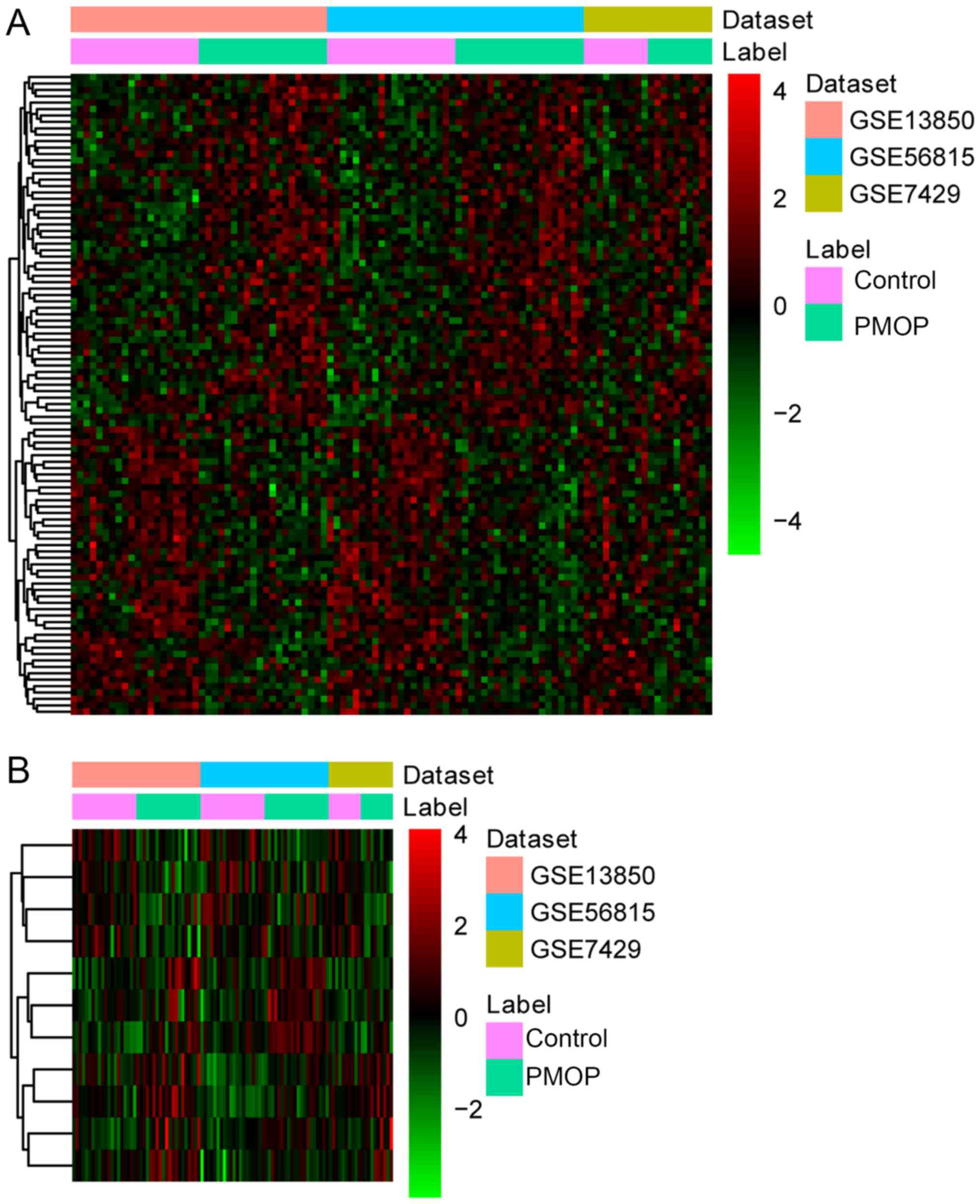
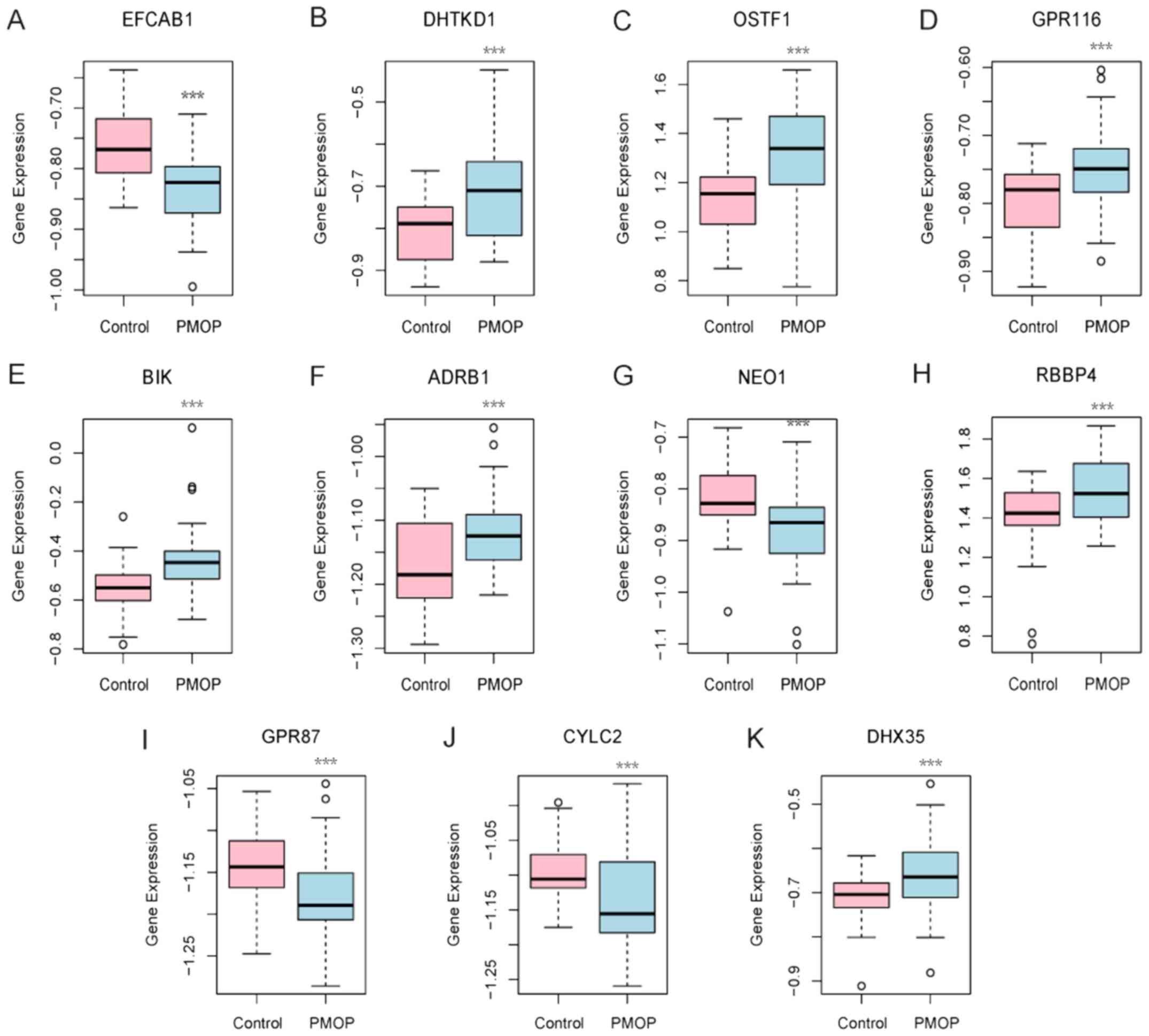
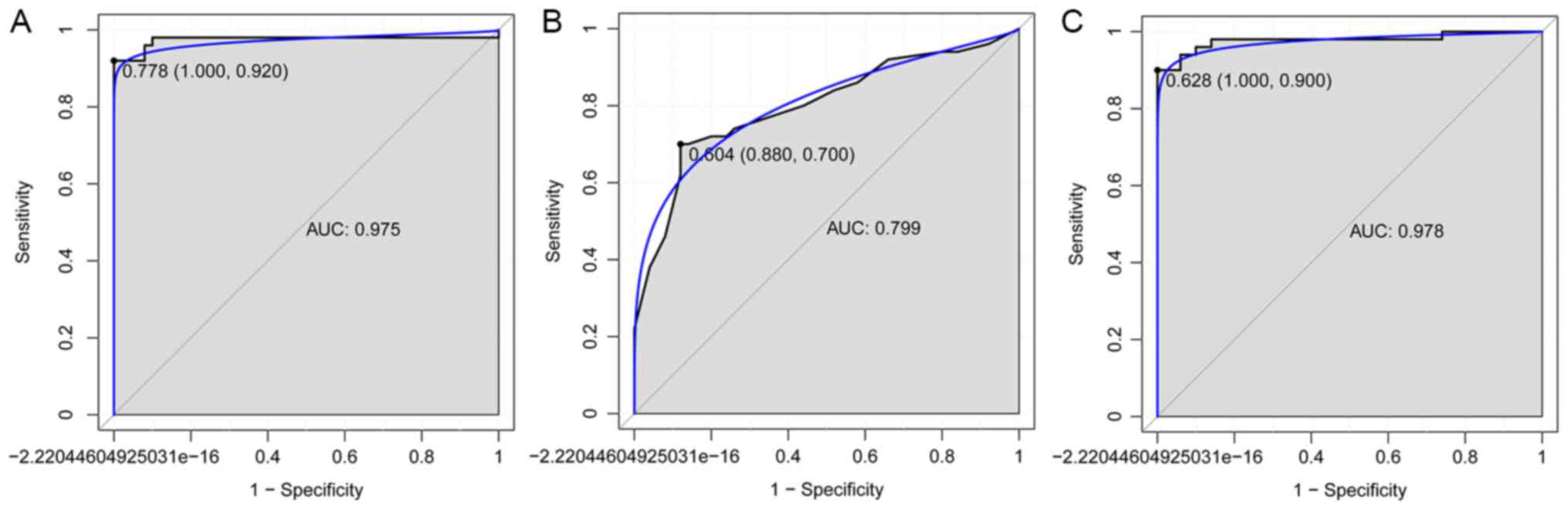
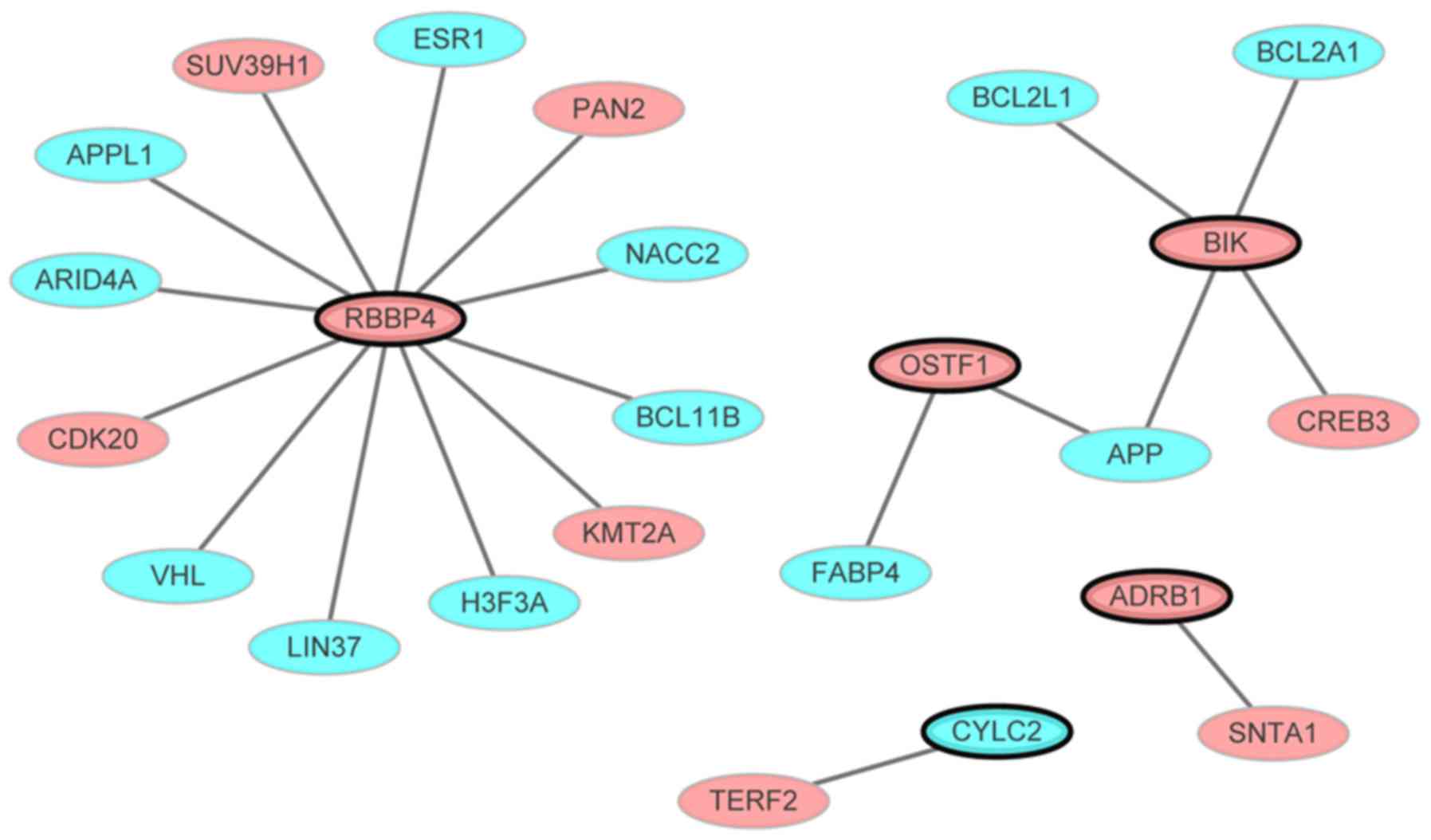
|
1
|
Kanis JA, McCloskey EV, Johansson H, Oden
A, Melton LJ III and Khaltaev N: A reference standard for the
description of osteoporosis. Bone. 42:467–475. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paschalis EP, Gamsjaeger S, Hassler N,
Fahrleitner-Pammer A, Dobnig H, Stepan JJ, Pavo I, Eriksen EF and
Klaushofer K: Vitamin D and calcium supplementation for three years
in postmenopausal osteoporosis significantly alters bone mineral
and organic matrix quality. Bone. 95:41–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meng J, Zhang D, Pan N, Sun N, Wang Q, Fan
J, Zhou P, Zhu W and Jiang L: Identification of mir-194-5p as a
potential biomarker for postmenopausal osteoporosis. PeerJ.
3:e9712015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sanders S and Geraci SA: Osteoporosis in
postmenopausal women: Considerations in prevention and treatment:
(women's health series). Southern Med J. 106:698–706. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan B, Li J and Zhang L: Identification of
B cells participated in the mechanism of postmenopausal women
osteoporosis using microarray analysis. Int J Clin Exp Med.
8:1027–1034. 2015.PubMed/NCBI
|
|
6
|
Liu Y, Wang Y, Yang N, Wu S, Lv Y and Xu
L: In silico analysis of the molecular mechanism of postmenopausal
osteoporosis. Mol Med Rep. 12:6584–6590. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma M, Luo S, Zhou W, Lu L, Cai J, Yuan F
and Yin F: Bioinformatics analysis of gene expression profiles in B
cells of postmenopausal osteoporosis patients. Taiwan J Obstet
Gynecol. 56:165–170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Breiman L: Random forests. Machine
Learning. 45:5–32. 2001. View Article : Google Scholar
|
|
11
|
Stoppiglia H, Dreyfus G, Dubois R and
Oussar Y: Ranking a random feature for variable and feature
selection. J Mach Learn Res. 3:1399–1414. 2003.
|
|
12
|
Xiao P, Chen Y, Jiang H, Liu YZ, Pan F,
Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR and Deng HW: In
vivo genome-wide expression study on human circulating B cells
suggests a novel esr1 and mapk3 network for postmenopausal
osteoporosis. J Bone Miner Res. 23:644–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tella SH and Gallagher JC: Prevention and
treatment of postmenopausal osteoporosis. J Steroid Biochem Mol
Biol. 155–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma M, Chen X, Lu L, Yuan F, Zeng W, Luo S,
Yin F and Cai J: Identification of crucial genes related to
postmenopausal osteoporosis using gene expression profiling. Aging
Clin Exp Res. 28:1067–1074. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vermeren M, Lyraki R, Wani S, Airik R,
Albagha O, Mort R, Hildebrandt F and Hurd T: Osteoclast stimulation
factor 1 (ostf1) knockout increases trabecular bone mass in mice.
Mamm Genome. 28:498–514. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Lin Y, Zhao H, Guo Q, Yan C and
Lin N: Revealing the effects of the herbal pair of euphorbia kansui
and glycyrrhiza on hepatocellular carcinoma ascites with
integrating network target analysis and experimental validation.
Int J Biol Sci. 12:594–606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takeda S, Elefteriou F, Levasseur R, Liu
X, Zhao L, Parker KL, Armstrong D, Ducy P and Karsenty G: Leptin
regulates bone formation via the sympathetic nervous system. Cell.
111:305–317. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Karsenty G: Leptin controls bone formation
through a hypothalamic relay. Recent Prog Horm Res. 56:401–415.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Elefteriou F, Ahn JD, Takeda S, Starbuck
M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, et al:
Leptin regulation of bone resorption by the sympathetic nervous
system and cart. Nature. 434:514–520. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Z, Xie J, Lee D, Liu Y, Jung J, Zhou
L, Xiong S, Mei L and Xiong WC: Neogenin regulation of bmp-induced
canonical smad signaling and endochondral bone formation. Dev cell.
19:90–102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brommage R, Liu J, Hansen GM, Kirkpatrick
LL, Potter DG, Sands AT, Zambrowicz B, Powell DR and Vogel P:
High-throughput screening of mouse gene knockouts identifies
established and novel skeletal phenotypes. Bone Res. 2:140342014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sharma D, Larriera AI, Palacio-Mancheno
PE, Gatti V, Fritton JC, Bromage TG, Cardoso L, Doty SB and Fritton
SP: The effects of estrogen deficiency on cortical bone
microporosity and mineralization. Bone. 110:1–10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Yao F, Liang X, Zhu X, Zheng R,
Jia B, Hou L and Zou X: Cloning and expression of
retinoblastoma-binding protein 4 gene in embryo diapause
termination and in response to salinity stress from brine shrimp
artemia sinica. Gene. 591:351–361. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Balboula AZ, Stein P, Schultz RM and
Schindler K: Rbbp4 regulates histone deacetylation and bipolar
spindle assembly during oocyte maturation in the mouse. Biol
Reprod. 92:1052015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saul D, Ninkovic M, Komrakova M, Wolff L,
Simka P, Gasimov T, Menger B, Hoffmann DB, Rohde V and Sehmisch S:
Effect of zileuton on osteoporotic bone and its healing, expression
of bone, and brain genes in rats. J Appl Physiol. 124:118–130.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shalan NA, Mustapha NM and Mohamed S: Noni
leaf and black tea enhance bone regeneration in estrogen-deficient
rats. Nutrition. 33:42–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu W, Zhu H, Gu M, Luo Q, Ding J, Yao Y,
Chen F and Wang Z: Dhtkd1 is essential for mitochondrial biogenesis
and function maintenance. FEBS Lett. 587:3587–3592. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Danhauser K, Sauer S, Haack TB, Wieland T,
Staufner C, Graf E, Zschocke J, Strom TM, Traub T, Okun JG, et al:
Dhtkd1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am J
Hum Genet. 91:1082–1087. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sherrill JD, Kc K, Wang X, Wen T,
Chamberlin A, Stucke EM, Collins MH, Abonia JP, Peng Y, Wu Q, et
al: Whole-exome sequencing uncovers oxidoreductases dhtkd1 and
ogdhl as linkers between mitochondrial dysfunction and eosinophilic
esophagitis. JCI Insight. 3:2018. View Article : Google Scholar
|
|
30
|
Kim JH and Lee DC: Mitochondrial DNA copy
number in peripheral blood is associated with femoral neck bone
mineral density in postmenopausal women. J Rheumatol. 39:1465–1472.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Glatt S, Halbauer D, Heindl S, Wrnitznig
A, Kozina D, Su KC, Puri C, Garin-Chesa P and Sommergruber W:
Hgpr87 contributes to viability of human tumor cells. Int J Cancer.
122:2008–2016. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y, Scoumanne A and Chen X: G
protein-coupled receptor 87: A promising opportunity for cancer
drug discovery. Mol Cell Pharmacol. 2:111–116. 2010.PubMed/NCBI
|
|
33
|
Wang L, Zhou W, Zhong Y, Huo Y, Fan P,
Zhan S, Xiao J, Jin X, Gou S, Yin T, et al: Overexpression of g
protein-coupled receptor gpr87 promotes pancreatic cancer
aggressiveness and activates nf-κb signaling pathway. Mol Cancer.
16:612017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hatzimichael E, Dasoula A, Kounnis V,
Benetatos L, Lo Nigro C, Lattanzio L, Papoudou-Bai A, Dranitsaris
G, Briasoulis E and Crook T: Bcl2-interacting killer cpg
methylation in multiple myeloma: A potential predictor of
relapsed/refractory disease with therapeutic implications. Leuk
Lymphoma. 53:1709–1713. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Langemeijer SM, Mariani N, Knops R,
Gilissen C, Woestenenk R, de Witte T, Huls G, van der Reijden BA
and Jansen JH: Apoptosis-related gene expression profiling in
hematopoietic cell fractions of MDS patients. PLoS One.
11:e01655822016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hoff AM, Johannessen B, Alagaratnam S,
Zhao S, Nome T, Løvf M, Bakken AC, Hektoen M, Sveen A, Lothe RA and
Skotheim RI: Novel rna variants in colorectal cancers. Oncotarget.
6:36587–36602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Figlioli G, Kohler A, Chen B, Elisei R,
Romei C, Cipollini M, Cristaudo A, Bambi F, Paolicchi E, Hoffmann
P, et al: Novel genome-wide association study-based candidate loci
for differentiated thyroid cancer risk. J Clin Endocrinol Metab.
99:E2084–E2092. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cha S, Lim JE, Park AY, Do JH, Lee SW,
Shin C, Cho NH, Kang JO, Nam JM, Kim JS, et al: Identification of
five novel genetic loci related to facial morphology by genome-wide
association studies. BMC Genomics. 19:4812018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ohara K, Arai E, Takahashi Y, Ito N,
Shibuya A, Tsuta K, Kushima R, Tsuda H, Ojima H, Fujimoto H, et al:
Genes involved in development and differentiation are commonly
methylated in cancers derived from multiple organs: A
single-institutional methylome analysis using 1007 tissue
specimens. Carcinogenesis. 38:241–251. 2017.PubMed/NCBI
|