Introduction
Breast cancer (BC) is the most common form of cancer
among women worldwide. In the United States, ~1 in 8 women will be
diagnosed with BC during their lifetime (1,2).
Despite advancements in antineoplastic strategies including
surgical treatment, adjuvant chemotherapy and radiotherapy,
prognosis remains poor (3–6). Furthermore, the use of various
prognostic markers including the cyclase associated actin
cytoskeleton regulatory protein 2, lactate dehydrogenase A,
AMP-activated protein kinase, Midline-2 and Claudin 12, have been
reported to improve BC patient outcomes (7–10).
However, in the United States, 66,120 new BC cases and 40,920 BC
mortalities are likely to occur in 2018, representing growth rates
of 30 and 14% relative to the previous year, respectively (11). The onset and progression of BC is a
multifactorial process, associated with genetic, endocrine and
external environmental factors (12). Hereditary phenomena appear in 5–10%
of BC patients, and germline gene mutations, particularly in breast
cancer type 1 susceptibility protein (BRCA)1 and BRCA2, closely
correlate with hereditary BC (13). Further investigation is essential
in understanding the complex molecular mechanisms underlying BC,
while concurrently identifying more potential target genes.
miRNAs are small, endogenous non-coding RNAs, 18–22
nucleotides in length (14,15).
By suppressing protein translation or enhancing the downregulation
of mRNA transcripts, miRNAs have a regulatory role in the
expression of target proteins (16,17).
Additionally, miRNAs have been reported to have effects on the
chemosensitivity, proliferation, migration, apoptosis, metastasis
and invasion of BC through targeting gene regulation (18–23).
The clinical features of miR-204-5p have been widely discussed in
the context of various cancers, such as hepatocellular carcinoma,
laryngeal squamous cell carcinoma, melanoma, oral squamous cell
carcinoma, colorectal cancer, papillary thyroid carcinoma and
endometrial carcinoma (24–30).
Nevertheless, the expression status and molecular mechanisms
underlying miR-204-5p in BC remains unclear. Therefore, in the
present study, the expression of the precursor miR-204 and mature
miR-204-5p in BC was investigated, using data downloaded from the
Cancer Genome Atlas (TCGA), University of California Santa Cruz
(UCSC) Xena and Gene Expression Omnibus (GEO) databases. In
addition to the data obtained from the TCGA and UCSC Xena
databases, a meta-analysis involving GEO microarrays was undertaken
to evaluate the expression of miR-204 and miR-204-5p in BC.
Furthermore, the putative genes selected from the intersection of
the predicted genes, differentially expressed genes (DEGs) from the
TCGA database, and DEGs from the GEO database were used to
determine Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) enrichments; in order to examine the molecular
mechanisms underlying miR-204-5p in BC, and to construct a
protein-protein interaction (PPI) network to draw interaction maps
of the identified DEGs.
Materials and methods
Breast cancer samples in the TCGA and
UCSC Xena databases
Precursor miR-204 expression data, as well as data
on several corresponding clinical parameters, including age,
gender, vital status, pathologic stage, tumor status, node status,
metastasis status, estrogen receptor (ER) status, the human
epidermal growth factor receptor (HER2) status and the progesterone
receptor (PR) status were obtained from the TCGA database
(http://cancergenome.nih.gov/) (31). Additionally, data on the expression
of mature miR-204-5p were acquired from the UCSC Xena database
(http://xena.ucsc.edu/).
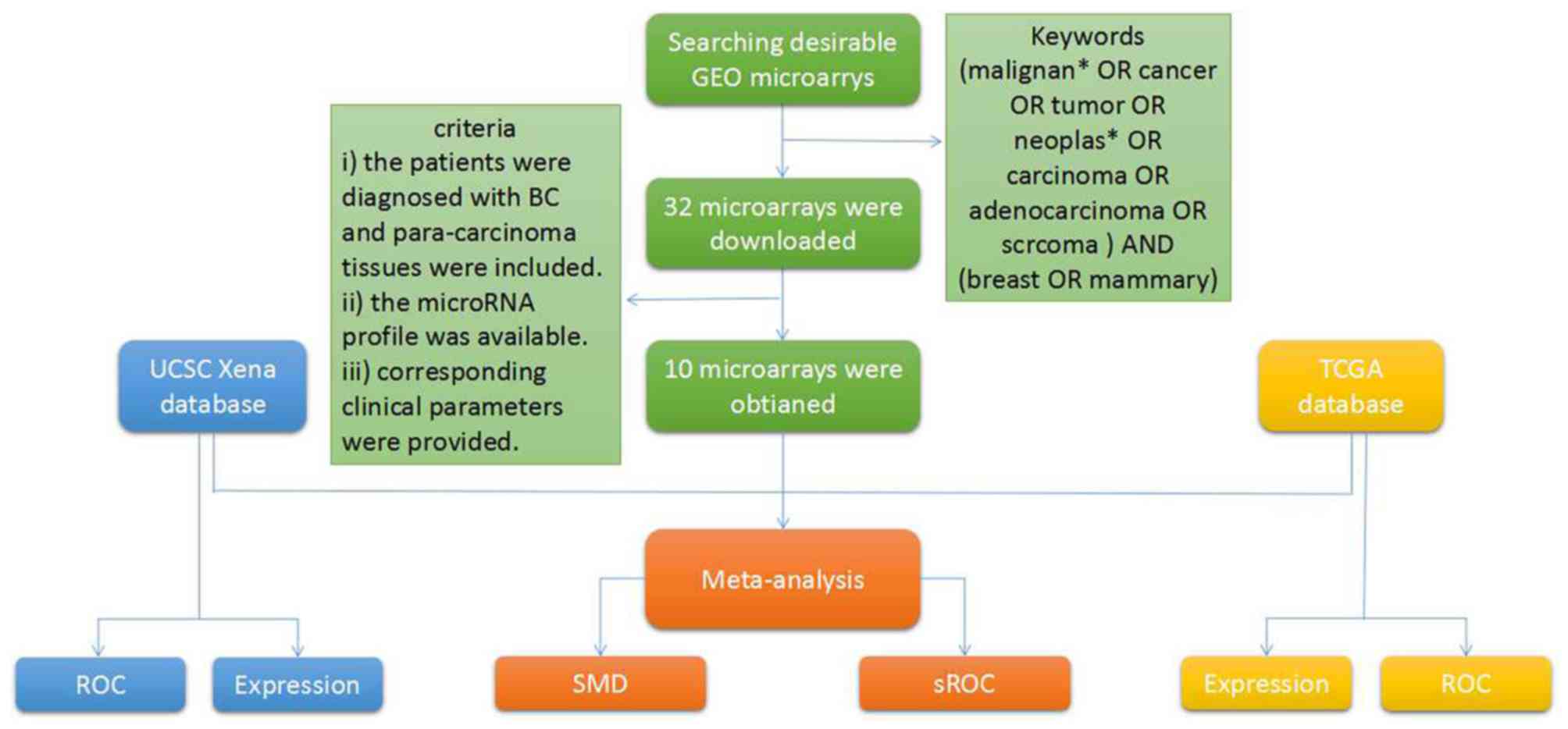
BC microarrays in the GEO
database
BC microarrays were drawn from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/)
(32), using the following search
terms: (neoplasm* OR cancer OR adenocarcinoma OR malignant* OR
carcinoma OR tumor OR tumor) and (breast OR mammary). Next, BC
microarrays were selected for further meta-analysis, based on the
following inclusion criteria: i) Patients were diagnosed with BC
tissue samples and para-carcinoma tissue samples; ii) the microRNA
profile was available; and iii) corresponding clinical parameters
were provided.
Predicted target genes and
differentially expressed genes of miR-204-5p in breast cancer
The target genes of miR-204-5p were acquired from
miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
(33), which contains 12 tools:
MicroT4, TargetScan, miRanda, miRNAMap, PICTAR2, RNA22, miRBridge,
miRDB, PITA, miRMap, RNAhybrid, and miRWalk. To ensure accuracy,
the potential predicted target genes of miR-204-5p were extracted,
if they emerged more than five times among those twelve tools. Gene
Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/index.html) (34) was used to download DEGs from the
TCGA database, using the following criteria: log2 |fold change
(FC)|>1 and P<0.05. In addition, the Gene-Cloud of
Biotechnology Information (35)
was used to analyze BC microarrays obtained from the GEO database,
using the following search logic: (Affymetrix) and (neoplasm* OR
cancer OR adenocarcinoma OR malignant* OR carcinoma OR tumor) and
(breast OR mammary). DEGs were selected from the GEO database,
again under the following criteria: log2 |FC|> 1 and
P<0.05.
Enrichment analyses and the
protein-protein interaction network
An intersection of potential target genes, DEGs from
the TCGA database and DEGs from the GEO database were plotted to
obtain putative genes. Subsequently, GO (http://www.geneontology.org/) (36,37)
and KEGG (https://www.genome.jp/kegg/)
(38,39) analyses were undertaken to identify
the potential biological processes and possible pathways of the
selected putative genes in BC. A functional network graph of GO was
drawn by the ORA Sample Run WEB-based Gene Set Analysis Toolkit
(http://www.webgestalt.org/option.php#). Moreover, the
PPI network was generated by Cytoscape 3.5.0, to build interaction
maps of the putative genes (40,41).
Additionally, a Spearman's correlation analysis was created to
identify the correlation between miR-204-5p and the hub gene.
Statistical analysis
SPSS 22.0 (IBM Corp., Armonk, NY, USA) statistical
software package was used to perform an independent-sample t-test
to estimate the expression of the precursor miR-204 and mature
miR-204-5p in BC tissue samples and para-carcinoma tissue samples,
and to determine the expression of the precursor miR-204 in groups
differentiated in terms of the aforementioned clinical parameters.
The area under the curve (AUC) of the receiver operating
characteristic (ROC) curve was evaluated to assess the
discriminatory capability of the precursor miR-204 for BC, in which
an AUC >0.7 was considered to denote a great discriminatory
capability. In addition, the Kaplan-Meier curve was undertaken to
evaluate the prognostic value of the precursor miR-204 in BC, and a
log-rank test was performed to compare the high and low miR-204
expression groups. STATA 12.0 (StataCorp LLC, College Station, TX,
USA) was used to undertake the meta-analysis. The standard mean
difference (SMD) was adopted to determine the expression of
miR-204-5p in BC and para-carcinoma tissue samples. Concurrently,
the heterogeneity of the BC microarrays, and the data obtained from
the TCGA and UCSC Xena databases, were estimated via a
heterogeneity test, with I2<50% signifying no
heterogeneity. A random-effects model may be conducted if the
heterogeneity existed. The sensitivity analysis was performed to
seek the source of the heterogeneity. Additionally, publication
bias was calculated using Deek's funnel plot dissymmetry tests,
with P<0.05 indicating an obvious publication bias.
Subsequently, summary (s)ROCs were determined, to calculate the
discriminatory capability of miR-204-5p in BC.
Results
Expression level of the precursor
miR-204 and mature miR-204-5p in breast cancer
A downregulation of miR-204 was detected in 1,077 BC
tissue samples in comparison to 104 para-carcinoma tissue samples
based on the TCGA database (2.284±1.983 vs. 5.775±1.391,
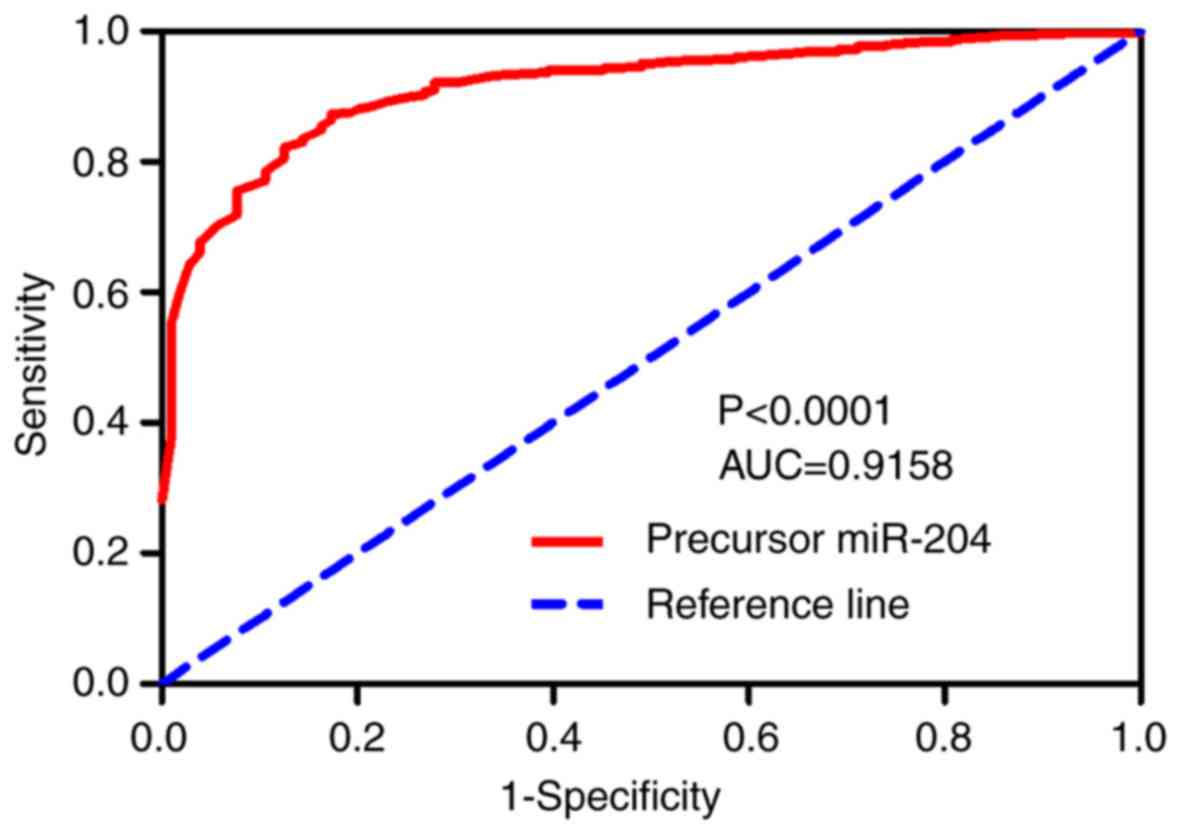
P<0.001, Table I). The AUC of
the miR-204 ROC curve was 0.9158, with a sensitivity of 87.28% and
specificity of 82.69%, thus indicating that precursor miR-204
possesses a great discriminatory capability for BC (P<0.0001;
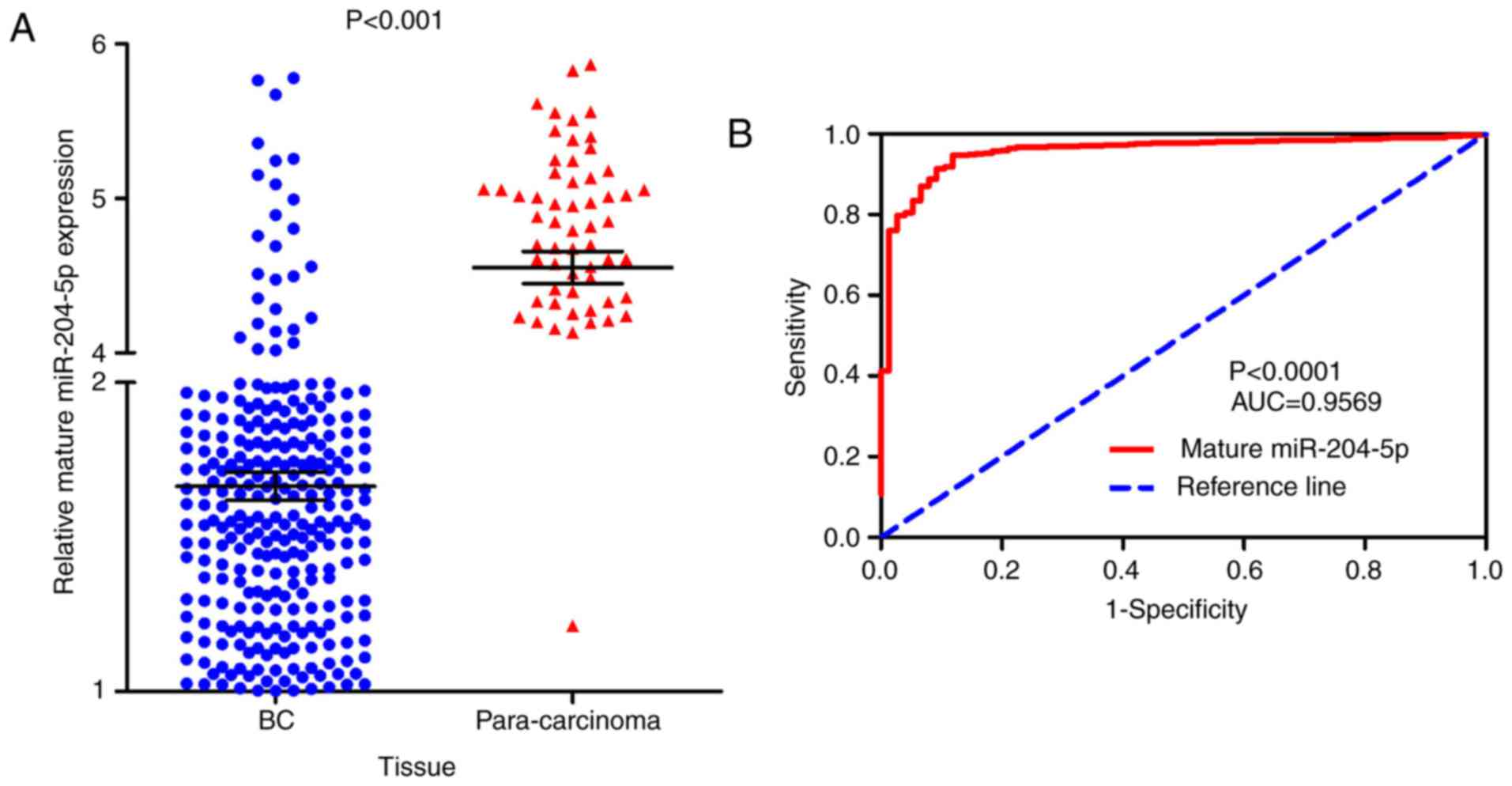
Fig. 1). In addition, the
expression of mature miR-204-5p was significantly decreased in 756
BC tissue samples compared with 76 para-carcinoma tissue samples
based on the UCSC Xena database (1.66422±1.251283 vs.
4.55208±0.905784; P<0.001; Fig.
2A). Additionally, mature miR-204-5p also featured a great
discriminatory capability for BC (P<0.0001; Fig. 2B). Additionally, downregulation of
the miR-204 was determined to be significant in several groups,
including individuals aged ≥60 years, those who were dead, negative
PR status, positive HER2 status and pathological stage IV (all
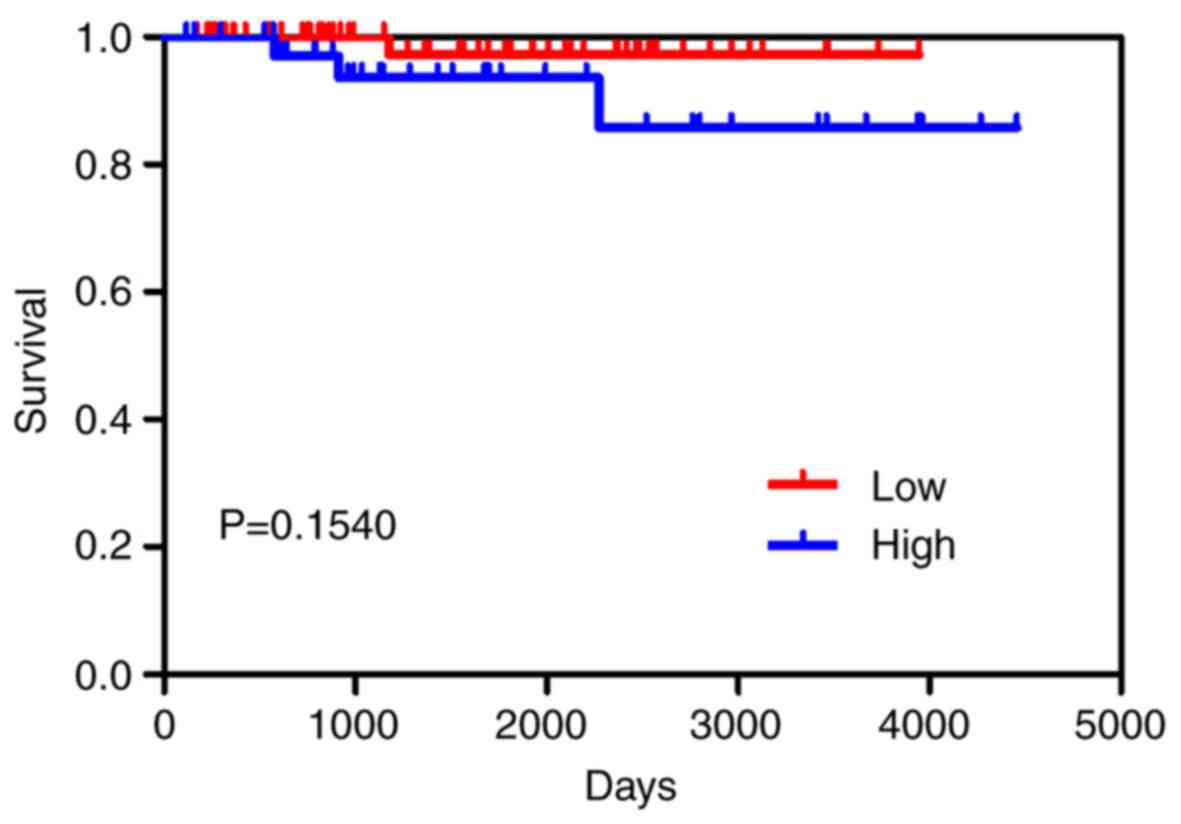
P<0.05, Table I). No
significant correlation was identified between the precursor
miR-204 and survival outcome in BC, as determined by the
Kaplan-Meier curve (P=0.154; Fig.
3).
 | Table I.Clinical parameters and the
expression level of precursor miR-204. |
Table I.
Clinical parameters and the
expression level of precursor miR-204.
|
|
| Expression level of
precursor microRNA-204 |
|---|
|
|
|
|
|---|
| Clinical
feature | n | Mean ± standard
deviation | t | P-value |
|---|
| Tissue |
| BC | 1,077 | 2.284±1.983 | −17.535 |
<0.001a |
|
Para-carcinoma | 104 | 5.775±1.391 |
|
|
| Age (years) |
|
<60 | 571 | 2.492±2.016 | 1.741 |
<0.001 |
|
≥60 | 506 | 2.049±1.921 |
|
|
| Sex |
|
Female | 1,065 | 2.297±1.987 | 3.148 | 0.043a |
|
Male | 12 | 1.129±1.267 |
|
|
| Vital status |
|
Alive | 975 | 2.325±2.005 | 2.100 | 0.036a |
|
Dead | 102 | 1.892±1.718 |
|
|
| Pathological
stage |
| Stages
I–II | 790 | 2.256±1.962 | −1.417 | 0.157 |
| Stages
III–IV | 264 | 2.456±2.061 |
|
|
| Tumor |
|
T1-2 | 899 | 2.236±1.965 | −1.772 | 0.077 |
|
T3-4 | 175 | 2.527±2.063 |
|
|
| Pathological
stage |
| Stage
I | 181 | 2.643±1.959 | F=4.179 | 0.006a |
| Stage
II | 609 | 2.140±1.950 |
|
|
| Stage
III | 244 | 2.499±2.048 |
|
|
| Stage
IV | 20 | 1.935±2.205 |
|
|
| Tumor |
| T1 | 279 | 2.643±1.906 | F=2.250 | 0.081 |
| T2 | 620 | 2.054±1.966 |
|
|
| T3 | 135 | 2.781±2.090 |
|
|
| T4 | 40 | 1.668±1.733 |
|
|
| Node |
| No | 508 | 2.207±1.937 | −1.395 | 0.163 |
|
Yes | 549 | 2.377±2.019 |
|
|
| Metastasis |
| No | 893 | 2.151±1.916 | 0.726 | 0.468 |
|
Yes | 21 | 1.843±1.843 |
|
|
| ER status |
|
Positive | 795 | 2.316±1.980 | 0.592 | 0.554 |
|
Negative | 232 | 2.228±2.001 |
|
|
| PR status |
|
Positive | 689 | 2.392±2.004 | 2.276 | 0.023a |
| HER2 status |
|
Negative | 335 | 2.092±1.929 | 4.831 |
<0.001a |
|
Positive | 159 | 1.565±0.137 |
|
|
|
Negative | 555 | 2.407±0.084 |
|
|
Meta-analysis
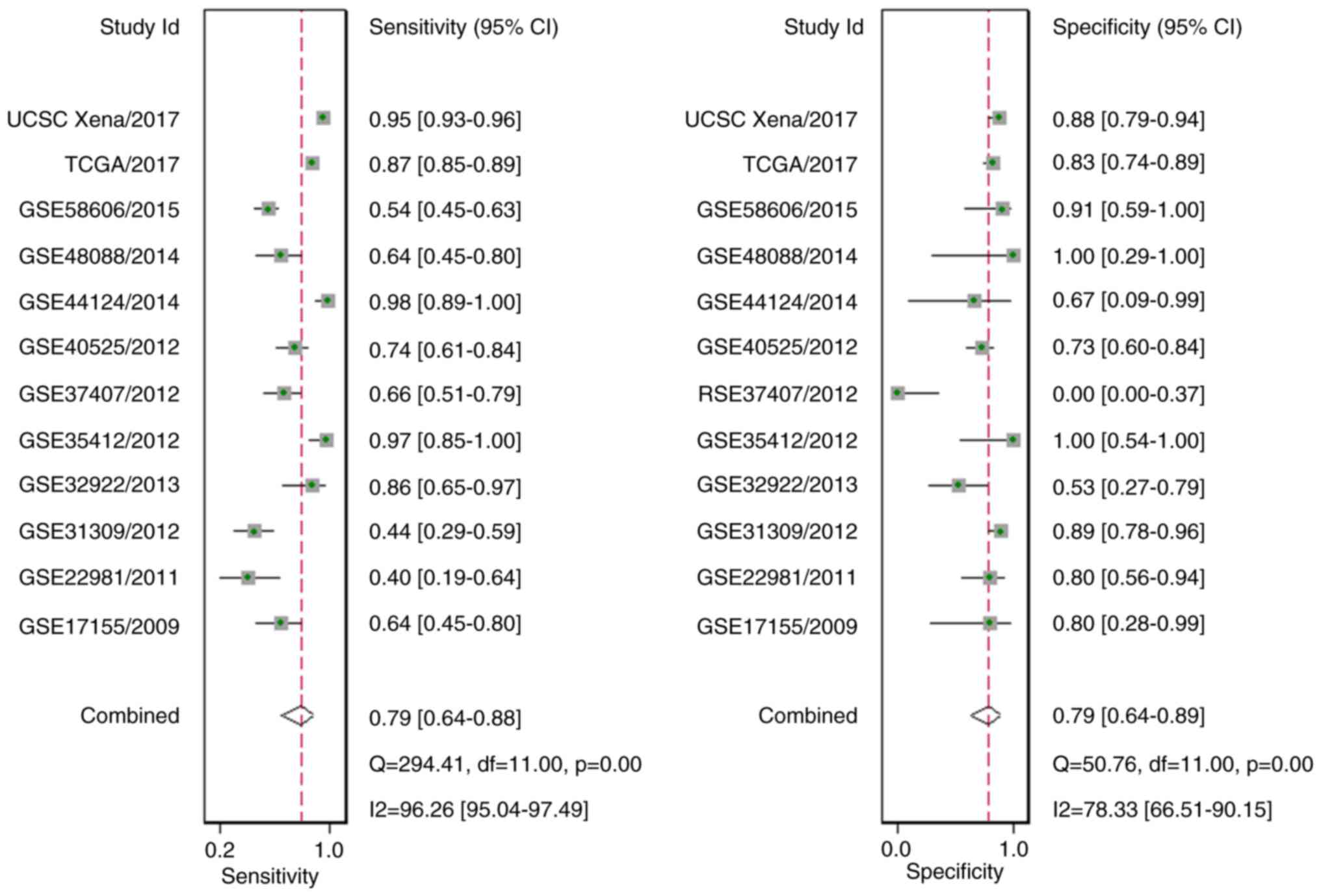
A total of 10 GEO microarrays containing 473 BC
tissue samples and 187 para-carcinoma tissue samples were acquired
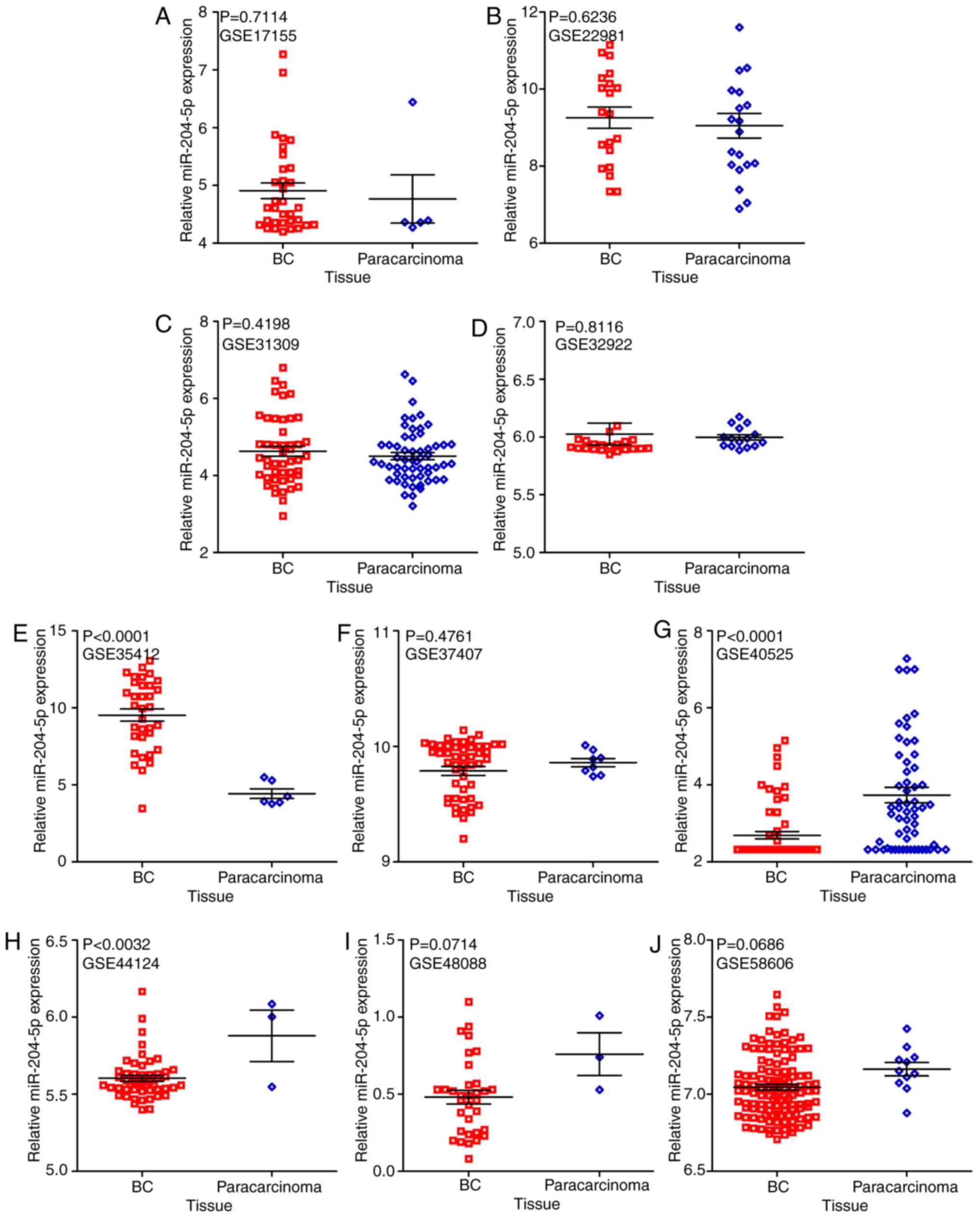
(Fig. 4; Table II) (42–51).
It was revealed that in two microarrays (GSE40525 and GSE44124),
the expression of miR-204-5p was markedly reduced in BC tissue
samples, in comparison to para-carcinoma tissue samples (both
P<0.05; Fig. 5). Additionally,
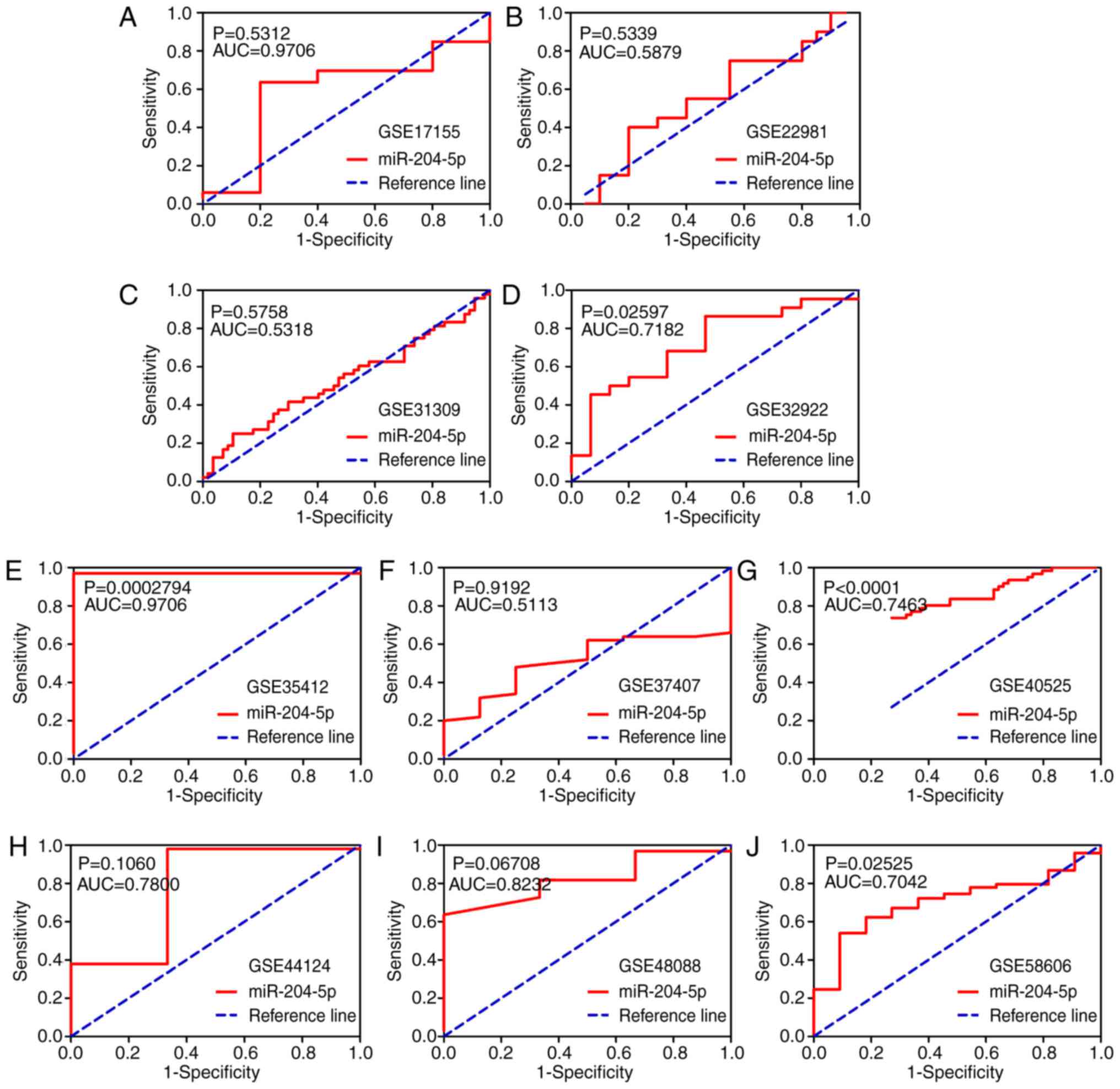
the ROC curve result implied that in four microarrays (GSE32922,
GSE35412, GSE40525 and GSE58606), miR-204-5p possessed a great
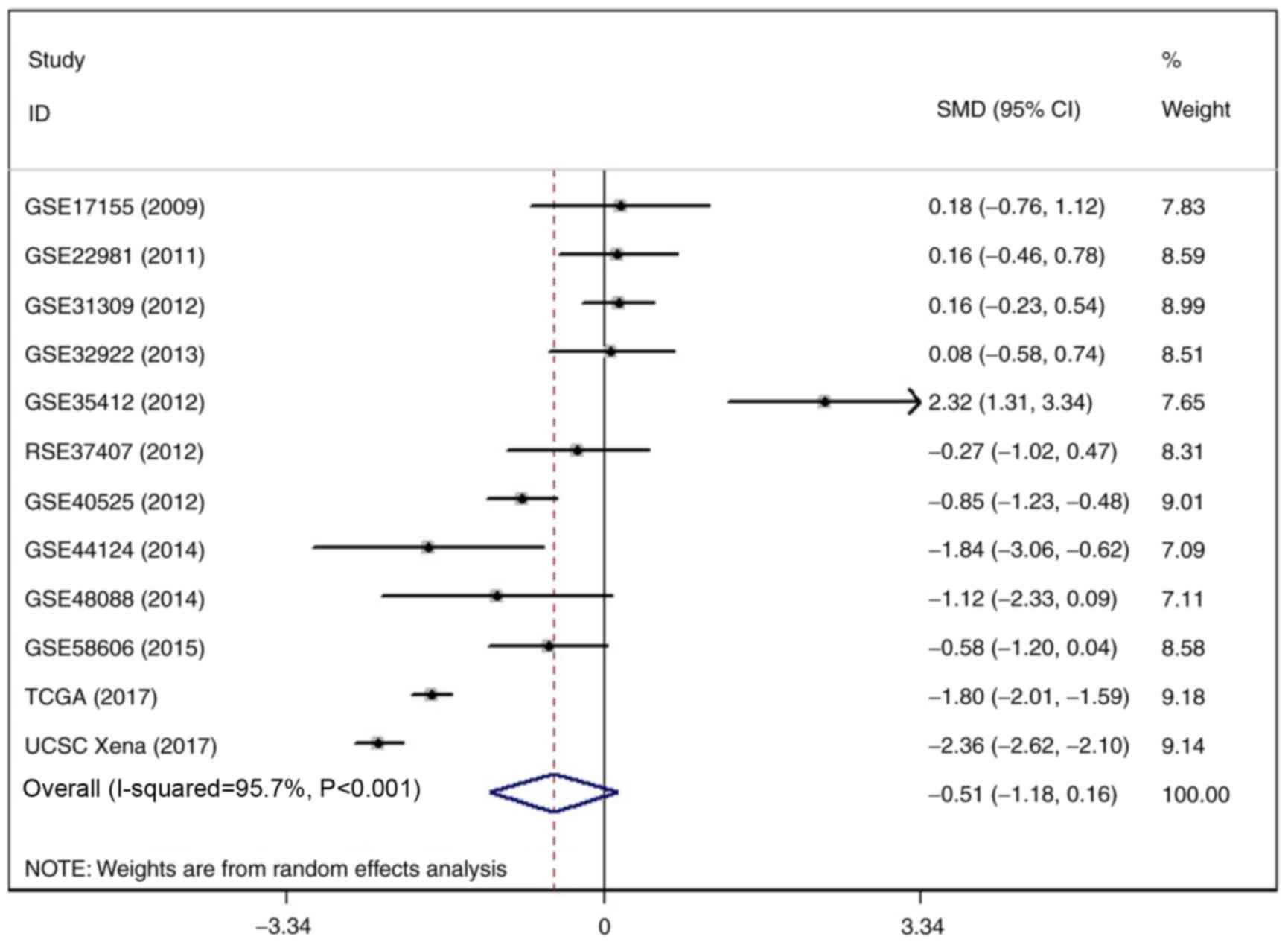
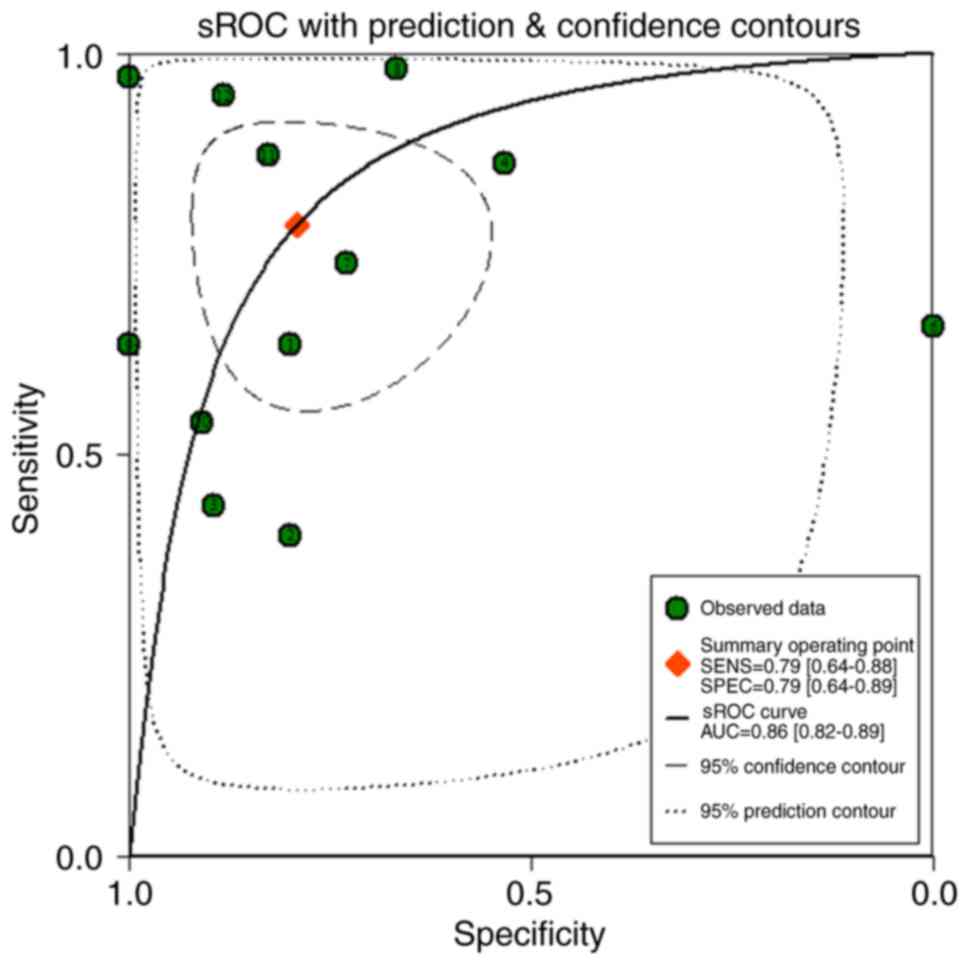
discriminatory capability for BC (all P<0.05; Fig. 6). Regarding the meta-analysis, a
significant heterogeneity outcome was achieved by the heterogeneity
test. Thus, a random-effects model was undertaken to calculate the
SMD and 95% confidence interval, the SMD outcome demonstrated that
the expression of miR-204-5p was reduced in BC tissue samples in
comparison to para-carcinoma tissue samples (I2=95.7%;
P<0.001; Fig. 7). The influence
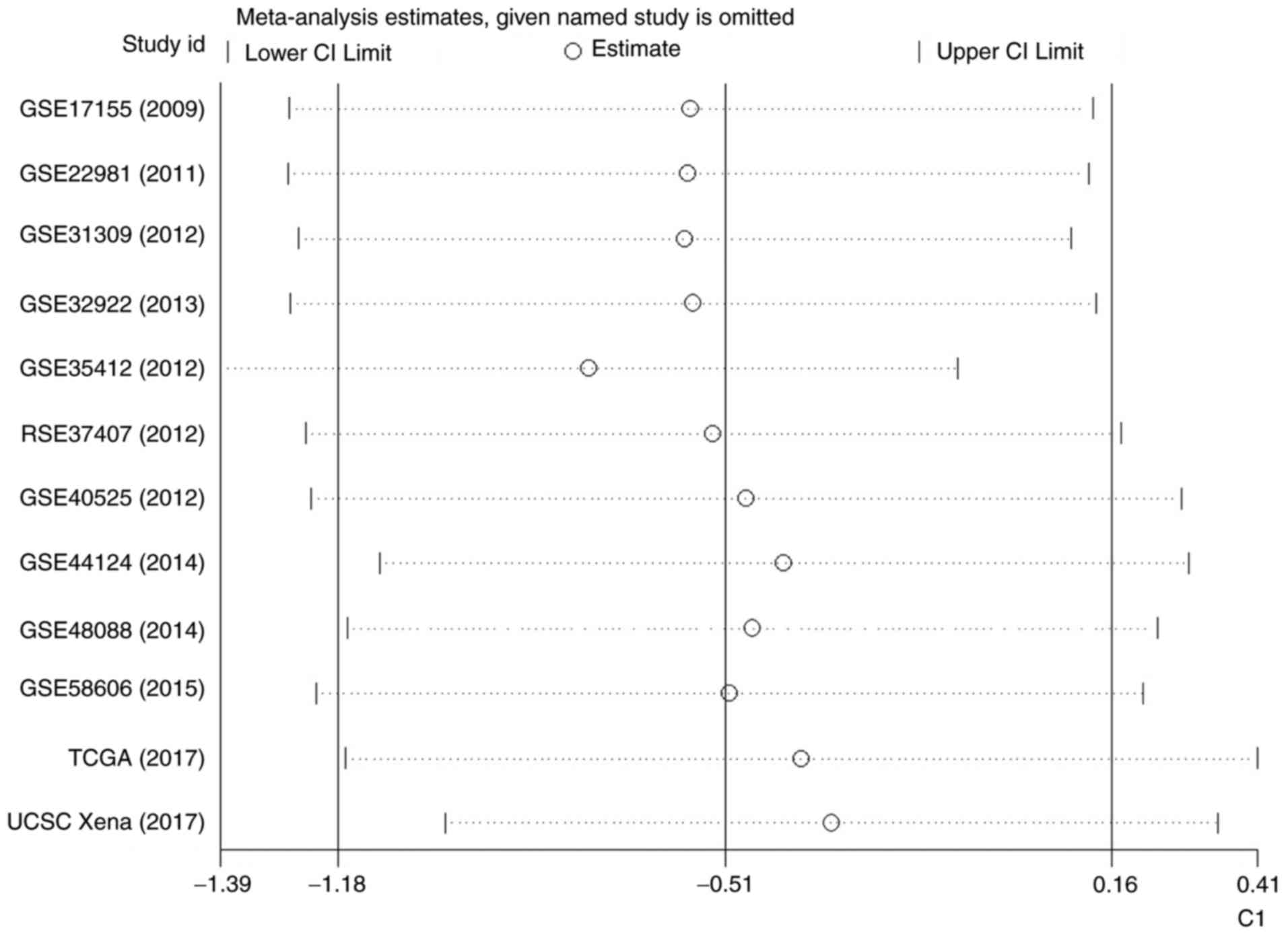
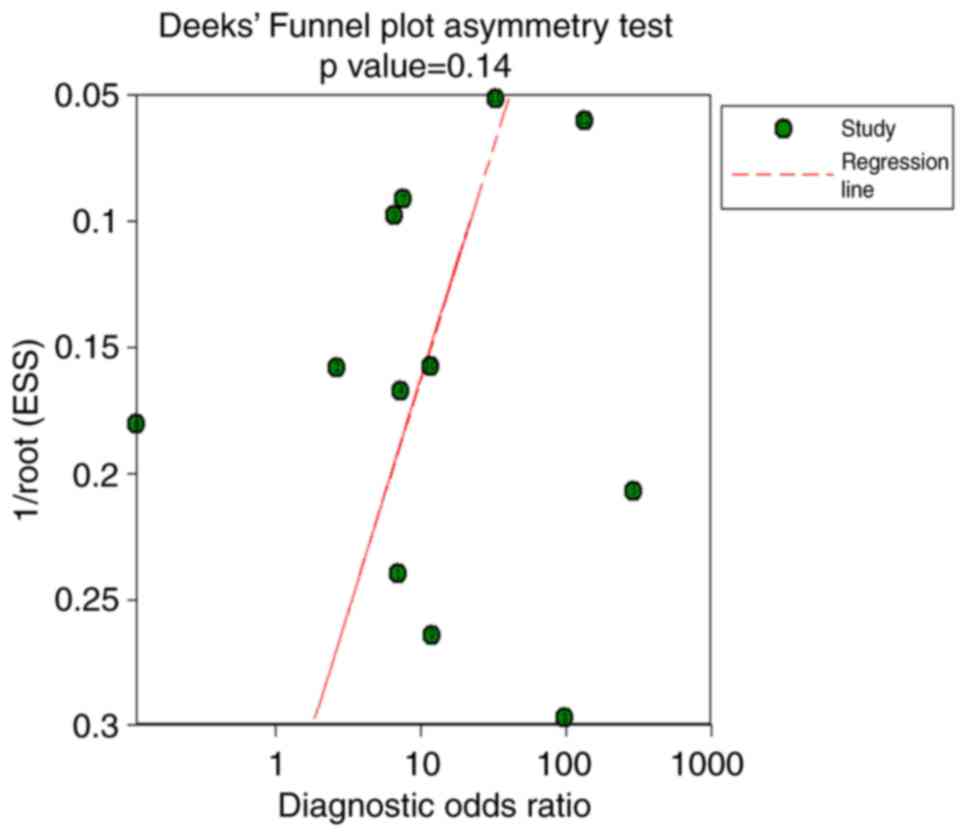
analysis demonstrated no significant difference (Fig. 8). Additionally, no significant
publication bias was detected via Deek's funnel plot asymmetry test
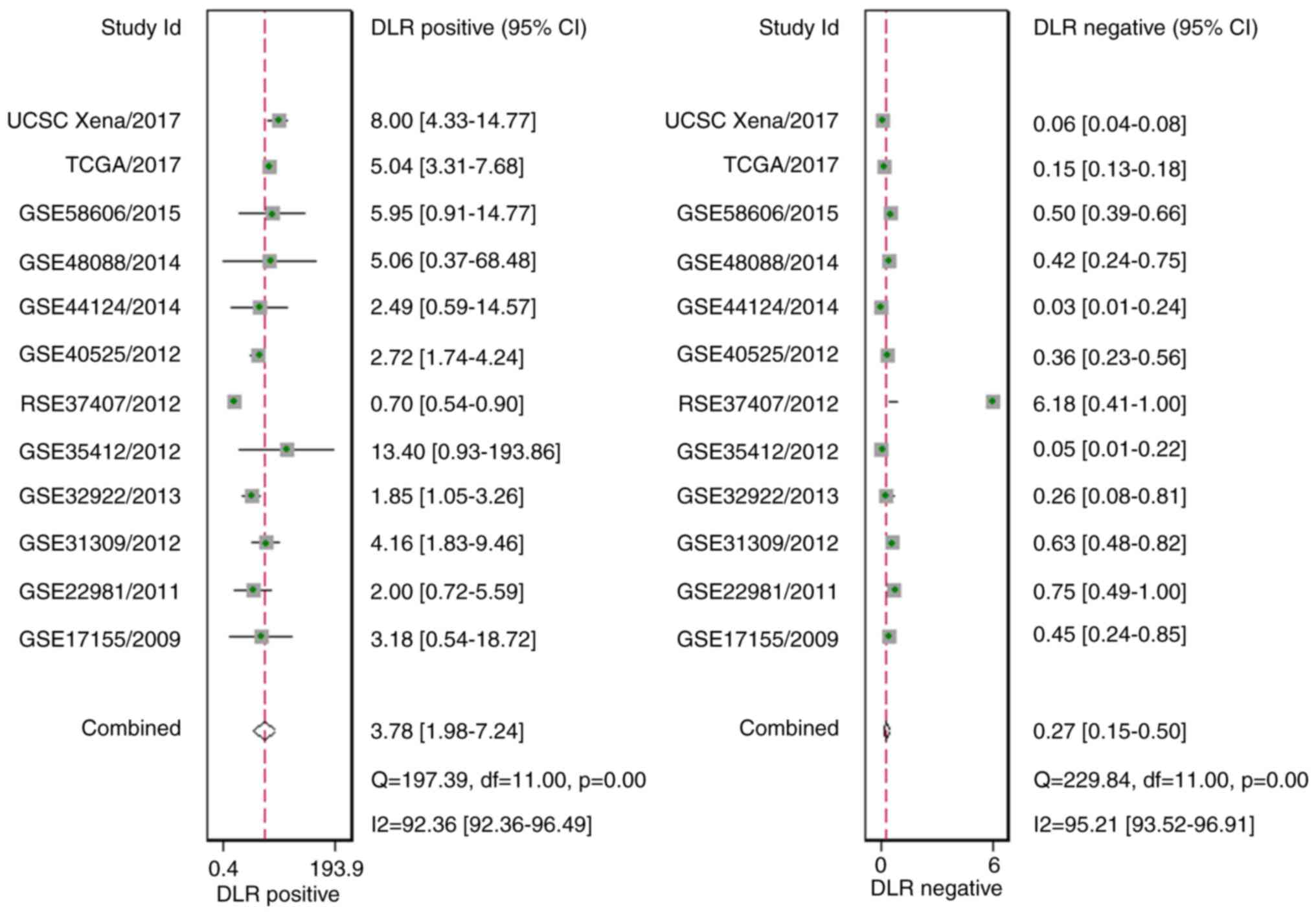
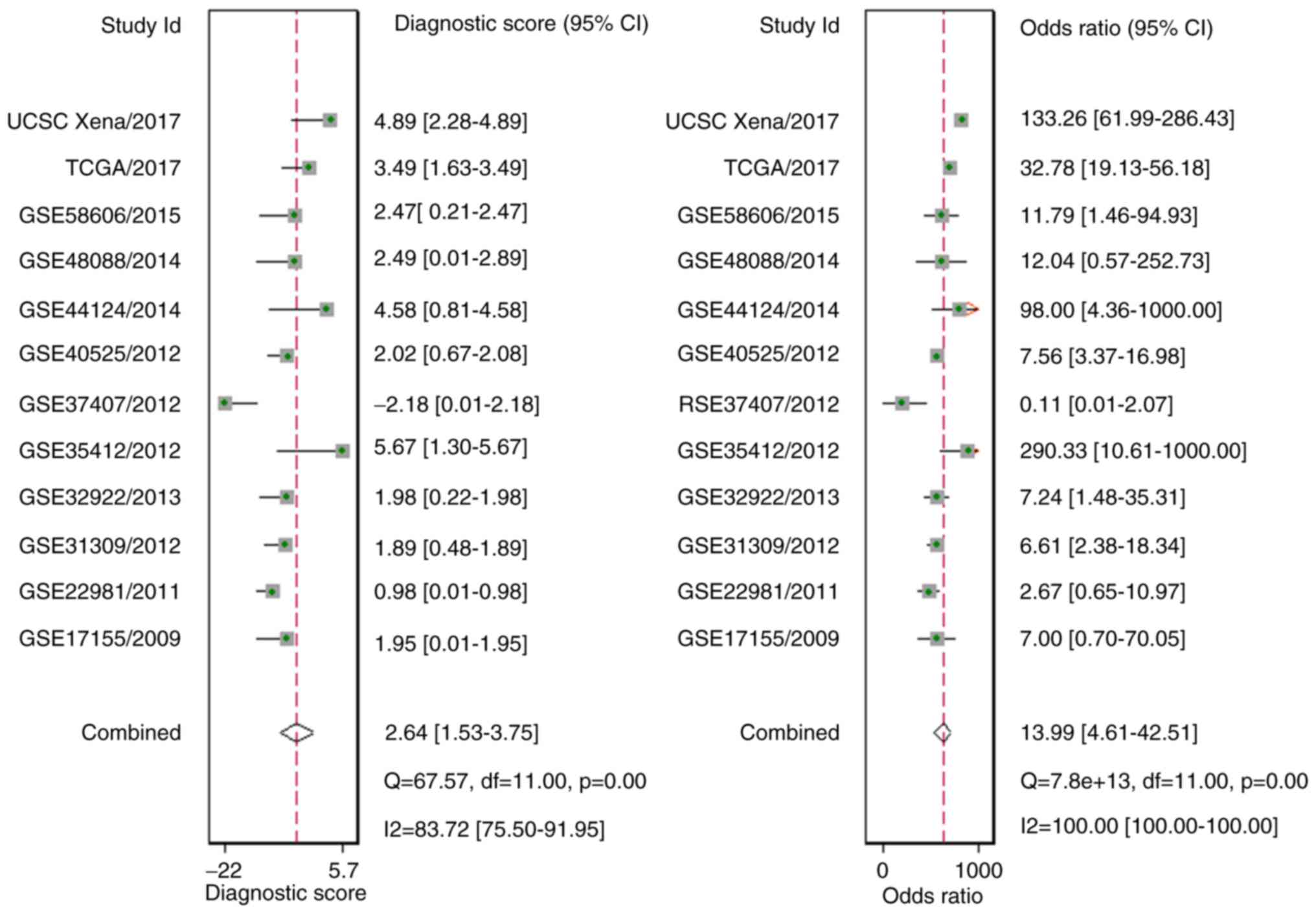
(P=0.14; Fig. 9). In addition, the
diagnostic likelihood ratio (DLR) positive, DLR negative,
diagnostic score, and odds ratio values were 3.78 (1.98–7.24), 0.27
(0.15–0.50), 2.64 (1.53–3.75) and 13.99 (4.61–42.51), respectively
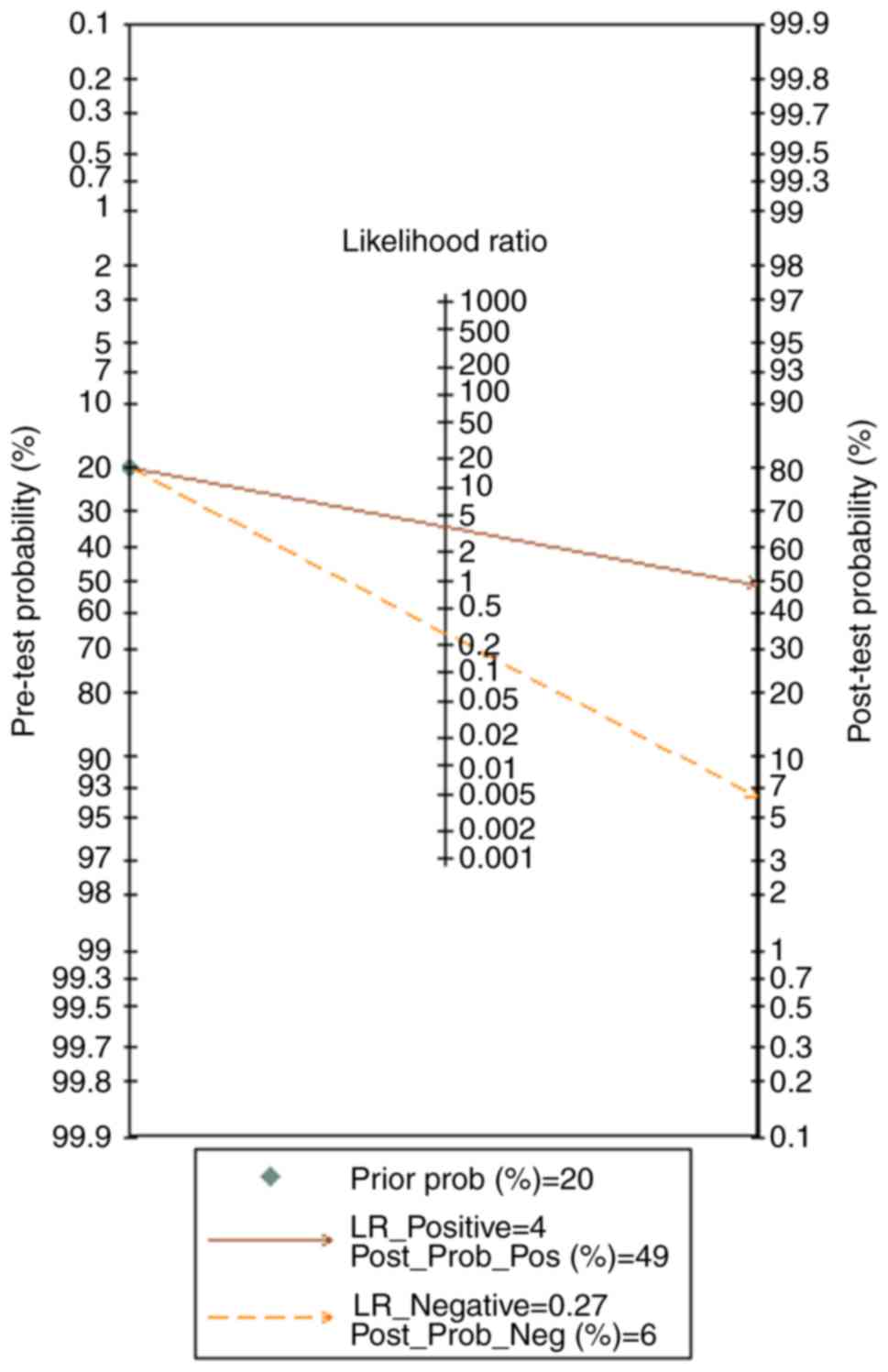
(Figs. 10 and 11). In addition, the prior probability
and post-probability positive and negative reached 20, 49 and 6%,
respectively (Fig. 12). Finally,
the AUC of the sROC was 0.86 (0.82–0.89), with a sensitivity of 79%
(64–88%) and a specificity of 79% (64–89%), thus indicating a great
discriminatory capability of miR-204-5p for BC (Figs. 13 and 14).
 | Table II.Introduction of the GEO
microarrays. |
Table II.
Introduction of the GEO
microarrays.
|
|
|
|
|
| BC | Para-carcinoma |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Author, year | Dataset | Country | Year | Platform | n | Mean | SD | n | Mean | SD | Area under the
curve | (Refs.) |
|---|
| Fassan et
al, 2009 | GSE17155 | Italy | 2009 | GPL8871 | 33 | 4.91 | 0.77 | 5 | 4.77 | 0.94 | 0.9706 | (42) |
| Zhao et al,
2010 | GSE22981 | USA | 2011 | GPL8179 | 20 | 9.26 | 1.23 | 20 | 9.05 | 1.43 | 0.5879 | (43) |
| Schrauder et
al, 2012 | GSE31309 | Germany | 2012 | GPL14132 | 48 | 4.63 | 0.90 | 57 | 4.50 | 0.70 | 0.5318 | (44) |
| Tanic et al,
2013 | GSE32922 | Spain | 2013 | GPL7723 | 22 | 6.03 | 0.44 | 15 | 6.00 | 0.09 | 0.7182 | (45) |
| Romero-Cordoba
et al, 2012 | GSE35412 | Mexico | 2012 | GPL9731 | 34 | 9.53 | 2.33 | 6 | 4.44 | 0.76 | 0.9706 | (46) |
| Gravgaard et
al, 2012 | GSE37407 | Sweden | 2012 | GPL13703 | 50 | 9.79 | 0.28 | 8 | 9.86 | 0.10 | 0.5113 | (47) |
| Biagioni et
al, 2012 | GSE40525 | Israel | 2012 | GPL8227 | 61 | 2.69 | 0.75 | 59 | 3.74 | 1.57 | 0.7463 | (48) |
| Feliciano et
al, 2013 | GSE44124 | Spain | 2014 | GPL14767 | 50 | 5.60 | 0.14 | 3 | 5.88 | 0.29 | 0.7800 | (49) |
| Peña-Chilet et
al, 2014 | GSE48088 | Spain | 2014 | GPL14613 | 33 | 0.48 | 0.25 | 3 | 0.76 | 0.24 | 0.8232 | (50) |
| Matamala et
al, 2015 | GSE58606 | Spain | 2015 | GPL18838 | 122 | 7.05 | 0.21 | 11 | 7.16 | 0.14 | 0.7042 | (51) |
Putative genes of miR-204-5p in breast
cancer
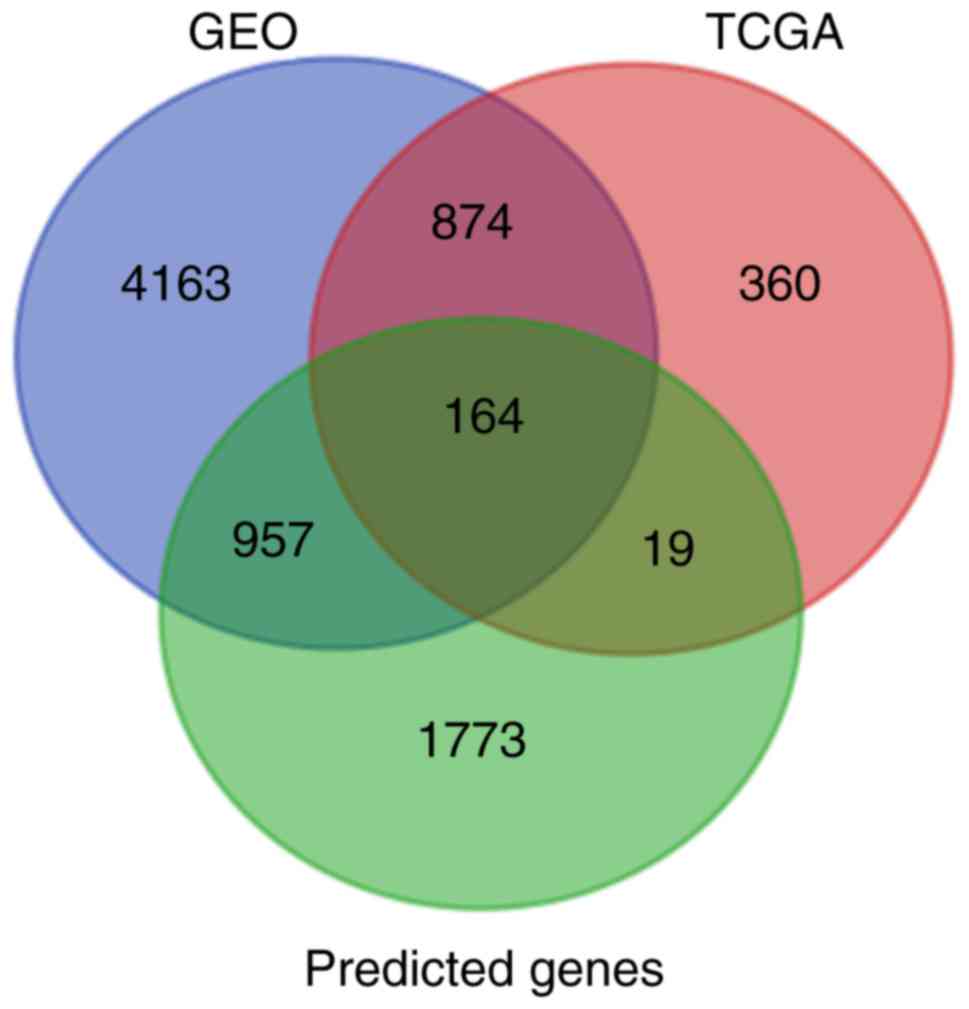
In total, 1,417 and 6,158 DEGs were acquired from
the TCGA and GEO databases, respectively. Additionally, 2,913
predicted target genes were obtained. The intersection was plotted
and 164 putative genes were obtained for use in further
bioinformatics analyses (Fig.
15).
Bioinformatics analyses
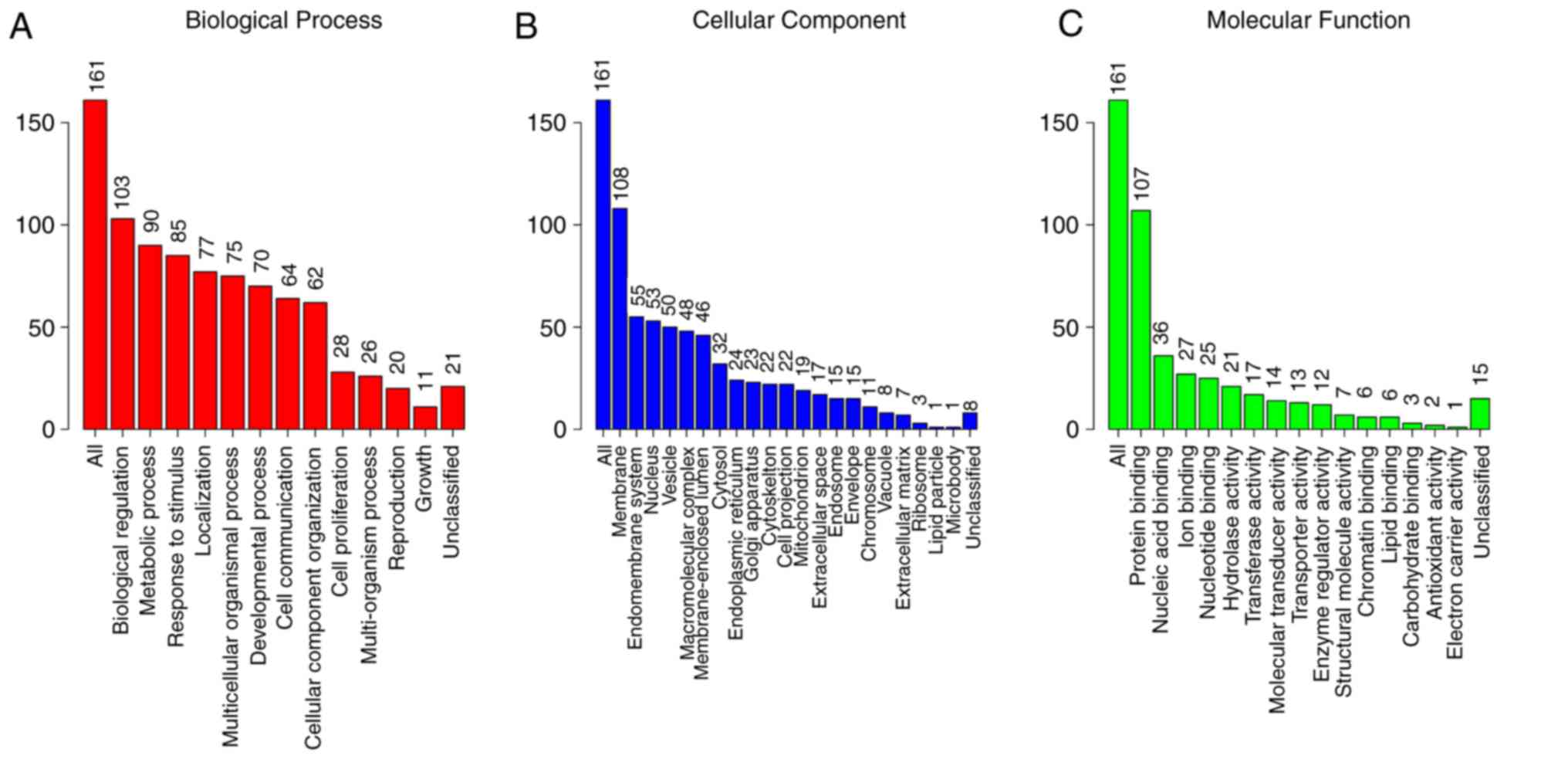
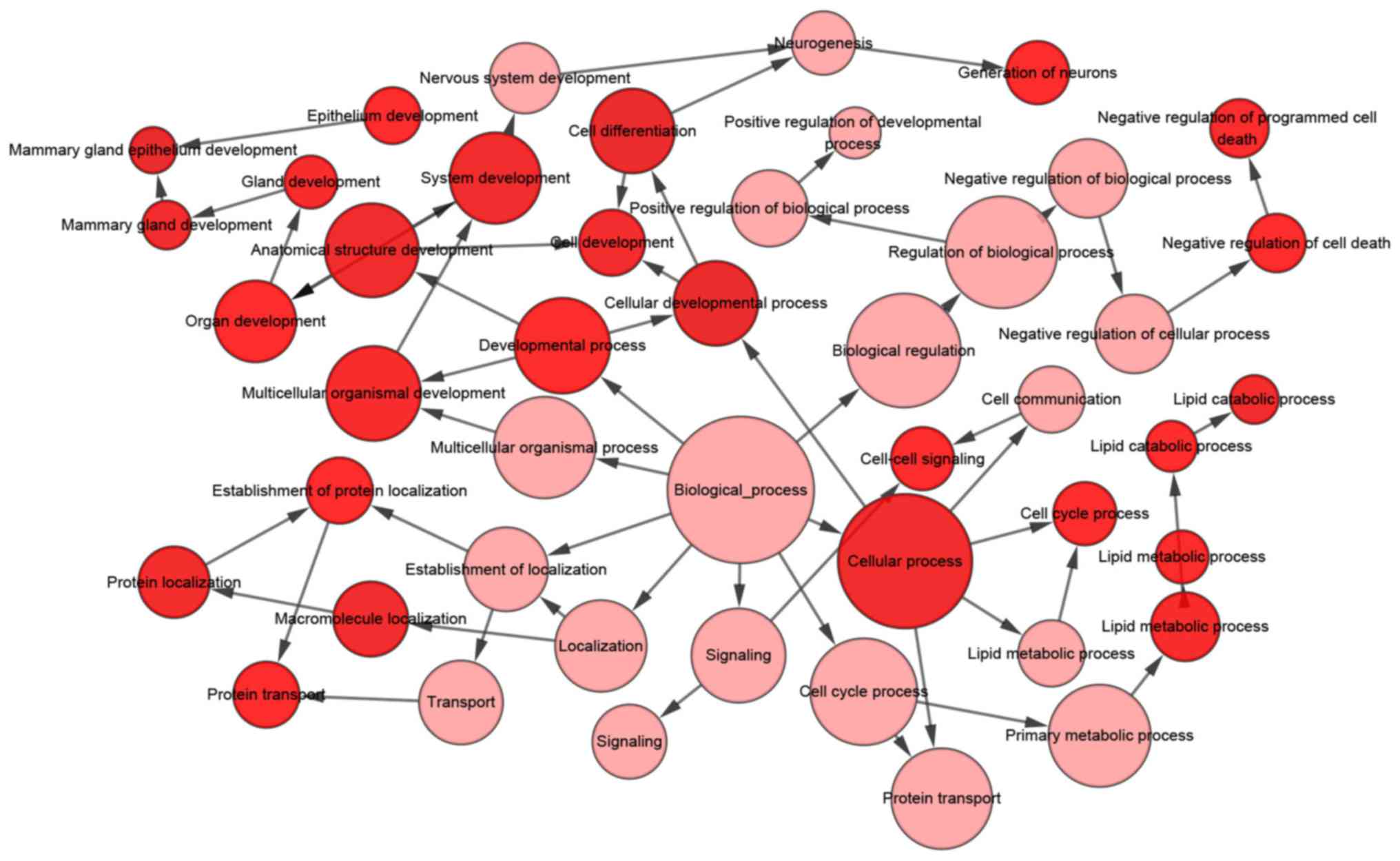
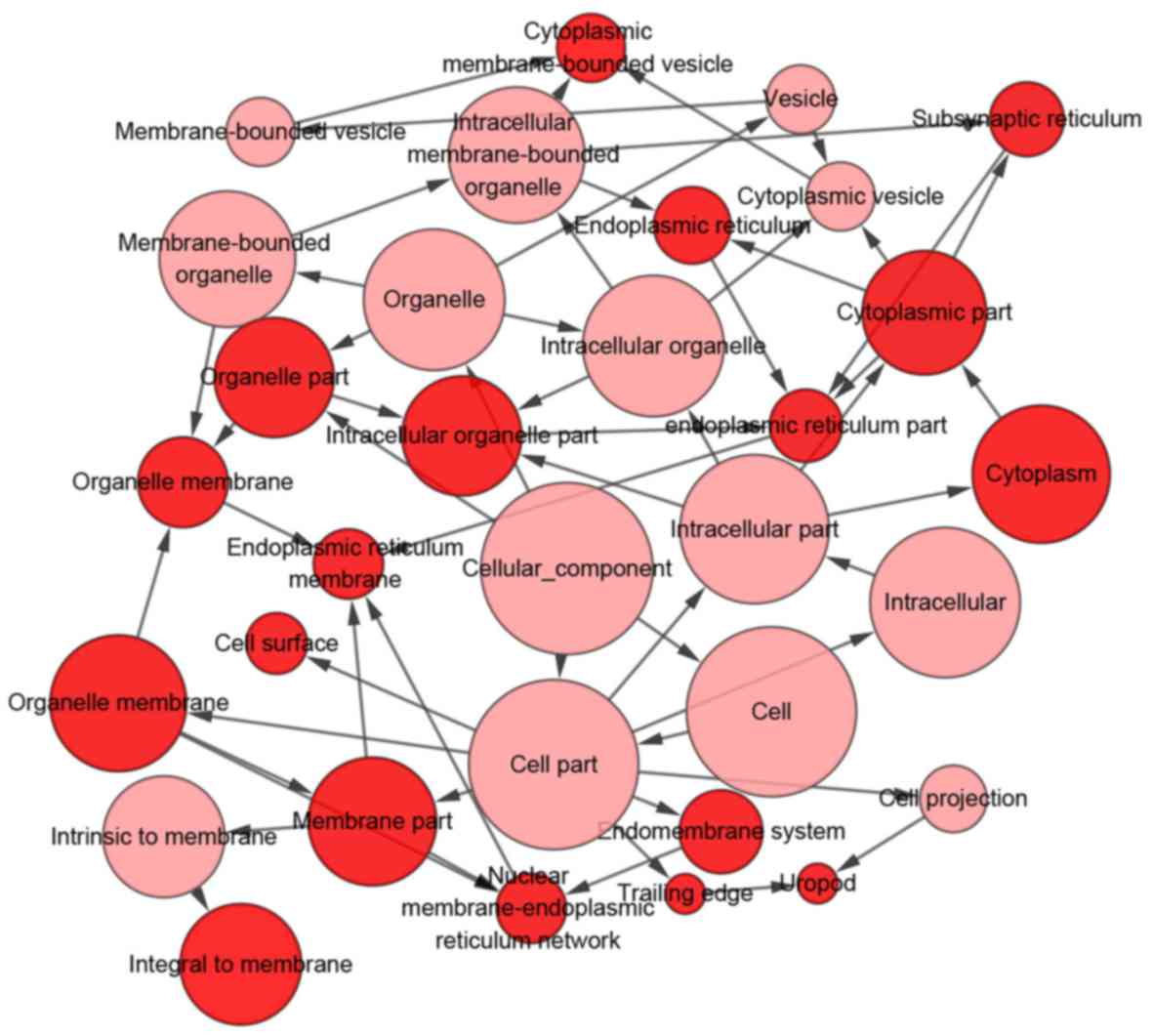
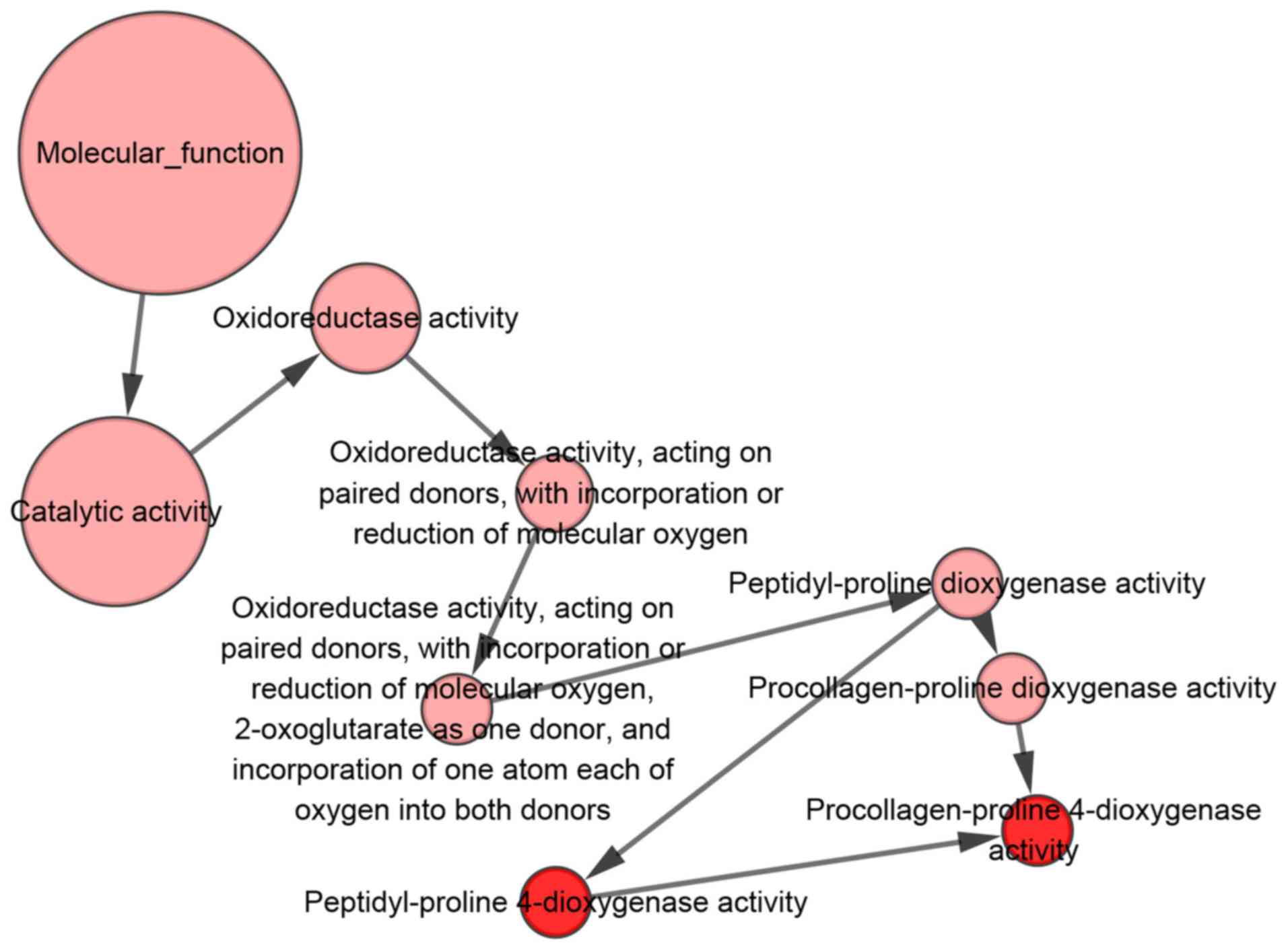
With regards to the GO analysis (Fig. 16), the putative genes of
miR-204-5p were identify to have mainly participated in ‘cell
development’ in biological process (BP) terms (Figs. 16A and 17), and were enriched in ‘cell surface’
in cellular component (CC) terms (Figs. 16B and 18). In addition, for molecular function
(MF), the putative genes were predominantly enriched in receptor
agonist activity (Figs. 16C and
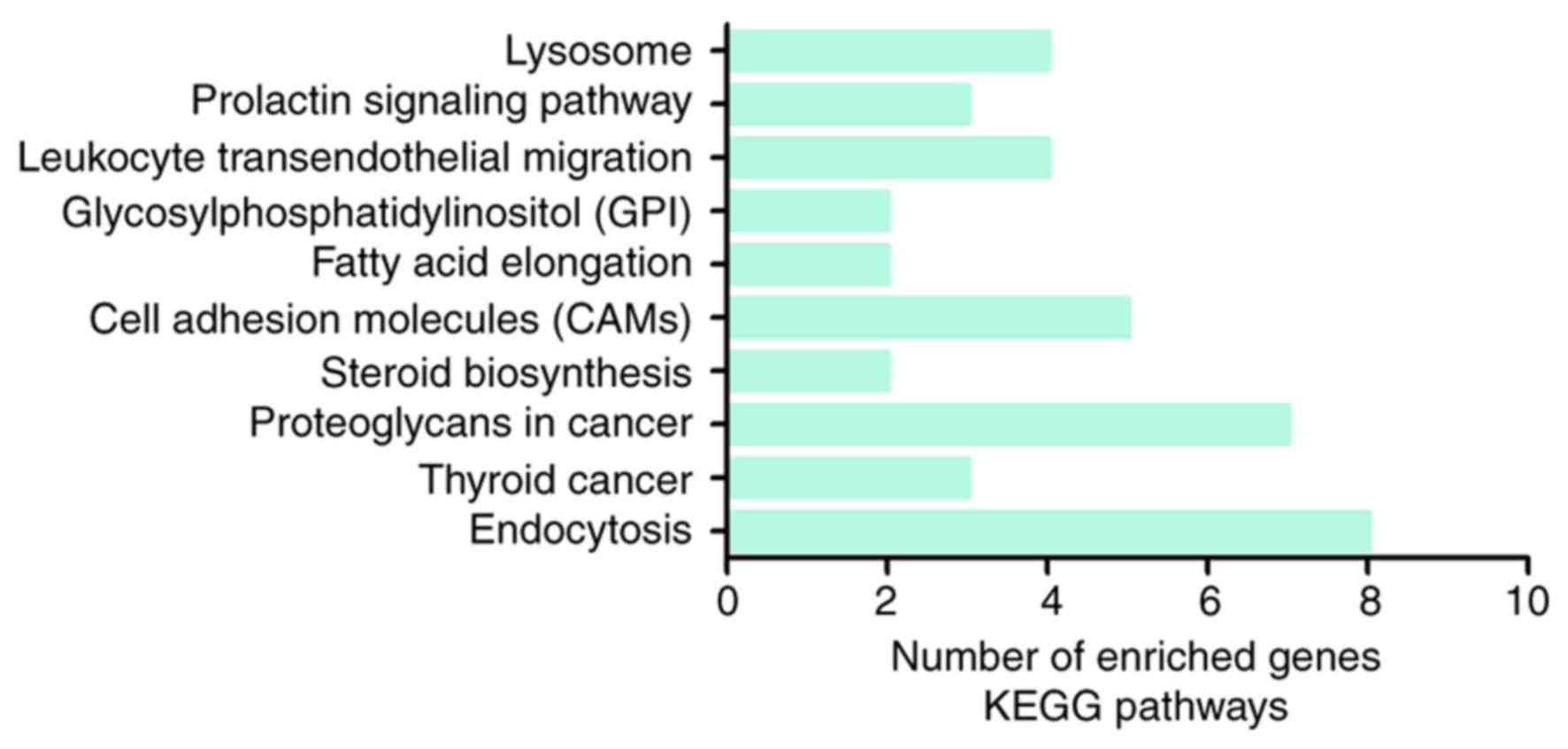
19). Regarding the KEGG pathway
analysis, the most enriched pathway of the putative genes was
‘endocytosis’ (Fig. 20). A total
of eight pathway-associated genes were obtained: ADP-ribosylation
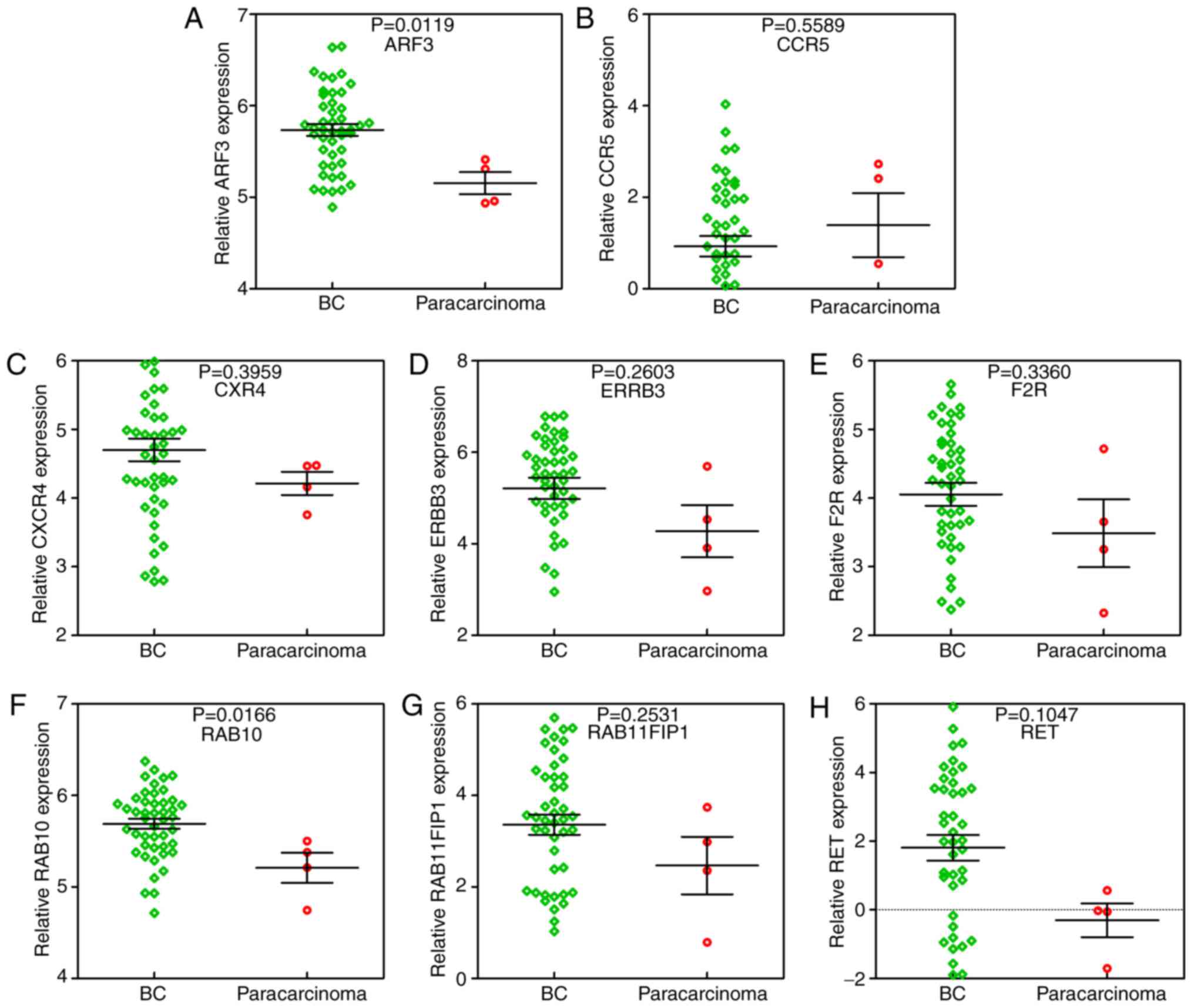
factor 3 (ARF3), C-C chemokine receptor type 5 (CCR5), C-X-C
chemokine receptor type 4 (CXCR4), receptor tyrosine-protein kinase
erbB-3 (ERBB3), proteinase-activated receptor 1 (F2R), Ras-related
protein Rab-10 (RAB10), Rab11 family-interacting protein 1
(RAB11FIP1) and proto-oncogene tyrosine-protein kinase receptor Ret
(RET).
Furthermore, it was identified that RAB10 was
significantly upregulated in BC tissue samples compared to
para-carcinoma tissue samples (Fig.
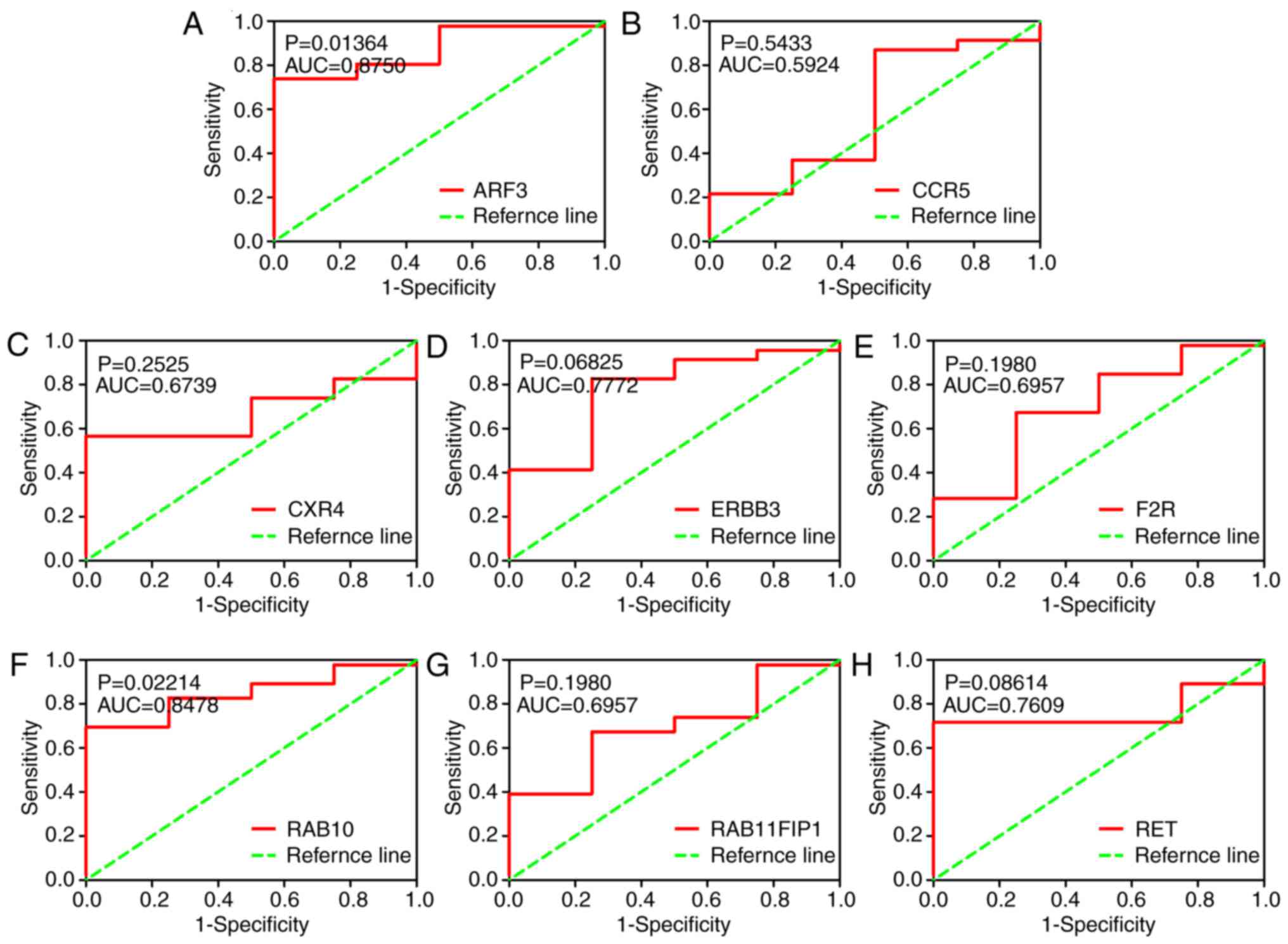
21). Additionally, ROC curve analysis suggested that RAB10
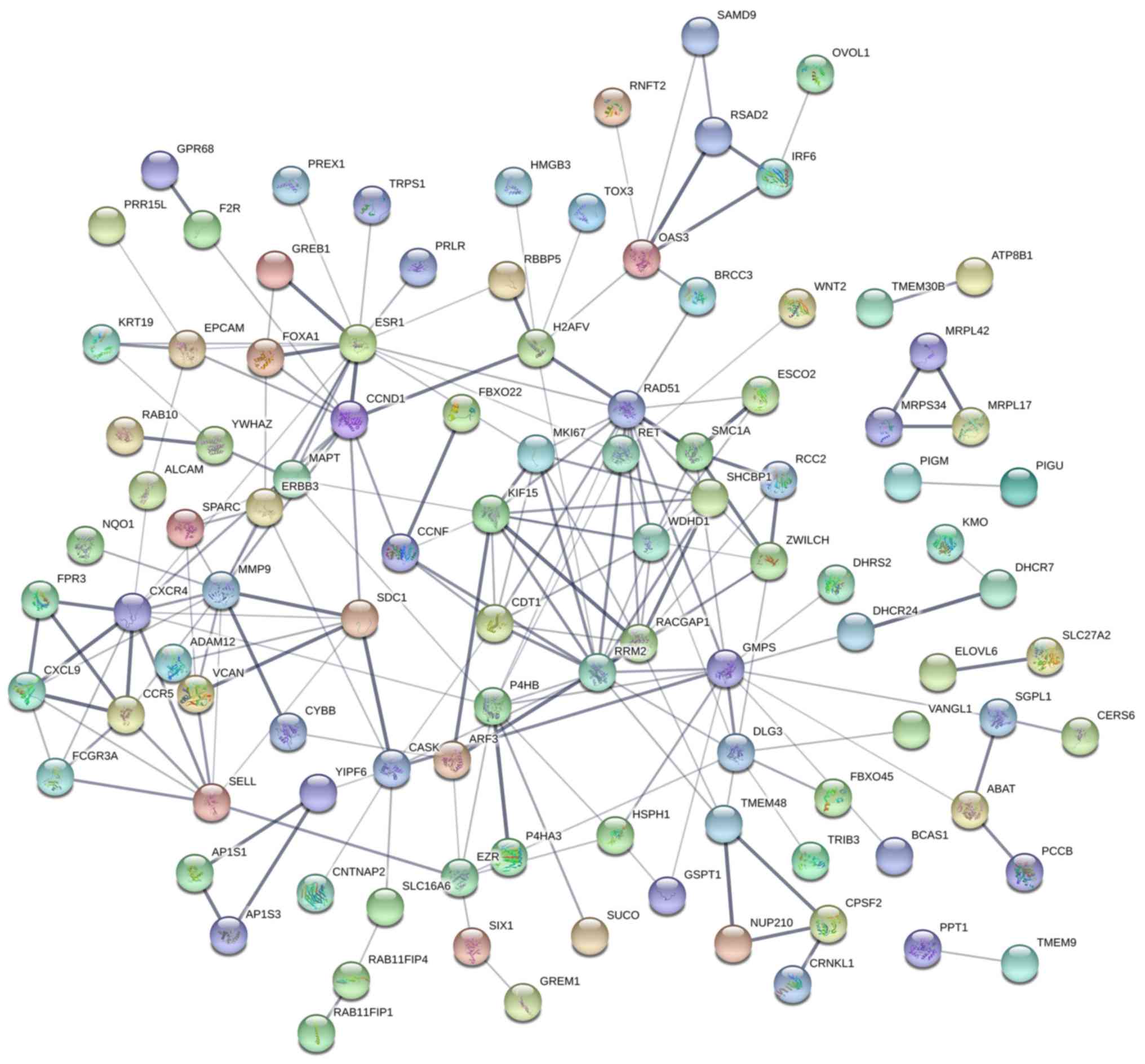
possessed a great discriminatory capability for BC (Fig. 22). In terms of the PPI network, in
the current study, estrogen receptor 1 (ESR1), ribonucleotide
reductase regulatory subunit M2 (RRM2) and Rac GTPase activating
protein 1 (RACGAP1) exhibited the highest degrees (Fig. 23). However, both ESR1 and RRM2 did
not have significant negative correlations with miR-204-5p, and
RRM2 mRNA expression levels were not significantly lower in
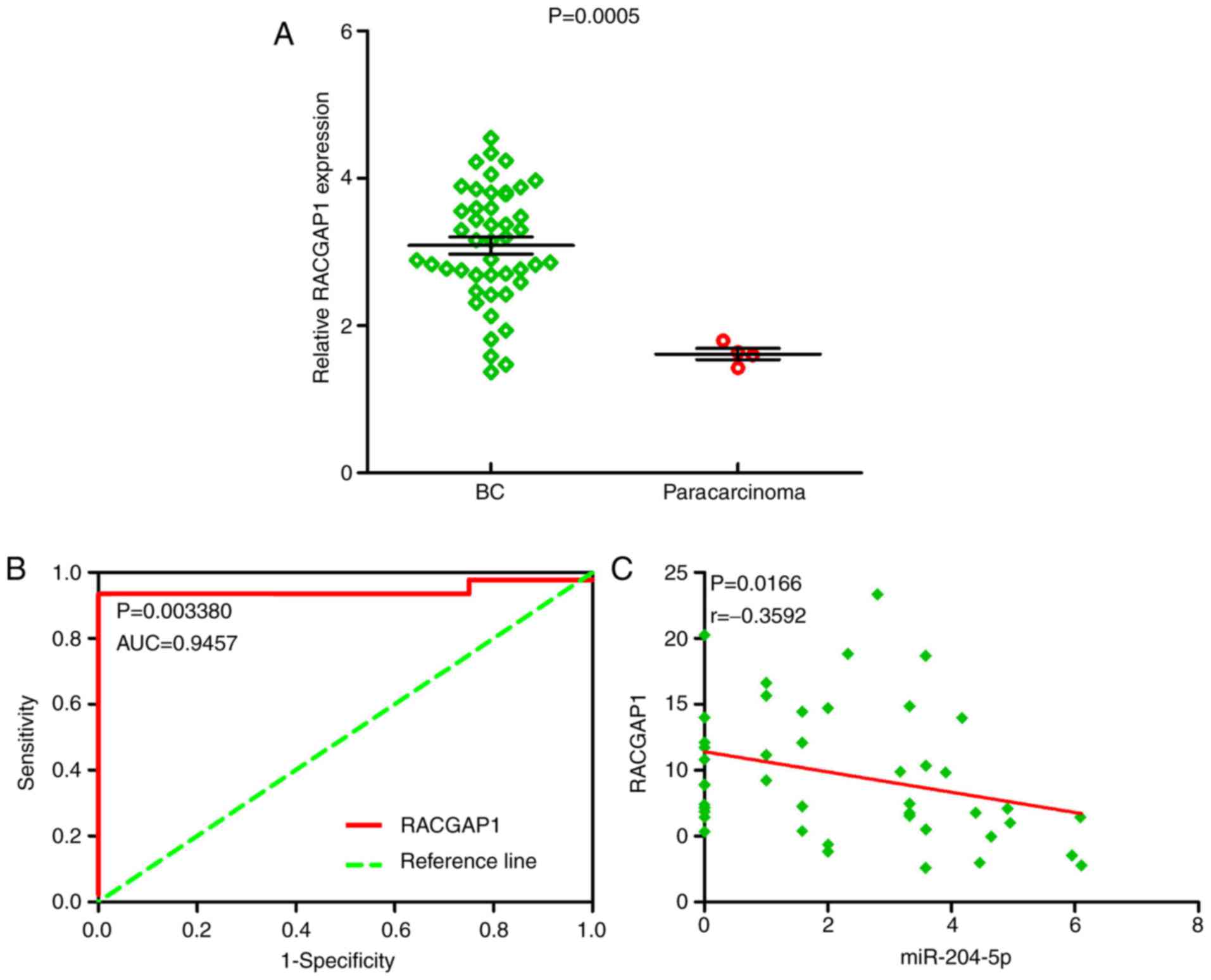
para-carcinoma tissue (data not shown). Therefore, RACGAP1 was
selected as the hub gene. RACGAP1 mRNA expression levels were
significantly increased in BC tissue samples in comparison to
para-carcinoma tissue samples (Fig.
24A). Furthermore, the ROC curve indicated that RACGAP1 had
great discriminatory capability for BC (Fig. 24B). Of note, a markedly negative
correlation trend was identified between RACGAP1 and miR-204-5p
(P=0.0166; r=−0.3592; Fig.
24C).
Discussion
To date, many studies have reported the decreased
expression of miR-204-5p in various cancers, including
hepatocellular carcinoma (24),
non-small cell lung cancer (52),
gastric cancer (53), oral
squamous cell carcinoma (27),
prostate cancer (54) and
esophageal cancer (55). Further,
recent studies have focused on the expression of miR-204-5p in BC.
For example, Wang et al (56) discovered that the expression level
of miR-204-5p was obviously reduced in 24 BC tissue samples in
comparison to corresponding normal tissue samples. In addition,
they reported a decrease in miR-204-5p expression in two BC cell
lines (MDA-MB-231 and MCF-7), compared with MCF-10A, a breast
epithelial cell line (56). Shen
et al (57) reported that
the expression of miR-204-5p was markedly suppressed in BC cells.
They also demonstrated that the expression of miR-204-5p was
markedly reduced in MCF-7 cells in comparison to HBL-100 cells,
which are normal breast epithelial cells. In addition, they
revealed that the upregulation of miR-204-5p inhibits the invasion,
proliferation and migration, and enhances the apoptosis of BC
cells.
Nonetheless, in-depth research featuring abundant
samples is still required. In the current study, the expression of
miR-204-5p in BC was evaluated in data obtained from the TCGA, GEO,
and UCSC Xena databases. In the TCGA database, a decreased trend in
precursor miR-204 expression was identified in 1,077 BC tissue
samples, in comparison with 104 para-carcinoma tissue samples. In
addition, in the UCSC Xena database, the expression of mature
miR-204-5p was notably reduced in 756 BC tissue samples, compared
with 76 para-carcinoma tissue samples. Furthermore, a number of the
GEO microarrays indicated that the expression of miR-204-5p was
downregulated in BC tissue samples, and the SMD in the
meta-analysis also showed that the expression of miR-204-5p was
notably lower in 2,306 BC tissue samples, compared with the 291
para-carcinoma tissue samples. The AUCs of the ROC and sROC curves
implied that t miR-204 and miR-204-5p exhibited great
discriminatory capacity in BC. Next, the prognostic value of
miR-204-5p in BC was determined. Prior analysis of BC samples
suggested that the decreased expression of miR-204-5p correlates
with poor overall survival and disease-free survival in BC
(58). Ye et al (59) demonstrated that miR-204-5p had no
prognostic value in BC through analyzing 563 BC tissue samples
obtained from the TCGA database. In the current study, no obvious
correlation was found between the precursor miR-204 and survival
outcome in BC based on analyzing 1,077 BC tissue samples from TCGA
database. Additionally, it was identified that miR-204
downregulation was significant in several groups, including age,
vital status, PR status, HER2 status and pathological stage. Taken
together with the results of these two aforementioned studies, it
was hypothesized that miR-204-5p may acts as a tumor suppressor in
the oncogenesis and progression of BC.
GO and KEGG analysis was performed to investigate
the potential biological processes and pathways of miR-204-5p in
BC. ‘Cell development’, ‘cell surface activity’ and ‘receptor
agonist activity’ were considered the most enriched processes in GO
analyses. Thus, it was suggested that miR-204-5p may participate
these processes in BC, by targeting its corresponding target genes.
However, further study is needed to verify the molecular mechanisms
underlying miR-204-5p and these three processes in BC.
Concurrently, in the KEGG analyses, ‘endocytosis’ was found to be
the most enriched, which was associated with ARF3, CCR5, CXCR4,
ERBB3, F2R, RAB10, RAB11FIP1 and RET. The expression and ROC curves
of the eight pathway-related genes were estimated, and it was
determined that RAB10 expression was significantly increased in BC
tissue samples, compared with para-carcinoma tissue samples. In
addition, RAB10 featured a great discriminatory capacity for BC
within non-cancerous breast tissue samples. Hence, it was proposed
that miR-204-5p may possess a vital effect on BC via genes
associated with endocytosis, including RAB10. However, the role of
endocytosis in BC is unclear and further investigation is urgently
required.
In researching the target genes of miR-204-5p in BC,
Flores-Peréz et al (60)
found that transforming growth factor β receptor 2 (TGFβR2) and
angiopoietin 1 (ANGPT1) are crucial in BC tumor angiogenesis; BC
cell migration and proliferation decreases when TGFβR2 is
suppressed, and the suppression of TGFβR2 and ANGPT1 inhibits
angiogenesis. Furthermore, Zeng et al (61) identified a negative correlation
trend between miR-204-5p and SIX homeobox 1 (Six1) expression in BC
tissue samples, and when miR-204-5p mimics or Six1 siRNA was
transfected, the expression of chromodomain helicase DNA binding
protein 1 was markedly increased, thus enhancing
epithelial-mesenchymal transition and affecting the invasion and
migration of BC cells (61).
Various target genes of miR-204-5p have been confirmed in previous
studies, including traditional serrated adenoma, MX dynamin like
GTPase 1, thioredoxin interacting protein, Src-associated in
mitosis 68 kDa protein and forkhead box A1 (57,62–64).
However, more target genes need to be determined. Accordingly, a
PPI network was generated in the present study. The hub gene
RACGAP1 was selected as an example for further investigation.
RACGAP1 is involved in cell cytokinesis, transformation, migration,
metastasis and growth (65,66).
In BC specifically, it has been reported that RACGAP1 is critical
in enhancing basal-like breast cancer proliferation and
oncogenicity (67,68). Furthermore, in untransformed cells,
RACGAP1 stimulates malignant phenotypes, and elevated RACGAP1
expression is correlated with poor BC outcomes (67,68).
In the current study, RACGAP1 expression was evaluated in BC tissue
sample data obtained from the TCGA database. RACGAP1 was
upregulated BC tissue samples, compared with para-carcinoma tissue
samples. In addition, a strong negative correlation trend was
identified between RACGAP1 and miR-204-5p. Thus, it was proposed
that miR-204-5p may serve a crucial role in BC by targeting
RACGAP1. However, these conclusions were made based on online
tools, so further in vivo and in vitro investigations
should be performed to verify the molecular mechanisms of RACGAP1
and miR-204-5p in BC.
There are several limitations to the present study.
First, a high degree of I2 existed in the heterogeneity
test. Thus, a random effects model was conducted to reduce the
degree of I2-however, it still exceeded 50%. This may be
a result of using various measures and platforms used to analyze
the data. The nine GEO microarrays were acquired from six
countries; GSE32922, GSE44124, GSE48088 and GSE58606 were obtained
from Spain, and GSE17155, GSE22981, GSE31309, GSE35412, GSE37407
and GSE40525 were obtained from Italy, the USA, Germany, Mexico,
Sweden and Israel, respectively. Second, a dual luciferase reporter
assay was not performed to verify the correlation between
miR-204-5p and the hub gene. Thus, in-depth investigations with
in vivo and in vitro experiments should be performed
in the future.
In conclusion, the results of the present study
identified that miR-204-5p expression was downregulated in BC
tissue samples in comparison to para-carcinoma tissue samples; this
suggested that miR-204-5p might function as a suppressor in the
oncogenesis and advancement of BC. Furthermore, it was revealed
that RACGAP1 may be a crucial target gene of miR-204-5p, and the
expression of RACGAP1 was markedly increased in BC tissue samples
in comparison to para-carcinoma tissue samples. Notably, a
significant negative correlation was identified between RACGAP1 and
miR-204-5p in BC. Therefore, it was concluded that miR-204-5p may
serve a crucial role in BC by targeting RACGAP1.
Acknowledgements
Not applicable.
Funding
The ‘Future Academic Star’ Fund of the Guangxi
Medical University (grant no. WLXSZX17050) supported the current
study.
Availability of data and materials
The data and materials of the present study are
available from the corresponding authors on reasonable request.
Authors' contributions
AGL and ZFW collected and analyzed the TCGA data.
HWJ and JJZ collected and analyzed the GEO data. RQH, GC and JM
collected and analyzed the UCSC data. KTC and JCZ conducted
meta-analysis and wrote the manuscript. All authors read the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sun W, Jiang YZ, Liu YR, Ma D and Shao ZM:
Nomograms to estimate long-term overall survival and breast
cancer-specific survival of patients with luminal breast cancer.
Oncotarget. 7:20496–20506. 2016.PubMed/NCBI
|
|
2
|
Samson M, Porter N, Orekoya O, Hebert JR,
Adams SA, Bennett CL and Steck SE: Progestin and breast cancer
risk: A systematic review. Breast Cancer Res Treat. 155:3–12. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chavez-MacGregor M, Clarke CA,
Lichtensztajn DY and Giordano SH: Delayed initiation of adjuvant
chemotherapy among patients with breast cancer. JAMA Oncol.
2:322–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Biglia N, D'Alonzo M, Sgro LG, Tomasi Cont
N, Bounous V and Robba E: Breast cancer treatment in mutation
carriers: Surgical treatment. Minerva Ginecol. 68:548–556.
2016.PubMed/NCBI
|
|
5
|
Monroy-Cisneros K, Esparza-Romero J,
Valencia ME, Guevara-Torres AG, Méndez-Estrada RO, Anduro-Corona I
and Astiazarán-García H: Antineoplastic treatment effect on bone
mineral density in Mexican breast cancer patients. BMC Cancer.
16:8602016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Frasier LL, Holden S, Holden T, Schumacher
JR, Leverson G, Anderson B, Greenberg CC and Neuman HB: Temporal
trends in postmastectomy radiation therapy and breast
reconstruction associated with changes in national comprehensive
cancer network guidelines. JAMA Oncol. 2:95–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu L, Peng S, Huang Q, Liu Y, Jiang H, Li
X and Wang J: Expression status of cyclaseassociated protein 2 as a
prognostic marker for human breast cancer. Oncol Rep. 36:1981–1988.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang X, Li X and Xie X, Ye F, Chen B,
Song C, Tang H and Xie X: High expressions of LDHA and AMPK as
prognostic biomarkers for breast cancer. Breast. 30:39–46. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Wu J, Yuan J, Zhu X, Wu H and Li
M: Midline2 is overexpressed and a prognostic indicator in human
breast cancer and promotes breast cancer cell proliferation in
vitro and in vivo. Front Med. 10:41–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iravani O, Yip GW, Thike AA, Chua PJ, Jane
Scully O, Tan PH and Bay BH: Prognostic significance of Claudin 12
in estrogen receptor-negative breast cancer. J Clin Pathol.
69:878–883. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Q, Zhao GS, Yuan XL, Li XH, Yang Z,
Cui YF, Guan QL, Sun XY, Shen W, Xu TA and Wang QS: Tumor necrosis
factor alpha-238G/A polymorphism and risk of breast cancer: An
update by meta-analysis. Medicine (Baltimore). 96:e74422017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baretta Z, Mocellin S, Goldin E, Olopade
OI and Huo D: Effect of BRCA germline mutations on breast cancer
prognosis: A systematic review and meta-analysis. Medicine
(Baltimore). 95:e49752016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Song Y and Zhu X: MicroRNA-181a
regulates apoptosis and autophagy process in Parkinson's disease by
inhibiting p38 mitogen-activated protein kinase (MAPK)/c-Jun
N-terminal kinases (JNK) signaling pathways. Med Sci Monit.
23:1597–1606. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Erbes T, Hirschfeld M, Rücker G, Jaeger M,
Boas J, Iborra S, Mayer S, Gitsch G and Stickeler E: Feasibility of
urinary microRNA detection in breast cancer patients and its
potential as an innovative non-invasive biomarker. BMC Cancer.
15:1932015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mesci A, Huang X, Taeb S, Jahangiri S, Kim
Y, Fokas E, Bruce J, Leong HS and Liu SK: Targeting of CCBE1 by
miR-330-3p in human breast cancer promotes metastasis. Br J Cancer.
116:1350–1357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song L, Zhang W, Chang Z, Pan Y, Zong H,
Fan Q and Wang L: miR-4417 targets tripartite motif-containing 35
(TRIM35) and regulates pyruvate kinase muscle 2 (PKM2)
phosphorylation to promote proliferation and suppress apoptosis in
hepatocellular carcinoma cells. Med Sci Monit. 23:1741–1750. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Q, He M, Guan S, Ma M, Wu H, Yu Z,
Jiang L, Wang Y, Zong X, Jin F and Wei M: MicroRNA-100 suppresses
the migration and invasion of breast cancer cells by targeting
FZD-8 and inhibiting Wnt/β-catenin signaling pathway. Tumour Biol.
37:5001–5011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Wang YW, Xing AY, Xiang S, Shi DB,
Liu L, Li YX and Gao P: Suppression of SPIN1-mediated PI3K-Akt
pathway by miR-489 increases chemosensitivity in breast cancer. J
Pathol. 239:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Lin S, Tseng KF, Han K, Wang Y,
Gan ZH, Min DL and Hu HY: Selumetinib suppresses cell
proliferation, migration and trigger apoptosis, G1 arrest in
triple-negative breast cancer cells. BMC Cancer. 16:8182016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia M, Li H, Wang JJ, Zeng HJ and Wang SH:
MiR-99a suppress proliferation, migration and invasion through
regulating insulin-like growth factor 1 receptor in breast cancer.
Eur Rev Med Pharmacol Sci. 20:1755–1763. 2016.PubMed/NCBI
|
|
22
|
Zhan Y, Liang X, Li L, Wang B, Ding F, Li
Y, Wang X, Zhan Q and Liu Z: MicroRNA-548j functions as a
metastasis promoter in human breast cancer by targeting Tensin1.
Mol Oncol. 10:838–849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mohammadi-Yeganeh S, Paryan M, Arefian E,
Vasei M, Ghanbarian H, Mahdian R, Karimipoor M and Soleimani M:
MicroRNA-340 inhibits the migration, invasion, and metastasis of
breast cancer cells by targeting Wnt pathway. Tumour Biol.
37:8993–9000. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo YH, Tang W, Zhang X, Tan Z, Guo WL,
Zhao N, Pang SM, Dang YW, Rong MH and Cao J: Promising significance
of the association of miR-204-5p expression with
clinicopathological features of hepatocellular carcinoma. Medicine
(Baltimore). 96:e75452017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao W, Wu Y, He X, Zhang C, Zhu M, Chen B,
Liu Q, Qu X, Li W, Wen S and Wang B: MicroRNA-204-5p inhibits
invasion and metastasis of laryngeal squamous cell carcinoma by
suppressing forkhead box C1. J Cancer. 8:2356–2368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J,
Ruan H, Ma S and Xu B: miR-204-5p acts as a tumor suppressor by
targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in
malignant melanoma. Onco Targets Ther. 10:1237–1246. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Li F and Zhou X: miR-204-5p
regulates cell proliferation and metastasis through inhibiting
CXCR4 expression in OSCC. Biomed Pharmacother. 82:202–207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu L, Wang J, Li X, Ma J, Shi C, Zhu H,
Xi Q, Zhang J, Zhao X and Gu M: MiR-204-5p suppresses cell
proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma.
Biochem Biophys Res Commun. 457:621–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li T, Pan H and Li R: The dual regulatory
role of miR-204 in cancer. Tumour Biol. 37:11667–11677. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Li G, Luo Q, Xie J and Gan C:
Integrated TCGA analysis implicates lncRNA CTB-193M12.5 as a
prognostic factor in lung adenocarcinoma. Cancer Cell Int.
18:272018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao K, Li Z and Tian H: Twenty-gene-based
prognostic model predicts lung adenocarcinoma survival. Onco
Targets Ther. 11:3415–3424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Q, Zhao G, Yang Z, Liu X and Xie P:
Downregulation of microRNA1243p suppresses the mTOR signaling
pathway by targeting DDIT4 in males with major depressive disorder.
Int J Mol Med. 41:493–500. 2018.PubMed/NCBI
|
|
34
|
Shang J, Wang F, Chen P, Wang X, Ding F,
Liu S and Zhao Q: Co-expression network analysis identified COL8A1
is associated with the progression and prognosis in human colon
adenocarcinoma. Dig Dis Sci. 63:1219–1228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Su L, Wang C, Zheng C, Wei H and Song X: A
meta-analysis of public microarray data identifies biological
regulatory networks in Parkinson's disease. BMC Med Genomics.
11:402018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu W, Li J and Wu B: Gene expression
profiling of the mouse gut: Effect of intestinal flora on
intestinal health. Mol Med Rep. 17:3667–3673. 2018.PubMed/NCBI
|
|
38
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun W, Qiu Z, Huang W and Cao M: Gene
expression profiles and proteinprotein interaction networks during
tongue carcinogenesis in the tumor microenvironment. Mol Med Rep.
17:165–171. 2018.PubMed/NCBI
|
|
40
|
Liu J, Li H, Sun L, Wang Z, Xing C and
Yuan Y: Aberrantly methylated-differentially expressed genes and
pathways in colorectal cancer. Cancer Cell Int. 17:752017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Y, Teng L, Liu W, Cao Y, Ding D, Wang
W, Chen H, Li C and An R: Identification of biological targets of
therapeutic intervention for clear cell renal cell carcinoma based
on bioinformatics approach. Cancer Cell Int. 16:162016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fassan M, Baffa R, Palazzo JP, Lloyd J,
Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM and
Rosenberg A: MicroRNA expression profiling of male breast cancer.
Breast Cancer Res. 11:R582009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao H, Shen J, Medico L, Wang D,
Ambrosone CB and Liu S: A pilot study of circulating miRNAs as
potential biomarkers of early stage breast cancer. PLoS One.
5:e137352010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7:e297702012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tanic M, Gómez-López G, Benítez J and
Martínez-Delgado B: Comparison between hereditary breast tumors and
normal breast tissue samples.
|
|
46
|
Romero-Cordoba S, Rodriguez-Cuevas S,
Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez
G, Bautista-Piña V, Arellano-Llamas R and Hidalgo-Miranda A:
Identification and pathway analysis of microRNAs with no previous
involvement in breast cancer. PLoS One. 7:e319042012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gravgaard KH, Lyng MB, Laenkholm AV,
Søkilde R, Nielsen BS, Litman T and Ditzel HJ: The miRNA-200 family
and miRNA-9 exhibit differential expression in primary versus
corresponding metastatic tissue in breast cancer. Breast Cancer Res
Treat. 134:207–217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Biagioni F, Bossel Ben-Moshe N, Fontemaggi
G, Canu V, Mori F, Antoniani B, Di Benedetto A, Santoro R, Germoni
S, De Angelis F, et al: miR-10b*, a master inhibitor of the cell
cycle, is down-regulated in human breast tumours. EMBO Mol Med.
4:1214–1229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Feliciano A, Castellvi J, Artero-Castro A,
Leal JA, Romagosa C, Hernández-Losa J, Peg V, Fabra A, Vidal F,
Kondoh H, et al: miR-125b acts as a tumor suppressor in breast
tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and
MEGF9. PLoS One. 8:e762472013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pena-Chilet M, Martínez MT, Pérez-Fidalgo
JA, Peiró-Chova L, Oltra SS, Tormo E, Alonso-Yuste E,
Martinez-Delgado B, Eroles P, Climent J, et al: MicroRNA profile in
very young women with breast cancer. BMC Cancer. 14:5292014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Matamala N, Vargas MT, Gonzalez-Campora R,
Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G,
Yanowsky K, Calvete-Candenas J, et al: Tumor microRNA expression
profiling identifies circulating microRNAs for early breast cancer
detection. Clin Chem. 61:1098–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang S, Gao L, Thakur A, Shi P, Liu F,
Feng J, Wang T, Liang Y, Liu JJ, Chen M and Ren H: miRNA-204
suppresses human non-small cell lung cancer by targeting ATF2.
Tumour Biol. 37:11177–11186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu Z, Long J, Du R, Ge C, Guo K and Xu Y:
miR-204 regulates the EMT by targeting snai1 to suppress the
invasion and migration of gastric cancer. Tumour Biol.
37:8327–8335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu G, Wang J, Chen G and Zhao X:
microRNA-204 modulates chemosensitivity and apoptosis of prostate
cancer cells by targeting zinc-finger E-box-binding homeobox 1
(ZEB1). Am J Transl Res. 9:3599–3610. 2017.PubMed/NCBI
|
|
55
|
Sun Y, Yu X and Bai Q: miR-204 inhibits
invasion and epithelial-mesenchymal transition by targeting FOXM1
in esophageal cancer. Int J Clin Exp Pathol. 8:12775–12783.
2015.PubMed/NCBI
|
|
56
|
Wang X, Qiu W, Zhang G, Xu S, Gao Q and
Yang Z: MicroRNA-204 targets JAK2 in breast cancer and induces cell
apoptosis through the STAT3/BCl-2/survivin pathway. Int J Clin Exp
Pathol. 8:5017–5025. 2015.PubMed/NCBI
|
|
57
|
Shen SQ, Huang LS, Xiao XL, Zhu XF, Xiong
DD, Cao XM, Wei KL, Chen G and Feng ZB: miR-204 regulates the
biological behavior of breast cancer MCF-7 cells by directly
targeting FOXA1. Oncol Rep. 38:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li W, Jin X, Zhang Q, Zhang G, Deng X and
Ma L: Decreased expression of miR-204 is associated with poor
prognosis in patients with breast cancer. Int J Clin Exp Pathol.
7:3287–3292. 2014.PubMed/NCBI
|
|
59
|
Ye ZH, Wen DY, Cai XY, Liang L, Wu PR, Qin
H, Yang H, He Y and Chen G: The protective value of miR-204-5p for
prognosis and its potential gene network in various malignancies: A
comprehensive exploration based on RNA-seq high-throughput data and
bioinformatics. Oncotarget. 8:104960–104980. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Flores-Peréz A, Marchat LA,
Rodriguez-Cuevas S, Bautista-Piña V, Hidalgo-Miranda A, Ocampo EA,
Martínez MS, Palma-Flores C, Fonseca-Sánchez MA, Astudillo-de la
Vega H, et al: Dual targeting of ANGPT1 and TGFBR2 genes by miR-204
controls angiogenesis in breast cancer. Sci Rep. 6:345042016.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zeng J, Wei M, Shi R, Cai C, Liu X, Li T
and Ma W: MiR-204-5p/Six1 feedback loop promotes
epithelial-mesenchymal transition in breast cancer. Tumour Biol.
37:2729–2735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu J and Li Y: Trichostatin A and
Tamoxifen inhibit breast cancer cell growth by miR-204 and ERα
reducing AKT/mTOR pathway. Biochem Biophys Res Commun. 467:242–247.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee H, Lee S, Bae H, Kang HS and Kim SJ:
Genome-wide identification of target genes for miR-204 and miR-211
identifies their proliferation stimulatory role in breast cancer
cells. Sci Rep. 6:252872016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang L, Tian H, Yuan J, Wu H, Wu J and Zhu
X: CONSORT: Sam68 Is directly regulated by MiR-204 and promotes the
self-renewal potential of breast cancer cells by activating the
Wnt/Beta-catenin signaling pathway. Medicine (Baltimore).
94:e22282015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Saigusa S, Tanaka K, Mohri Y, Ohi M,
Shimura T, Kitajima T, Kondo S, Okugawa Y, Toiyama Y, Inoue Y and
Kusunoki M: Clinical significance of RacGAP1 expression at the
invasive front of gastric cancer. Gastric Cancer. 18:84–92. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Imaoka H, Toiyama Y, Saigusa S, Kawamura
M, Kawamoto A, Okugawa Y, Hiro J, Tanaka K, Inoue Y, Mohri Y and
Kusunoki M: RacGAP1 expression, increasing tumor malignant
potential, as a predictive biomarker for lymph node metastasis and
poor prognosis in colorectal cancer. Carcinogenesis. 36:346–354.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lawson CD and Der CJ: Filling GAPs in our
knowledge: ARHGAP11A and RACGAP1 act as oncogenes in basal-like
breast cancers. Small GTPases. 9:290–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sahin S, Işık Gönül İ, Çakır A, Seçkin S
and Uluoğlu Ö: Clinicopathological significance of the
proliferation markers Ki67, RacGAP1, and topoisomerase 2 alpha in
breast cancer. Int J Surg Pathol. 24:607–613. 2016. View Article : Google Scholar : PubMed/NCBI
|