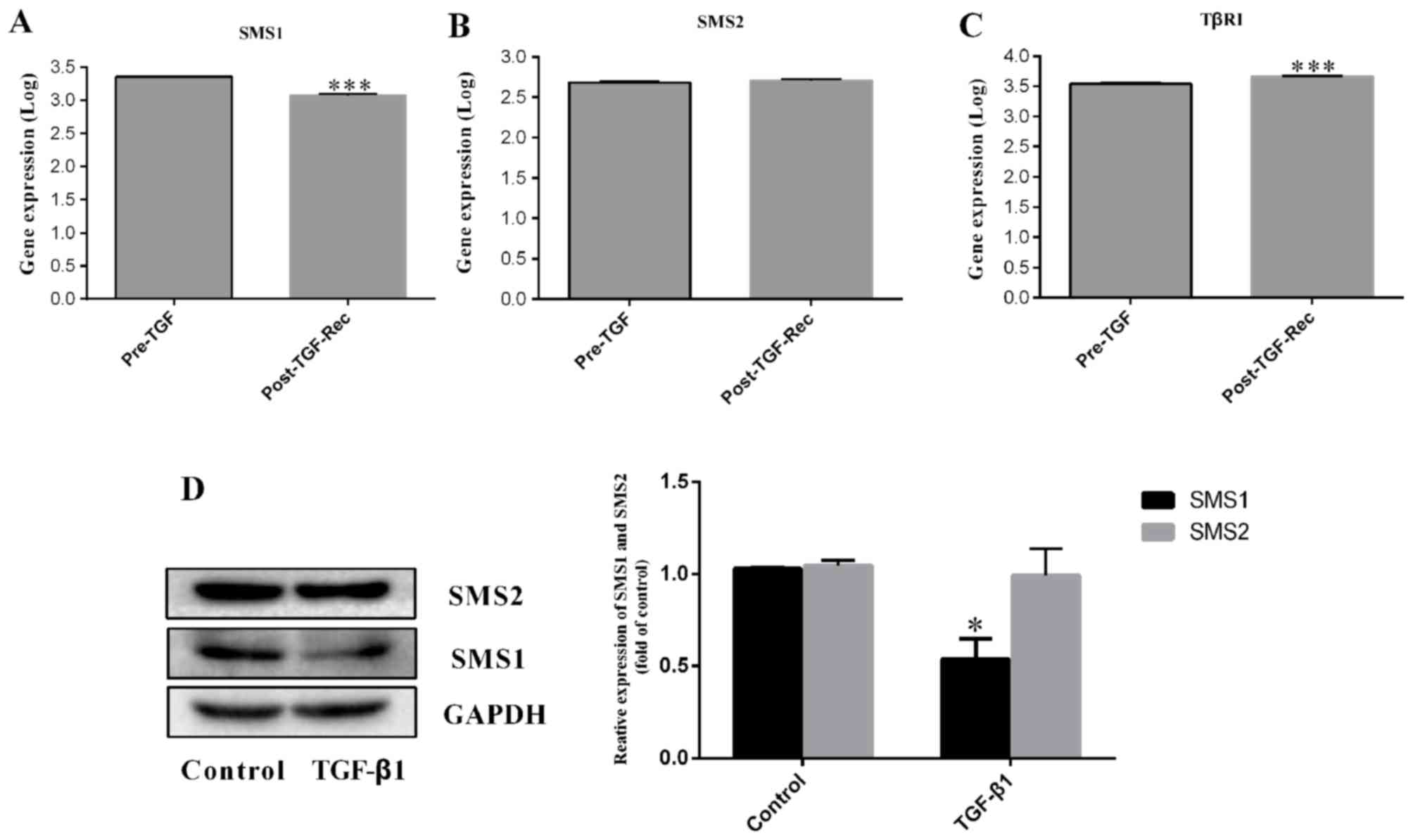
expression of SMS1 decreased by 0.48-fold (P<0.001; n=3;
Fig. 1A), whereas, the expression
of SMS2 was not significantly different (P=0.731; n=3; Fig. 1B), in the Post-TGF-Rec group
compared with the Pre-TGF group. However, TβRI (Fig. 1C) was increased by 0.32-fold
(P<0.001; n=3). To validate the bioinformatics results, the
expression of SMS1 and SMS2 in MDA-MB-231 cells following treatment
with 10 ng/ml TGF-β1 was measured. The results confirmed that the
expression of SMS1 was significantly decreased (Fig. 1D; P<0.05; n=3), whereas SMS2 was
not significantly different between groups (Fig. 1D; P>0.05; n=3); these results
were consistent with the bioinformatics data.
Expression of SMS1 is significantly
decreased in the TGF-β1-induced EMT process in MDA-MB-231
cells
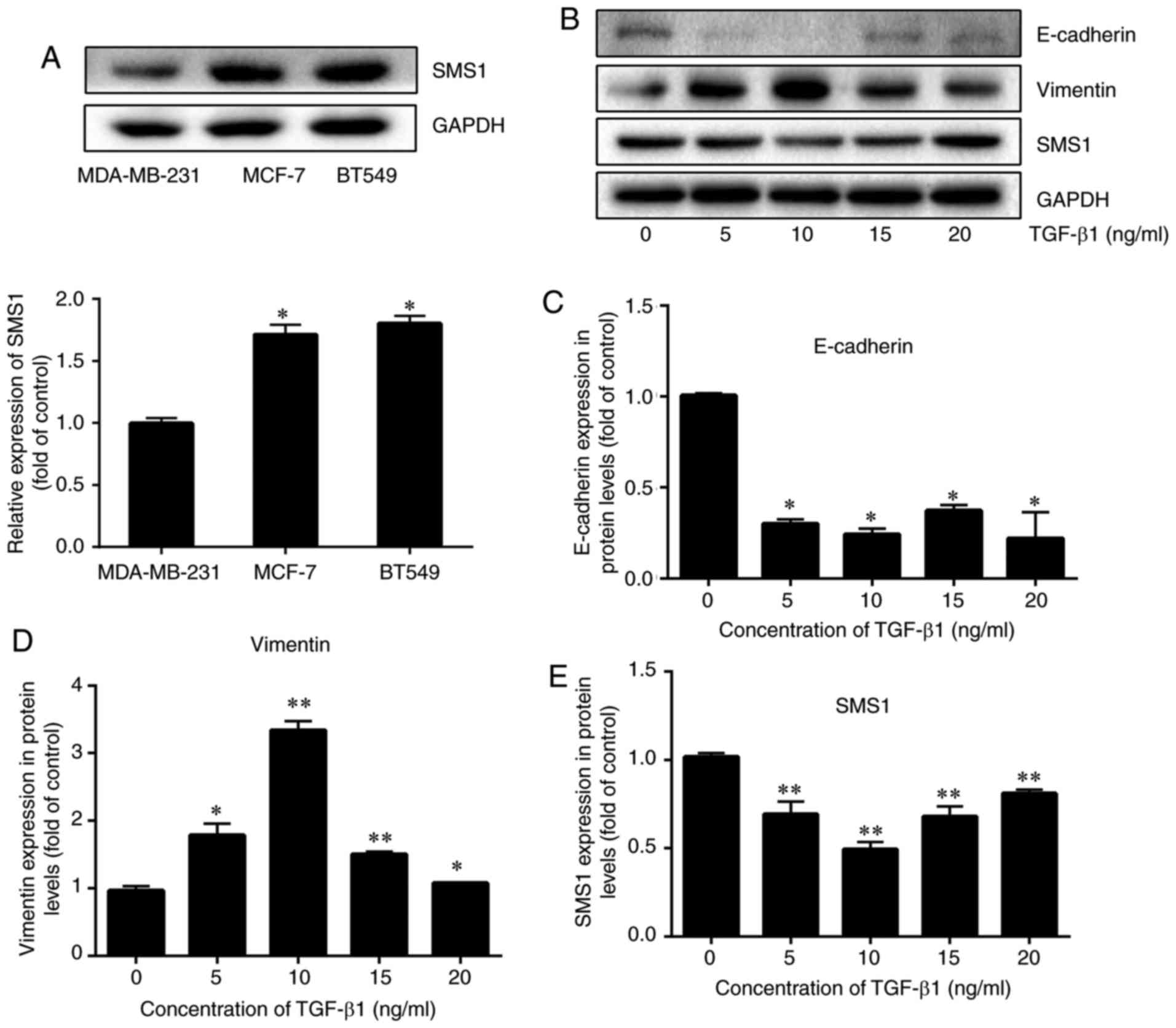
To examine whether the expression of SMS1 is
different in breast cancer cells, the expression of SMS1 in three
breast cancer cells, including MDA-MB-231, MCF-7 and BT549 was
investigated. The results demonstrated that the expression of SMS1
was the lowest in MDA-MB-231, followed by MCF-7 and BT549 (Fig. 2A; P<0.05, n=3). Therefore, in
the present study, MDA-MB-231 was selected.
MDA-MB-231 cells were cultured with different
concentrations of TGF-β1 (0, 5, 10, 15 and 20 ng/ml) for 72 h and
the expression of SMS1, E-cadherin and vimentin was examined. An
increase in vimentin and a decrease in E-cadherin expression were
evident when MDA-MB-231 cells were treated with 10 ng/ml TGF-β1
(P<0.05 and P<0.01, respectively; n=3; Fig. 2B-D, respectively). Similarly, the
decrease in SMS1 expression was evident when MDA-MB-231 cells were
treated with 10 ng/ml TGF-β1 (P<0.05 and P<0.01,
respectively; n=3; Fig. 2E).
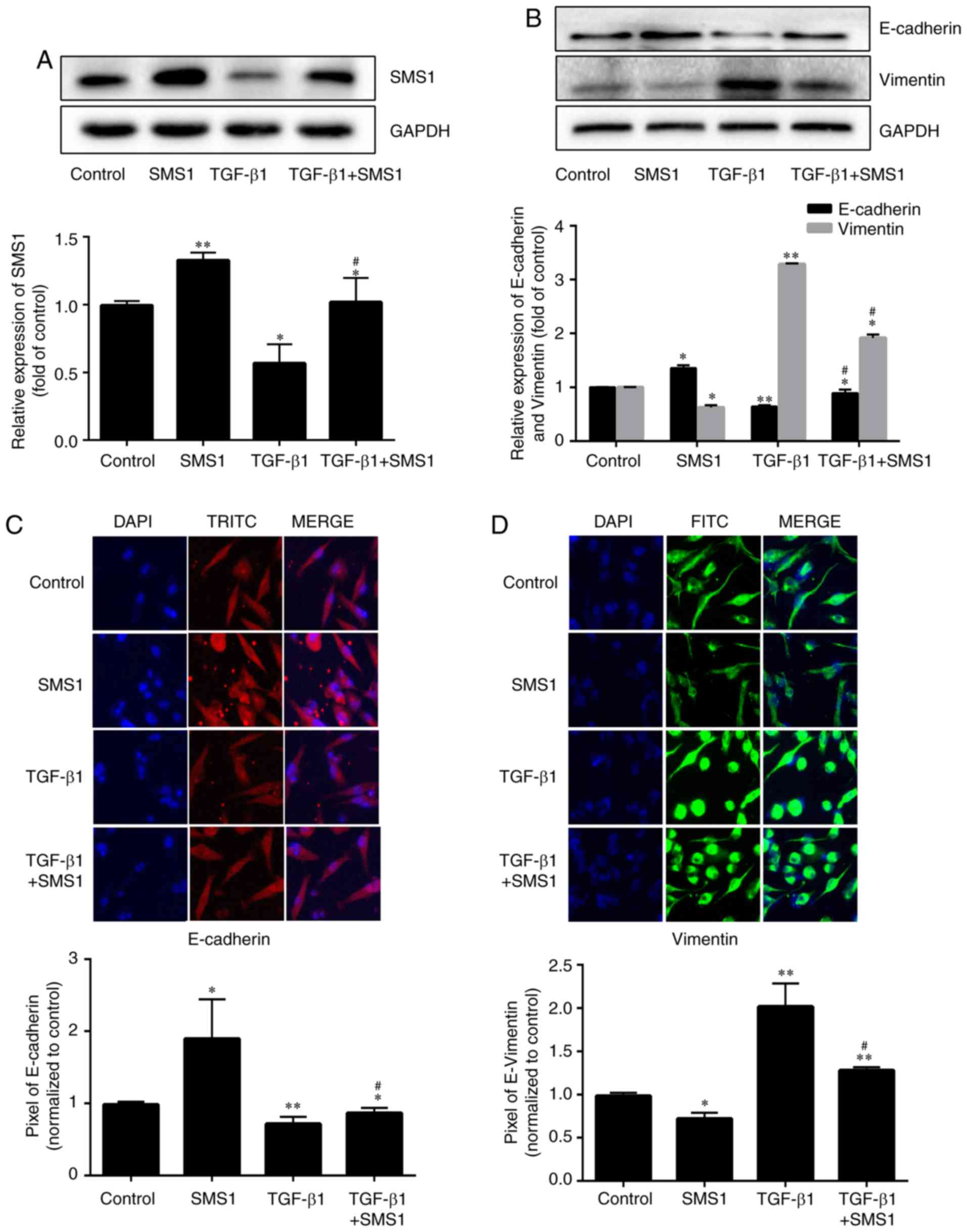
Overexpression of SMS1 inhibits
TGF-β1-induced EMT
Protein expression levels of the EMT markers
E-cadherin and vimentin were measured to evaluate the influence of
the overexpression of SMS1 on the TGF-β1-induced EMT process
(Fig. 3A; P<0.05 and P<0.01,
respectively, n=3). Following treatment with TGF-β1, the expression
of E-cadherin was decreased by 0.39-fold, and vimentin was
increased by 2.3-fold compared with the control group (Fig. 3B; P<0.05 or P<0.01; n=3). In
contrast, overexpression of SMS1 together with TGF-β1 treatment
blocked TGF-β1-induced EMT, in which the expression of E-cadherin
was decreased by 0.23-fold and vimentin was increased by 0.92-fold.
The immunofluorescence and confocal microscopy imaging results
verified these results to some extent (Fig. 3C and D).
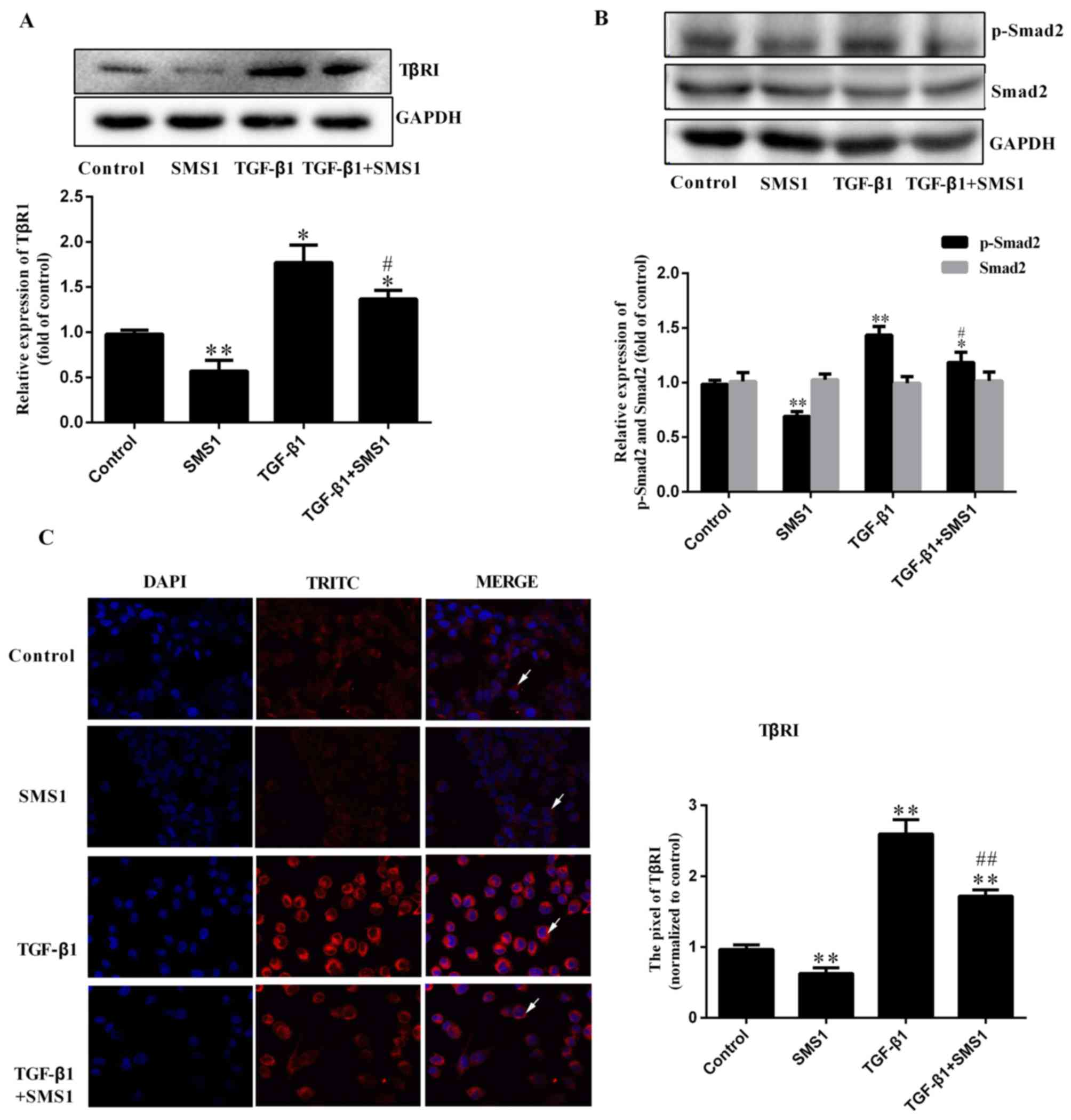
Overexpression of SMS1 regulates
TGF-β1-induced EMT via TGF-β type I receptor
Previous studies have demonstrated that the
Smad-dependent signaling pathway is induced by TGF-β1, including
the activation of TβRI and phosphorylated Smad2 and Smad3 (10,13).
To further investigate the mechanism of SMS1 in regulating the
TGF-β1 induced EMT process, the effect on Smad-dependent signaling
pathway proteins was investigated. Western blot analysis revealed
that TGF-β1 treatment induced the expression of TβRI and the
phosphorylation of Smad2, which were increased by 0.77 and
0.43-fold, respectively. The expression of Smad2 demonstrated no
significant difference (Fig. 4B;
P>0.05, respectively; n=3). However, SMS1 overexpression
downregulated TβRI expression and blocked Smad2 phosphorylation
(Fig. 4A and B; P<0.05 and
P<0.01, respectively, n=3). The expression of TβRI was
investigated by immunofluorescence. The results demonstrated that
the overexpression of SMS1 altered the expression of TβRI on the
cell membrane, which may be associated with the endocytosis of TβRI
(Fig. 4C) (16,17).
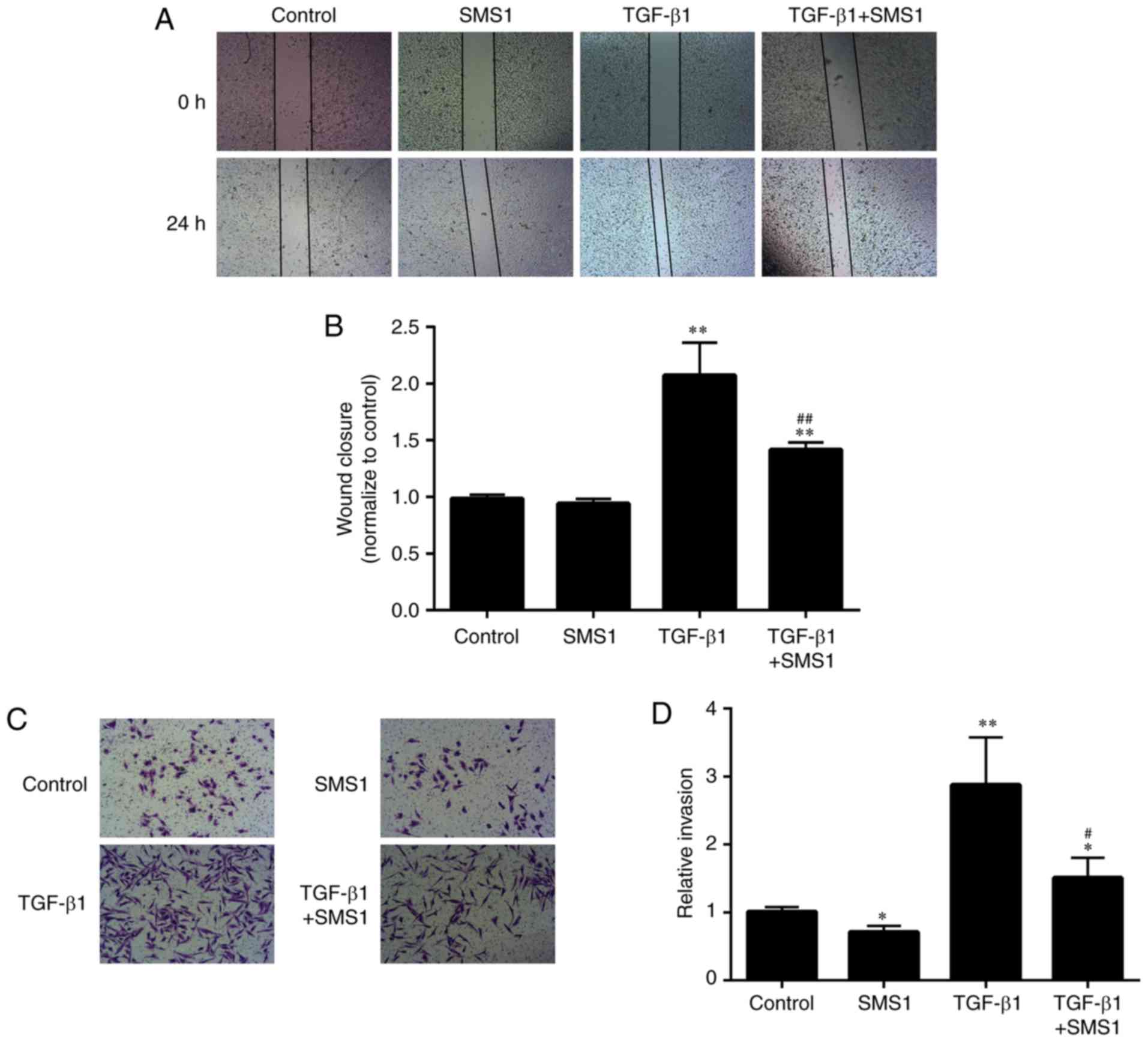
Overexpression of SMS1 inhibits
MDA-MB-231 cell migration and invasion induced by TGF-β1
To further clarify the role of SMS1 in the
TGF-β1-induced EMT in MDA-MB-231 cells, changes in the migration
and invasion abilities of cells treated with or without TGF-β1
following transfection with an SMS1 plasmid, were investigated.
Treatment with TGF-β1 significantly increased the cell migration
abilities in MDA-MB-231 cells by 1.1-fold, as demonstrated by wound
healing analysis. In addition, the increased migration abilities of
MDA-MB-231 cells were significantly inhibited (by 0.32-fold) by
transfection with an SMS1 plasmid (Fig. 5A and B; P<0.01; n=3).
Furthermore, treatment with TGF-β1 for 72 h significantly increased
the invasion abilities of MDA-MB-231 cells by 1.9-fold in a
Matrigel invasion assay. Overexpression of SMS1 significantly
suppressed the TGF-β1-induced invasion abilities of MDA-MB-231
cells by 0.47-fold (Fig. 5C and D;
P<0.05 and P<0.01, respectively; n=3).
Discussion
The aim of the present study was to assess the
importance of SMS1 in TGF-β1-induced EMT development. The results
showed that SMS1 overexpression can downregulate TβRI expression
and interfere with TGF-β1-induced Smad2 phosphorylation, it can
increase the expression of E-cadherin and decrease the expression
of vimentin. Finally, the migration and invasion of MDA-MB-231
cells were suppressed following SMS1 overexpression together with
treatment with TGF-β1. These results demonstrated that SMS1
overexpression can inhibit the EMT in MDA-MB-231 cells.
MS1 and SMS2 are the key enzymes in SM biosynthesis,
but SMS1 is found on the Golgi apparatus, and SMS2 exists in the
Golgi apparatus and plasma membranes (20). The expression levels of SMS1 and
TβRI were negatively associated with each other when breast cancer
cells were treated with TGF-β1; however, the expression of SMS2 was
not altered and was not correlated with that of TβRI. This suggests
that SMS1 and SMS2 may have different functions in the EMT.
Additionally, three breast cancer cell lines expressed SMS1;
however, the expression of SMS1 was lower in MDA-MB-231 cells.
Furthermore, MDA-MB-231 is a triple-negative breast cancer cell
line, which has high metastatic and invasive ability (25). Therefore, in the present study,
MDA-MB-231 cells was used to demonstrate that SMS1 can regulate the
EMT in MDA-MB-231 cells.
Previous studies have demonstrated that TGF-β1
induces EMT to promote tumor invasion and metastasis (11–14).
During EMT development, the morphological alterations are
characterized by upregulated expression of the mesenchymal marker
vimentin and downregulated expression of the epithelial marker
E-cadherin. Our findings were consistent with their findings,
however, SMS1 can inhibit these changes, which shown that
overexpression SMS1 can inhibit EMT (Figs. 3 and 5).
Similar to other cell surface receptors (26), TGF-β receptors are mainly
internalized via clathrin-dependent endocytosis, which is an
essential regulatory event in signal transduction (27,28).
In addition, lipid rafts/caveolae negatively regulate TGF-β1
signaling pathway through lipid raft-induced internalization of
TGF-β receptors, promoting receptor degradation (29). Therefore, altering the contents of
the main components of lipid raft would affect the distribution of
TGF-β receptors in lipid raft/caveolae and no lipid raft, and the
signal transduction of TGF-β receptors. In the present study,
overexpression of SMS1 negatively regulated TβRI expression,
moreover, SMS1 overexpression may block the TGF-β1-induced
phosphorylation of Smad 2 and Smad2-mediated transcriptional
activity without affecting total Smad 2 expression levels and
additionally suppressed the TGFβ-1-induced EMT and cell migration.
As demonstrated in previous studies (27–29),
caveolin-1 and TβRI have distributed colocalization in the cell
membrane. A possible explanation for this might be that SMS1
promotes lipid raft/caveolae-mediated internalization and the
interaction between TβRI and caveolin-1, resulting in decreased
surface expression of TβRI and Smad activation. Unlike
clathrin-dependent endocytosis, lipid raft/caveolae-mediated
internalization facilitates the degradation of TβRI receptors and
therefore inhibits TGF-β1 signaling pathway (15,29).
In fact, a number of previous studies identified
that altering the components of lipid rafts/caveolae may affect the
TGF-β/Smad signaling pathway (30,31).
In addition to SM, cholesterol is additionally a primary component
of lipid rafts (30). Cholesterol
precursors and cholesterol biosimilars can regulate signal
transduction and the TGF-β/Smad pathway. For example, 7-DHC
(precursor) and Euphol (biosimilar) can suppress TGF-β-stimulated
luciferase activity by promoting lipid raft/caveolae formation and
subsequently recruiting cell-surface TGF-β receptors from non-lipid
raft microdomains to lipid rafts/caveolae, where TGF-β receptors
become inactive in transducing canonical signaling and undergo
rapid degradation upon TGF-β binding (29–31).
In contrast, methyl-β-cyclodextrin, a sterol-chelating agent,
reverses 7-DHC-induced suppression of TGF-β-stimulated luciferase
activity by extrusion of 7-DHC from resident lipid rafts/caveolae
(31). Furthermore, other factors
that can affect the distribution of TGF-β receptors between lipid
rafts/caveolae and non-lipid raft microdomains can also regulate
the TGF-β/Smad pathway (27–29).
Previous studies identified that cholest-4-en-3-one and dimethyl
sulfoxide may increase lipid raft and/or caveolae accumulation of
TGF-β receptors and facilitate the rapid degradation of TGF-β, thus
suppressing TGF-β-induced signaling (32,33).
In addition, ethanol also disrupts the location of other membrane
proteins in lipid rafts/caveolae that utilize lipid rafts/caveolae
as signaling platforms and enhances canonical TGF-β signaling by
increasing the non-lipid raft microdomain localization of TGF-β
receptors (34). Altogether,
canonical TGF-β signaling pathway is tightly associated with lipid
rafts and their primary components.
Although the studies presented thus far have
indicated that receptor endocytosis is not essential for TGF-β1
signaling, lipid raft-mediated endocytosis of TGF-β receptors
facilitates receptor degradation and thus turns off the signaling
pathway (32,35). However, this is the first study, to
the best of the authors' knowledge to investigate that the
attenuation of the TGF-β1-induced EMT by SMS1, which could
influence the formation of lipid rafts via TβRI expression and
could be an important mechanism for the controlled progression of
developmental events. The present study provides novel insight into
the impact of SMS1 in signal transduction, and it has a number of
important implications for future targeted therapy for breast
cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from
National Natural Science Foundation of China (grant no.
81560151).
Availability of data and materials
The datasets used and/or generated during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
SL, HH and PZ performed the experiments and
bioinformatics analysis, and were major contributors in writing the
manuscript. YW analyzed and interpreted the data of the manuscript,
and was responsible for the design and drafting of the manuscript.
XH and HL were responsible for the statistical analysis. NY
designed the study and analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fitzmaurice C, Akinyemiju TF, Al Lami FH,
Alam T, Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N,
Amini E, Anderson BO, et al: Global, regional, and national cancer
incidence, mortality, years of life lost, years lived with
disability, and disability-adjusted life-years for 29 cancer
groups, 1990 to 2016: A systematic analysis for the global burden
of disease study. JAMA Oncol. 4:1553–1568. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–81. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Q, Yang W, Wang X, Li X, Qi S, Zhang
Y and Gao MQ: TGF-beta1 induces EMT in bovine mammary epithelial
cells through the TGFbeta1/Smad signaling pathway. Cell Physiol
Biochem. 43:82–93. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yeh YC, Wei WC, Wang MG, Lin SC, Sung JM
and Tang MJ: Transforming growth factor-β1 induces smad3-dependent
β 1, integrin gene expression in epithelial-to-mesenchymal
transition during chronic tubulointerstitial fibrosis. Am J Pathol.
177:743–1754. 2010. View Article : Google Scholar
|
|
6
|
Attisano L and Wrana JL: Signal
transduction by the TGF-beta superfamily. Science. 296:1646–1647.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hwangbo C, Tae N, Lee S, Kim O, Park OK,
Kim J, Kwon SH and Lee JH: Syntenin regulates TGF-β1-induced Smad
activation and the epithelial-to-mesenchymal transition by
inhibiting caveolin-mediated TGF-β type I receptor internalization.
Oncogene. 35:389–401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–42. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miettinen PJ, Ebner R, Lopez AR and
Derynck R: TGF-beta induced transdifferentiation of mammary
epithelial cells to mesenchymal cells: Involvement of type I
receptors. J Cell Biol. 127:2021–2036. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Derynck R and Zhang YE: Smad-dependent and
smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin XL, Liu M, Liu Y, Hu H, Pan Y, Zou W,
Fan X and Hu X: Transforming growth factor beta1 promotes migration
and invasion in HepG2 cells: Epithelialtomesenchymal transition via
JAK/STAT3 signaling. Int J Mol Med. 41:129–136. 2017.PubMed/NCBI
|
|
12
|
Kim JY, An HJ, Kim WH, Gwon MG, Gu H, Park
YY and Park KK: Anti-fibrotic effects of synthetic
oligodeoxynucleotide for TGF-beta1 and smad in an animal model of
liver cirrhosis. Mol Ther Nucleic Acids. 8:250–263. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boldbaatar A, Lee S, Han S, Jeong AL, Ka
HI, Buyanravjikh S, Lee JH, Lim JS, Lee MS and Yang Y: Eupatolide
inhibits the TGF-beta1-induced migration of breast cancer cells via
downregulation of SMAD3 phosphorylation and transcriptional
repression of ALK5. Oncol Lett. 14:6031–6039. 2017.PubMed/NCBI
|
|
14
|
Wang Y, Liu X, Zheng H, Wang Q, An L and
Wei G: Suppression of CUL4A attenuates TGF-beta1-induced
epithelial-to-mesenchymal transition in breast cancer cells. Int J
Mol Med. 40:1114–1124. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Le Roy C and Wrana JL: Clathrin- and
non-clathrin-mediated endocytic regulation of cell signalling. Nat
Rev Mol Cell Biol. 6:112–26. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Midgley AC, Rogers M, Hallett MB, Clayton
A, Bowen T, Phillips AO and Steadman R: Transforming growth
factor-beta1 (TGF-beta1)-stimulated fibroblast to myofibroblast
differentiation is mediated by hyaluronan (HA)-facilitated
epidermal growth factor receptor (EGFR) and CD44 co-localization in
lipid rafts. J Biol Chem. 288:14824–14838. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen YG: Endocytic regulation of TGF-beta
signaling. Cell Res. 19:58–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Xia K, Xiong M, Wang X and Yan N:
Effects of sepsis on the metabolism of sphingomyelin and
cholesterol in mice with liver dysfunction. Exp Ther Med.
14:5635–5640. 2017.PubMed/NCBI
|
|
19
|
Taniguchi M and Okazaki T: The role of
sphingomyelin and sphingomyelin synthases in cell death,
proliferation and migration-from cell and animal models to human
disorders. Biochim Biophys Acta. 1841:692–703. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yeang C, Ding T, Chirico WJ and Jiang XC:
Subcellular targeting domains of sphingomyelin synthase 1 and 2.
Nutr Metab (Lond). 8:892011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Torki S, Soltani A, Shirzad H, Esmaeil N
and Ghatrehsamani M: Synergistic antitumor effect of NVP-BEZ235 and
CAPE on MDA-MB-231 breast cancer cells. Biomed Pharmacother.
92:39–45. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo S, Pan Z, Liu S, Yuan S and Yan N:
Sphingomyelin synthase 2 overexpression promotes cisplatin-induced
apoptosis of HepG2 cells. Oncol Lett. 15:483–488. 2018.PubMed/NCBI
|
|
23
|
Yan N, Ding T, Dong J, Li Y and Wu M:
Sphingomyelin synthase overexpression increases cholesterol
accumulation and decreases cholesterol secretion in liver cells.
Lipids Health Dis. 10:462011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu S, Ding Y, Gong J and Yan N:
Sphingomyelin synthase 2 affects CD14 associated induction of
NFkappaB by lipopolysaccharides in acute lung injury in mice. Mol
Med Rep. 14:3301–3306. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kwiatkowska E, Wojtala M, Gajewska A,
Soszynski M, Bartosz G and Sadowska-Bartosz I: Effect of
3-bromopyruvate acid on the redox equilibrium in non-invasive MCF-7
and invasive MDA-MB-231 breast cancer cells. J Bioenerg Biomembr.
48:23–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Xia J, Liu S, Stein S, Ramon C, Xi
H, Wang L, Xiong X, Zhang L, He D, et al: Endocytosis and membrane
receptor internalization: Implication of F-BAR protein carom. Front
Biosci (Landmark Ed). 22:1439–1457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Conner SD and Schmid SL: Regulated portals
of entry into the cell. Nature. 422:37–44. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mitchell H, Choudhury A, Pagano RE and
Leof EB: Ligand-dependent and -independent transforming growth
factor-beta receptor recycling regulated by clathrin-mediated
endocytosis and Rab11. Mol Biol Cell. 15:4166–4178. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Razani B, Zhang XL, Bitzer M, von
Gersdorff G, Bottinger EP and Lisanti MP: Caveolin-1 regulates
transforming growth factor (TGF)-beta/SMAD signaling through an
interaction with the TGF-beta type I receptor. J Biol Chem.
276:6727–6738. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang B, Paffett ML, Naik JS, Jernigan NL,
Walker BR and Resta TC: Cholesterol regulation of pulmonary
endothelial calcium homeostasis. Curr Top Membr. 82:53–91. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang SS, Liu IH, Chen CL, Chang JM,
Johnson FE and Huang JS: 7-Dehydrocholesterol (7-DHC), but not
cholesterol, causes suppression of canonical TGF-beta signaling and
is likely involved in the development of Atherosclerotic
Cardiovascular Disease (ASCVD). J Cell Biochem. 118:1387–1400.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen CL, Wu DC, Liu MY, Lin MW, Huang HT,
Huang YB, Chen LC, Chen YY, Chen JJ, Yang PH, et al:
Cholest-4-en-3-one attenuates TGF-beta responsiveness by inducing
TGF-beta receptors degradation in Mv1Lu cells and colorectal
adenocarcinoma cells. J Recept Signal Transduct Res. 37:189–199.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang SS, Chen CL, Huang FW, Hou WH and
Huang JS: DMSO enhances TGF-beta activity by recruiting the type II
TGF-beta receptor from intracellular vesicles to the plasma
membrane. J Cell Biochem. 117:1568–1579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang SS, Chen CL, Huang FW, Johnson FE
and Huang JS: Ethanol enhances TGF-beta activity by recruiting
TGF-beta receptors from intracellular vesicles/lipid rafts/caveolae
to non-lipid raft microdomains. J Cell Biochem. 117:860–871. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen CL, Huang SS and Huang JS: Cellular
heparan sulfate negatively modulates transforming growth
factor-beta1 (TGF-beta1) responsiveness in epithelial cells. J Biol
Chem. 281:11506–11514. 2006. View Article : Google Scholar : PubMed/NCBI
|