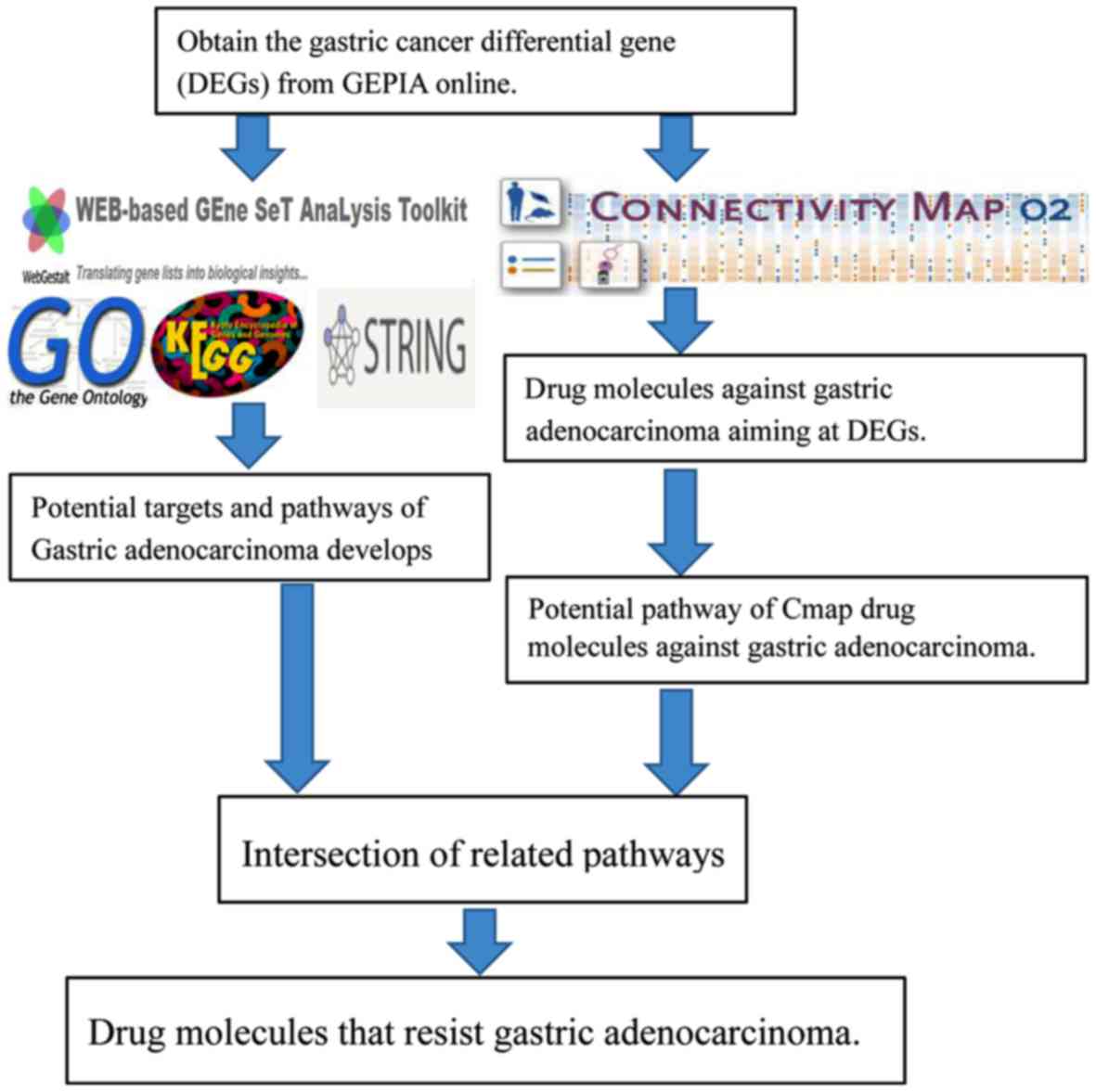
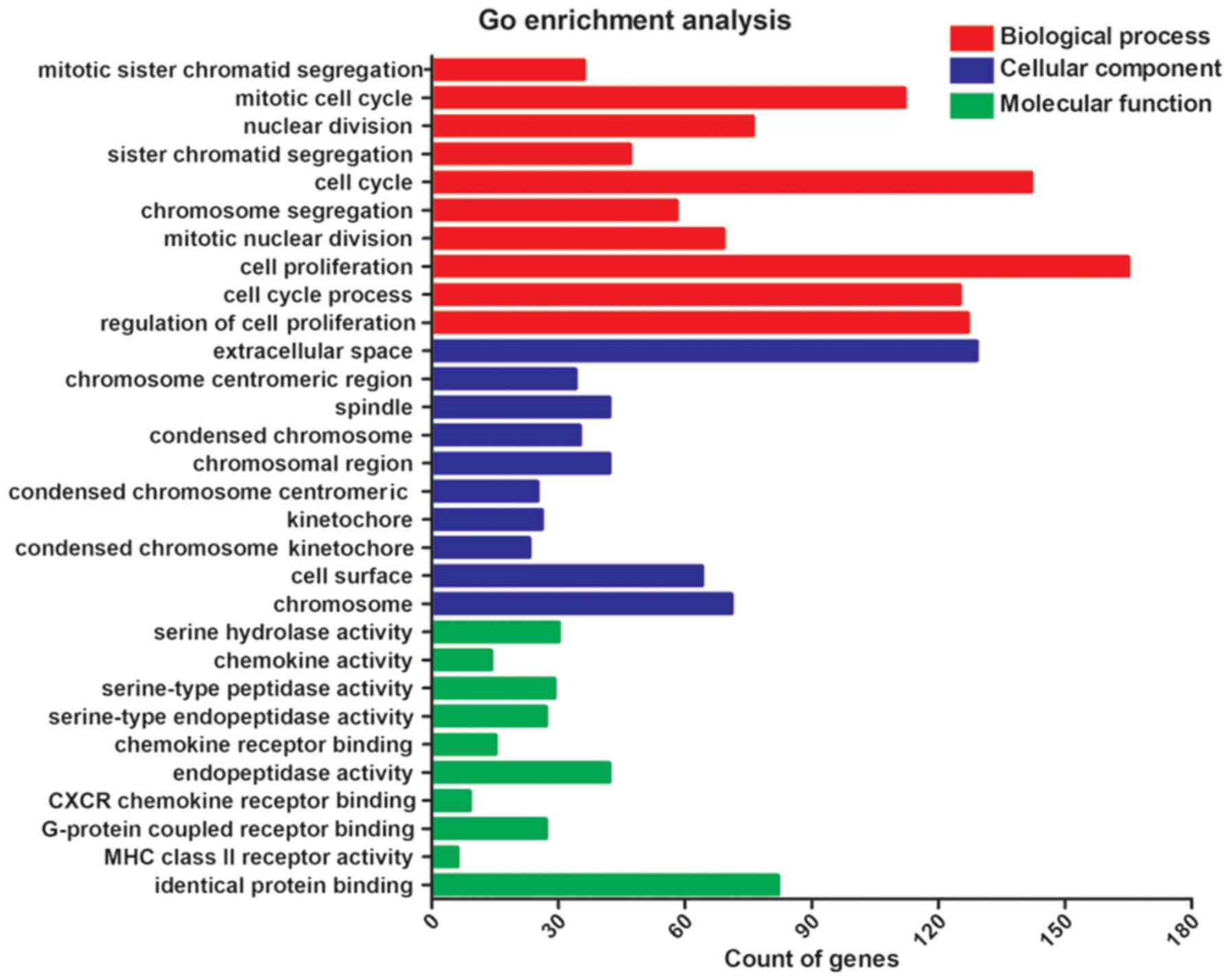
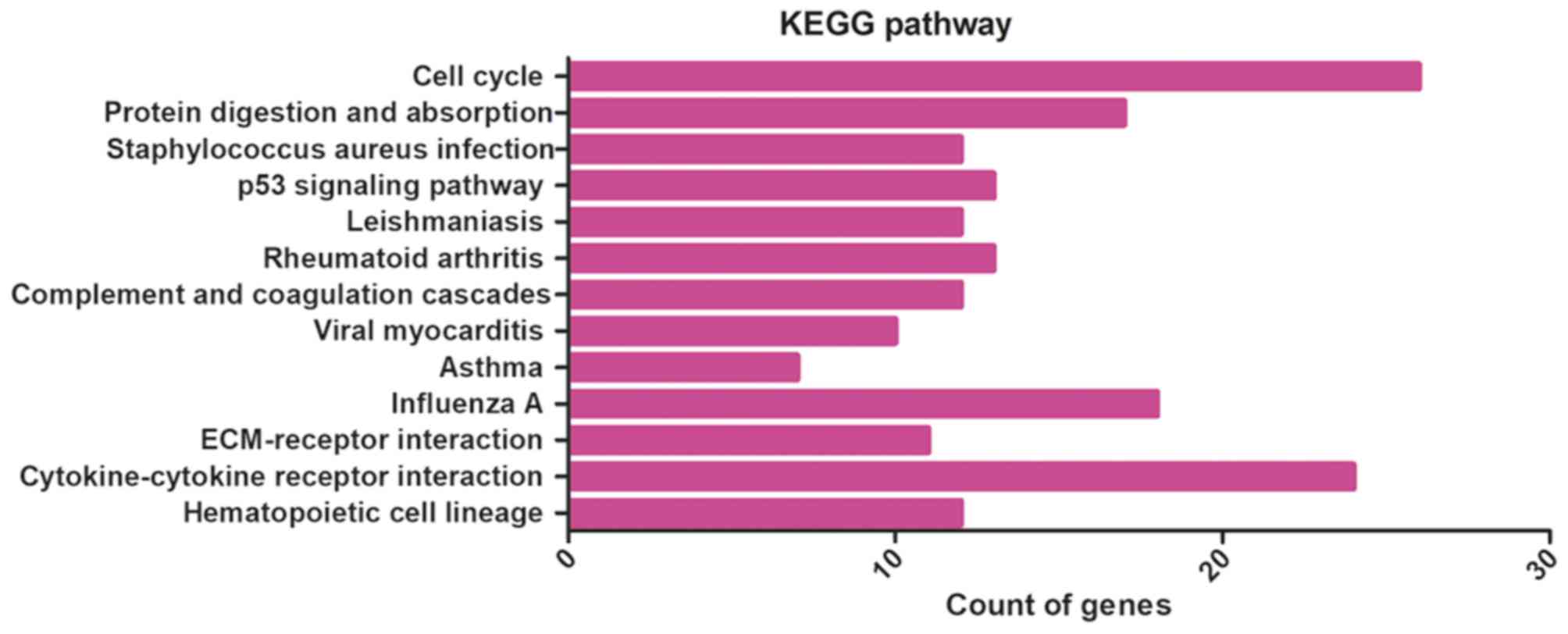
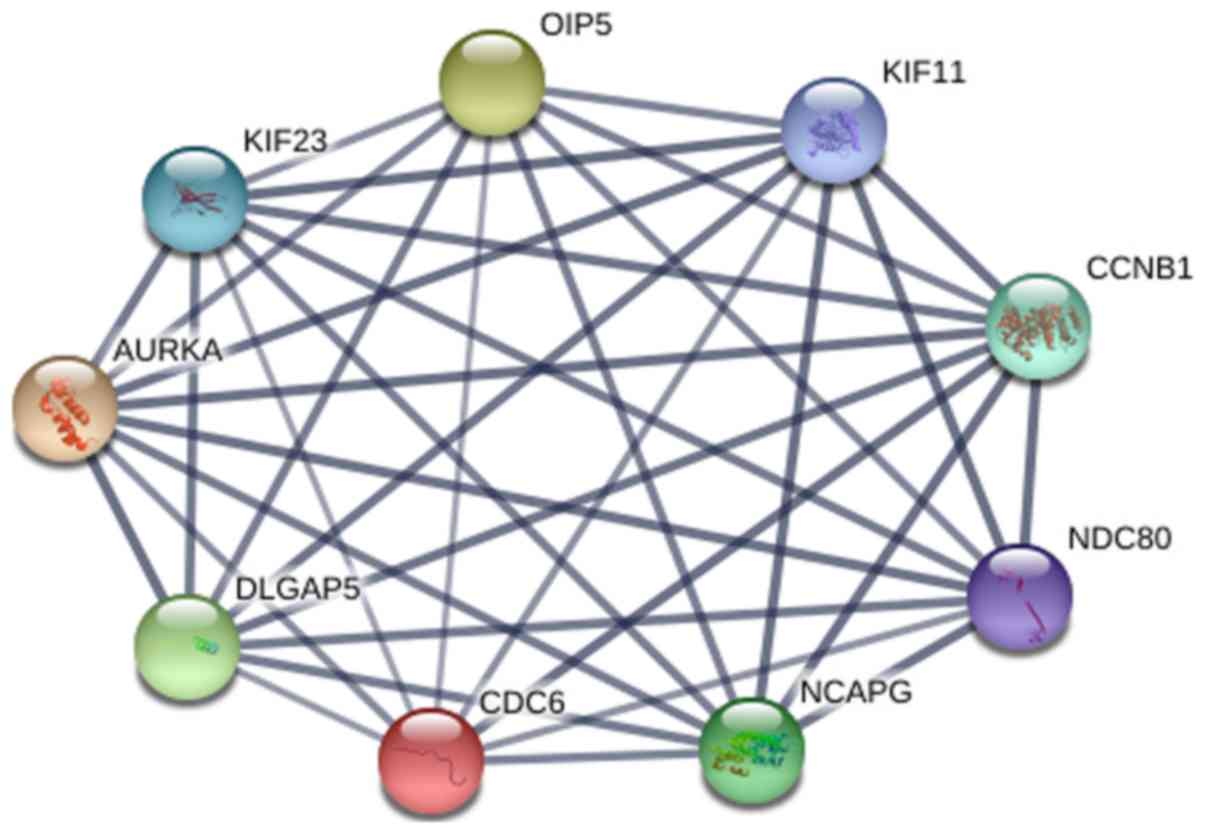
|
1
|
Wippel HH, Santos MDM, Clasen MA, Kurt LU,
Nogueira FCS, Carvalho CE, McCormick TM, Neto GPB, Alves LR, da
Gloria da Costa Carvalho M, et al: Comparing intestinal versus
diffuse gastric cancer using a PEFF-oriented proteomic pipeline. J
Proteomics. 171:63–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li W, Song D, Li H, Liang L, Zhao N and
Liu T: Reduction in peripheral CD19+CD24hCD27+ B cell frequency
predicts favourable clinical course in XELOX-treated patients with
advanced gastric cancer. Cell Physiol Biochem. 41:2045–2052. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
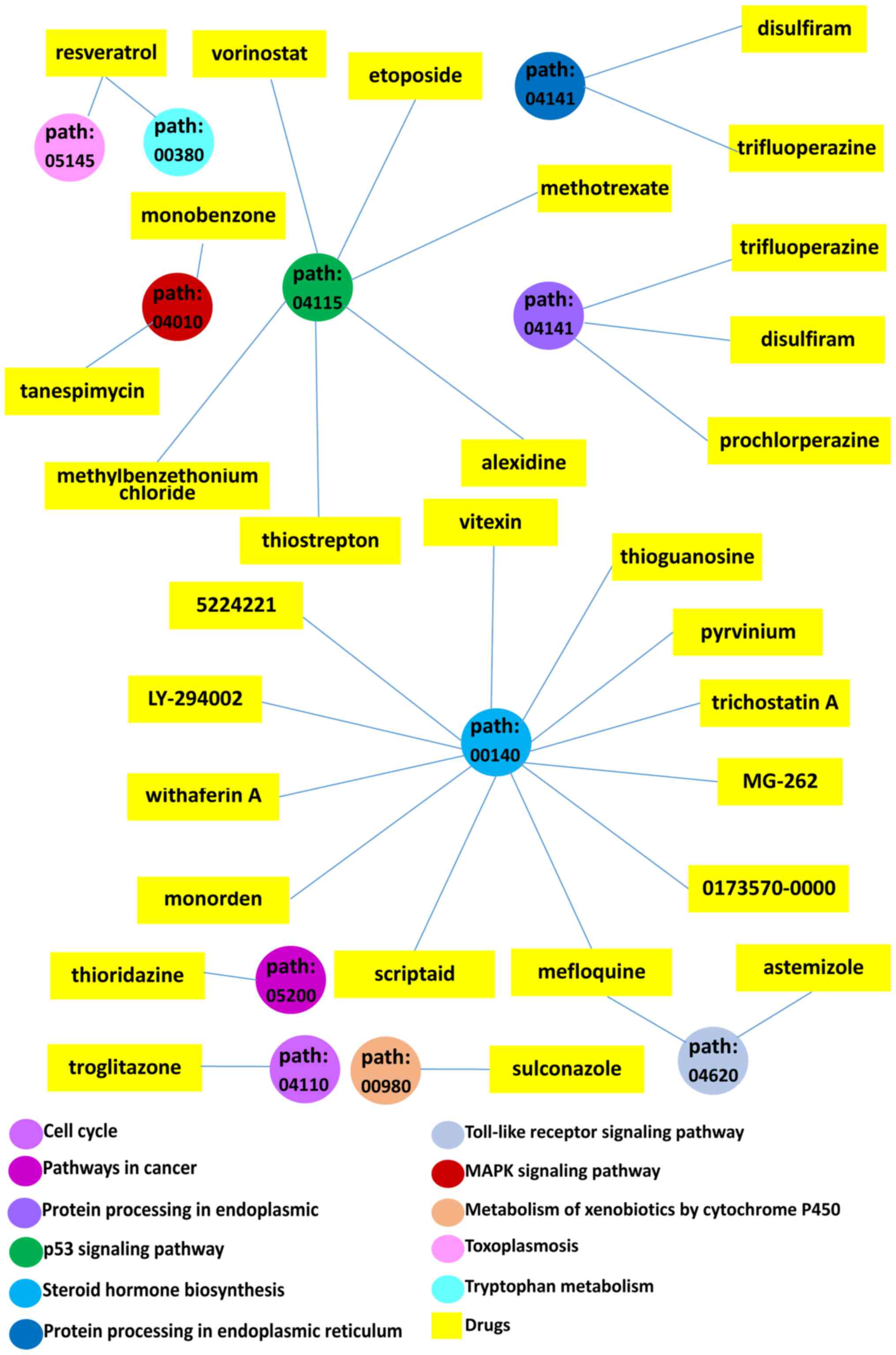
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Costa NR, Gil da Costa RM and Medeiros R:
A viral map of gastrointestinal cancers. Life Sci. 199:188–200.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coburn N, Cosby R, Klein L, Knight G,
Malthaner R, Mamazza J, Mercer CD and Ringash J: Staging and
surgical approaches in gastric cancer: A systematic review. Cancer
Treat Rev. 63:104–115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen W, Zhou H, Ye L and Zhan B:
Overexpression of SULT2B1b promotes angiogenesis in human gastric
cancer. Cell Physiol Biochem. 38:1040–1054. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng H, Hong H, Zhang L, Cai X, Hu M, Cai
Y, Zhou B, Lin J, Zhao C and Hu W: Nifuratel, a novel STAT3
inhibitor with potent activity against human gastric cancer cells.
Cancer Manag Res. 9:565–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pavlakis N, Sjoquist KM, Martin AJ,
Tsobanis E, Yip S, Kang YK, Bang YJ, Alcindor T, O'Callaghan CJ,
Burnell MJ, et al: Regorafenib for the treatment of advanced
gastric cancer (INTEGRATE): A multinational placebo-controlled
phase II trial. J Clin Oncol. 34:2728–2735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Belizário JE, Sangiuliano BA, Perez-Sosa
M, Neyra JM and Moreira DF: Using pharmacogenomic databases for
discovering patient-target genes and small molecule candidates to
cancer therapy. Front Pharmacol. 7:3122016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang L, Zeng JH, Wang JY, He RQ, Ma J,
Chen G, Cai XY and Hu XH: Down-regulation of miR-26a-5p in
hepatocellular carcinoma: A qRT-PCR and bioinformatics study.
Pathol Res Pract. 213:1494–1509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiong DD, Lv J, Wei KL, Feng ZB, Chen JT,
Liu KC, Chen G and Luo DZ: A nine-miRNA signature as a potential
diagnostic marker for breast carcinoma: An integrated study of
1,110 cases. Oncol Rep. 37:3297–3304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR,
Pan WY, He ZL, Lv J, Feng ZB and Chen G: Long non-coding RNA HOTTIP
promotes hepatocellular carcinoma tumorigenesis and development: A
comprehensive investigation based on bioinformatics, qRT-PCR and
meta-analysis of 393 cases. Int J Oncol. 51:1705–1721. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Dang YW, Wang X, Yang X, Zhang R,
Lv ZL and Chen G: Comprehensive analysis of long non-coding RNA
PVT1 gene interaction regulatory network in hepatocellular
carcinoma using gene microarray and bioinformatics. Am J Transl
Res. 9:3904–3917. 2017.PubMed/NCBI
|
|
15
|
He RQ, Li XJ, Liang L, Xie Y, Luo DZ, Ma
J, Peng ZG, Hu XH and Chen G: The suppressive role of miR-542-5p in
NSCLC: The evidence from clinical data and in vivo
validation using a chick chorioallantoic membrane model. BMC
Cancer. 17:6552017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao L, Li SH, Tian YX, Zhu QQ, Chen G,
Pang YY and Hu XH: Role of downregulated miR-133a-3p expression in
bladder cancer: A bioinformatics study. Onco Targets Ther.
10:3667–3683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dang YW, Lin P, Liu LM, He RQ, Zhang LJ,
Peng ZG, Li XJ and Chen G: In silico analysis of the potential
mechanism of telocinobufagin on breast cancer MCF-7 cells. Pathol
Res Pract. 214:631–643. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Li M, Wang Y and Liu X:
Integrating subpathway analysis to identify candidate agents for
hepatocellular carcinoma. Onco Targets Ther. 9:1221–1230. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Musa A, Ghoraie LS, Zhang SD, Glazko G,
Yli-Harja O, Dehmer M, Haibe-Kains B and Emmert-Streib F: A review
of connectivity map and computational approaches in
pharmacogenomics. Brief Bioinform. 19:506–523. 2018.PubMed/NCBI
|
|
20
|
Brum AM, van de Peppel J, Nguyen L, Aliev
A, Schreuders-Koedam M, Gajadien T, van der Leije CS, van Kerkwijk
A, Eijken M, van Leeuwen JPTM and van der Eerden BCJ: Using the
connectivity map to discover compounds influencing human osteoblast
differentiation. J Cell Physiol. 233:4895–4906. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Busby J, Murray L, Mills K, Zhang SD,
Liberante F and Cardwell CR: A combined connectivity mapping and
pharmacoepidemiology approach to identify existing medications with
breast cancer causing or preventing properties. Pharmacoepidemiol
Drug Saf. 27:78–86. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiao SJ, Zhu XC, Deng H, Zhou WP, Yang WY,
Yuan LK, Zhang JY, Tian S, Xu L, Zhang L and Xia HM: Gene
expression profiling coupled with connectivity map database mining
reveals potential therapeutic drugs for Hirschsprung disease. J
Pediatr Surg. 53:1716–1721. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Varadan P, Ganesh A, Konindala R,
Nagendrababu V, Ashok R and Deivanayagam K: Comparison of the
antibacterial efficacy of alexidine and chlorhexidine against
enterococcus faecalis: An in vitro study. Cureus.
9:e18052017.PubMed/NCBI
|
|
24
|
Feng R, Rios JA, Onishi T, Lokshin A,
Gorelik E and Lentzsch S: Cell-based and cytokine-directed chemical
screen to identify potential anti-multiple myeloma agents. Leuk
Res. 34:917–924. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Conesa C, Doss MX, Antzelevitch C,
Sachinidis A, Sancho J and Carrodeguas JA: Identification of
specific pluripotent stem cell death-inducing small molecules by
chemical screening. Stem Cell Rev. 8:116–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang H, Han Y, Yang X, Li M, Zhu R, Hu J,
Zhang X, Wei R, Li K and Gao R: HNRNPK inhibits gastric cancer cell
proliferation through p53/p21/CCND1 pathway. Oncotarget.
8:103364–103374. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu H, Wang C, Yang D, Wei Z, Xu J, Hu Z,
Zhang Y, Wang W, Yan R and Cai Q: Curcumin regulates proliferation,
autophagy, and apoptosis in gastric cancer cells by affecting PI3K
and P53 signaling. J Cell Physiol. 233:4634–4642. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun M, Si G, Sun HS and Si FC: Inhibition
of CREPT restrains gastric cancer growth by regulation of cycle
arrest, migration and apoptosis via ROS-regulated p53 pathway.
Biochem Biophys Res Commun. 496:1183–1190. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang C, Wang J and Bai P: Troglitazone
induces apoptosis in gastric cancer cells through the NAG-1
pathway. Mol Med Rep. 4:93–97. 2011.PubMed/NCBI
|
|
30
|
Li X, Qiu W, Liu B, Yao R, Liu S, Yao Y
and Liang J: Forkhead box transcription factor 1 expression in
gastric cancer: FOXM1 is a poor prognostic factor and mediates
resistance to docetaxel. J Transl Med. 11:2042013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yoo C, Ryu MH, Na YS, Ryoo BY, Lee CW,
Maeng J, Kim SY, Koo DH, Park I and Kang YK: Phase I and
pharmacodynamic study of vorinostat combined with capecitabine and
cisplatin as first-line chemotherapy in advanced gastric cancer.
Invest New Drugs. 32:271–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ebrahimifar M, Hasanzadegan Roudsari M,
Kazemi SM, Ebrahimi Shahmabadi H, Kanaani L, Alavi SA and Izadi
Vasfi M: Enhancing effects of curcumin on cytotoxicity of
paclitaxel, methotrexate and vincristine in gastric cancer cells.
Asian Pac J Cancer Prev. 18:65–68. 2017.PubMed/NCBI
|
|
33
|
Duo-Ji MM, Ci-Ren BS, Long ZW, Zhang XH
and Luo DL: Short-term efficacy of different chemotherapy regimens
in the treatment of advanced gastric cancer: A network
meta-analysis. Oncotarget. 8:37896–37911. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
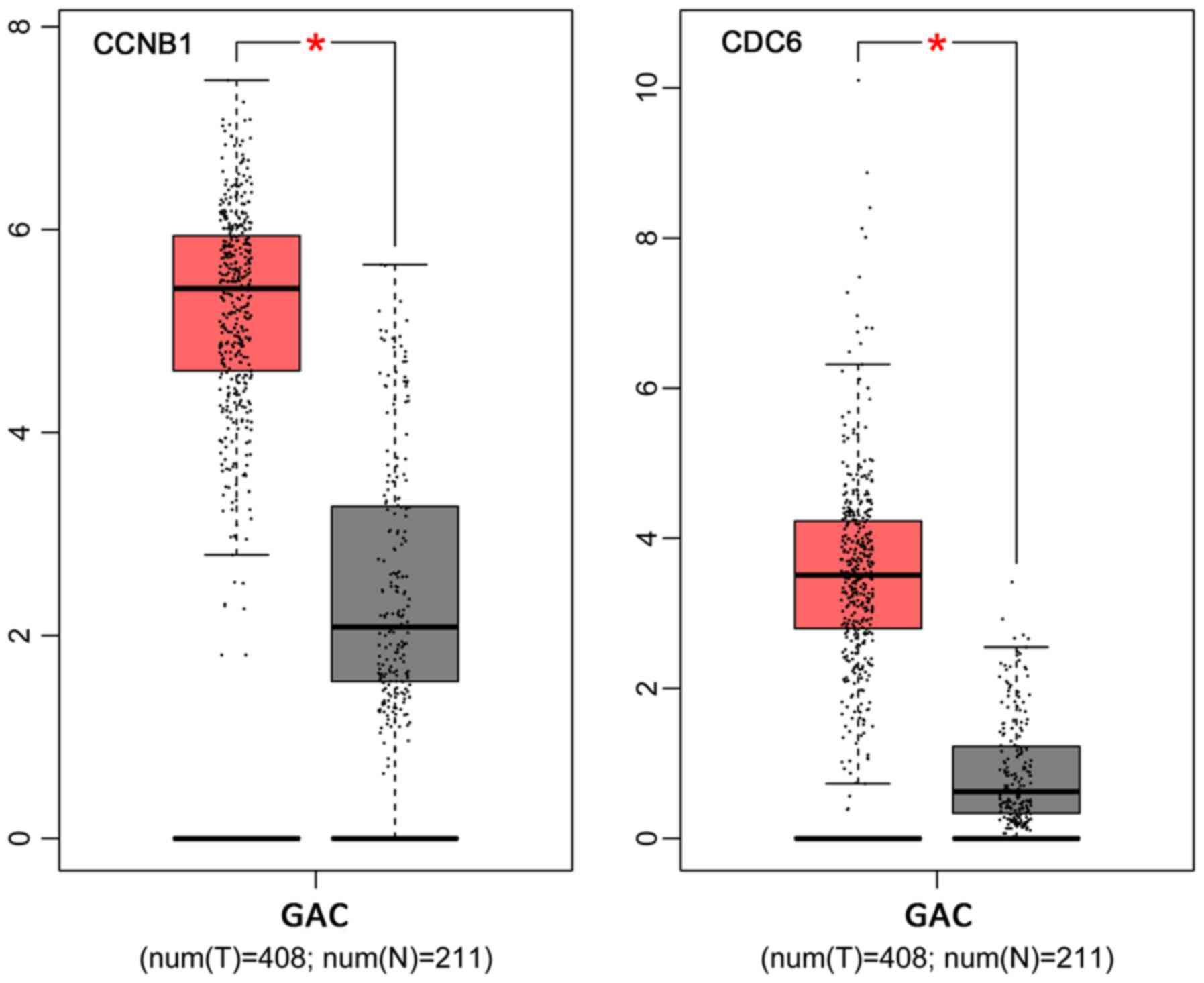
Shi Q, Wang W, Jia Z, Chen P, Ma K and
Zhou C: ISL1, a novel regulator of CCNB1, CCNB2 and c-MYC genes,
promotes gastric cancer cell proliferation and tumor growth.
Oncotarget. 7:36489–36500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Ji S, Fu LY, Jiang T, Wu D and Meng
FD: Knockdown of cyclin-dependent kinase inhibitor 3 inhibits
proliferation and invasion in human gastric cancer cells. Oncol
Res. 25:721–731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang QQ, Chen J, Zhou DL, Duan YF, Qi CL,
Li JC, He XD, Zhang M, Yang YX and Wang L: Dipalmitoylphosphatidic
acid inhibits tumor growth in triple-negative breast cancer. Int J
Biol Sci. 13:471–479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karanika S, Karantanos T, Li L, Wang J,
Park S, Yang G, Zuo X, Song JH, Maity SN, Manyam GC, et al:
Targeting DNA damage response in prostate cancer by inhibiting
androgen receptor-CDC6-ATR-Chk1 signaling. Cell Rep. 18:1970–1981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deng Y, Jiang L, Wang Y, Xi Q, Zhong J,
Liu J, Yang S, Liu R, Wang J, Huang M, et al: High expression of
CDC6 is associated with accelerated cell proliferation and poor
prognosis of epithelial ovarian cancer. Pathol Res Pract.
212:239–246. 2016. View Article : Google Scholar : PubMed/NCBI
|