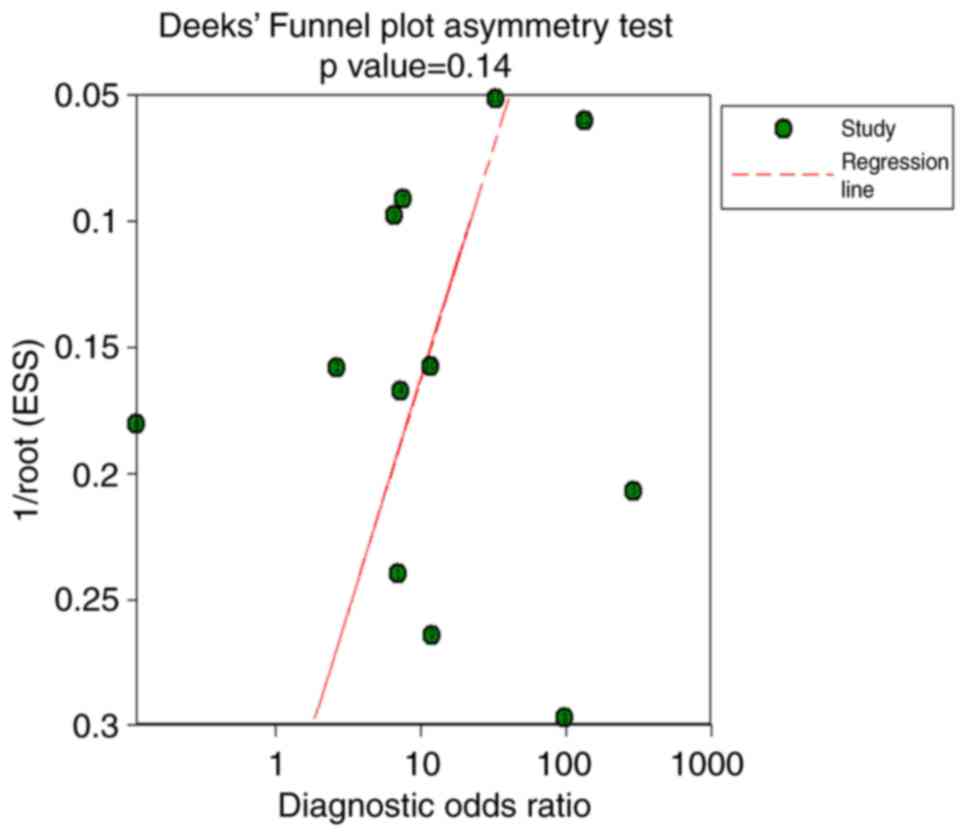
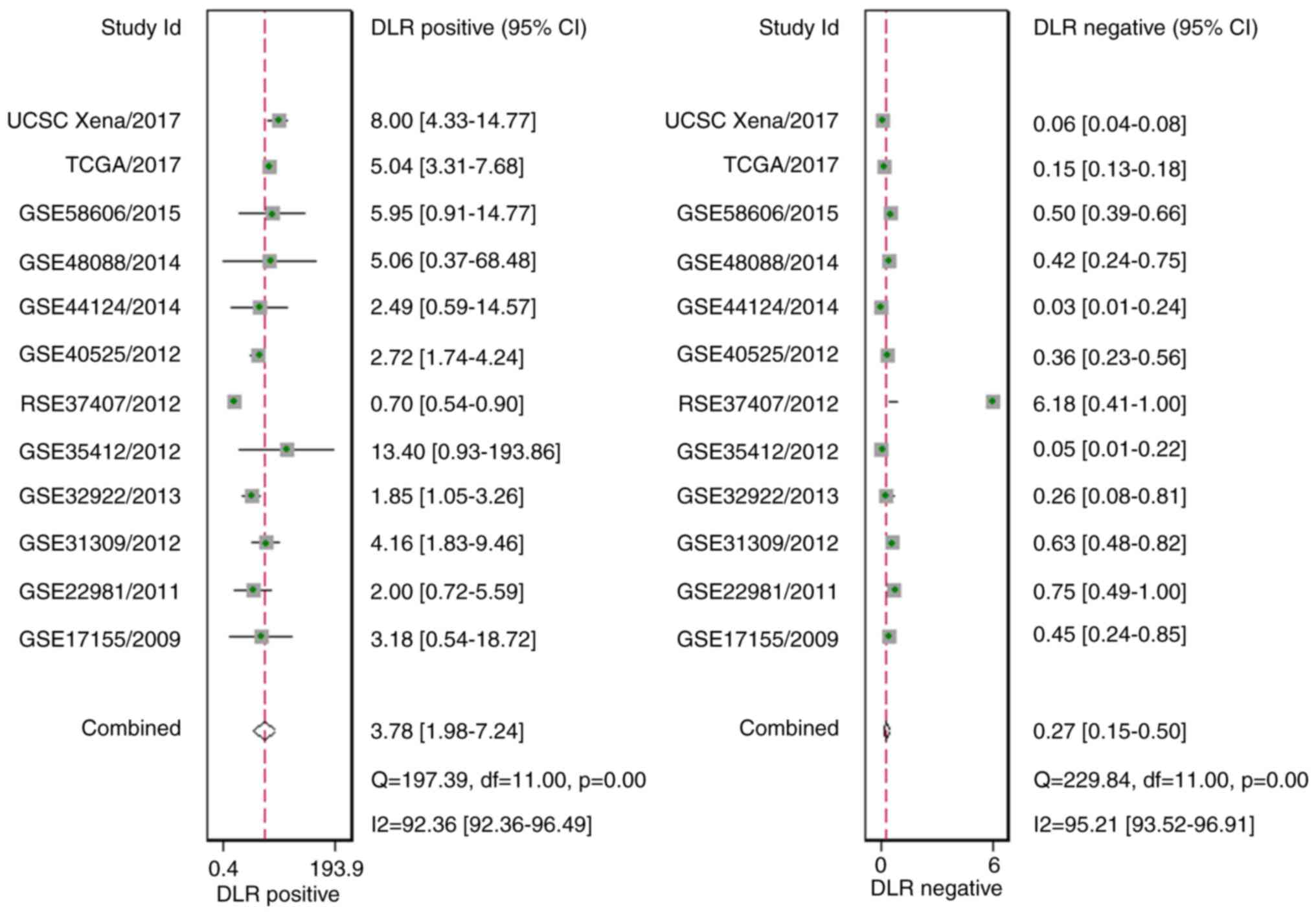
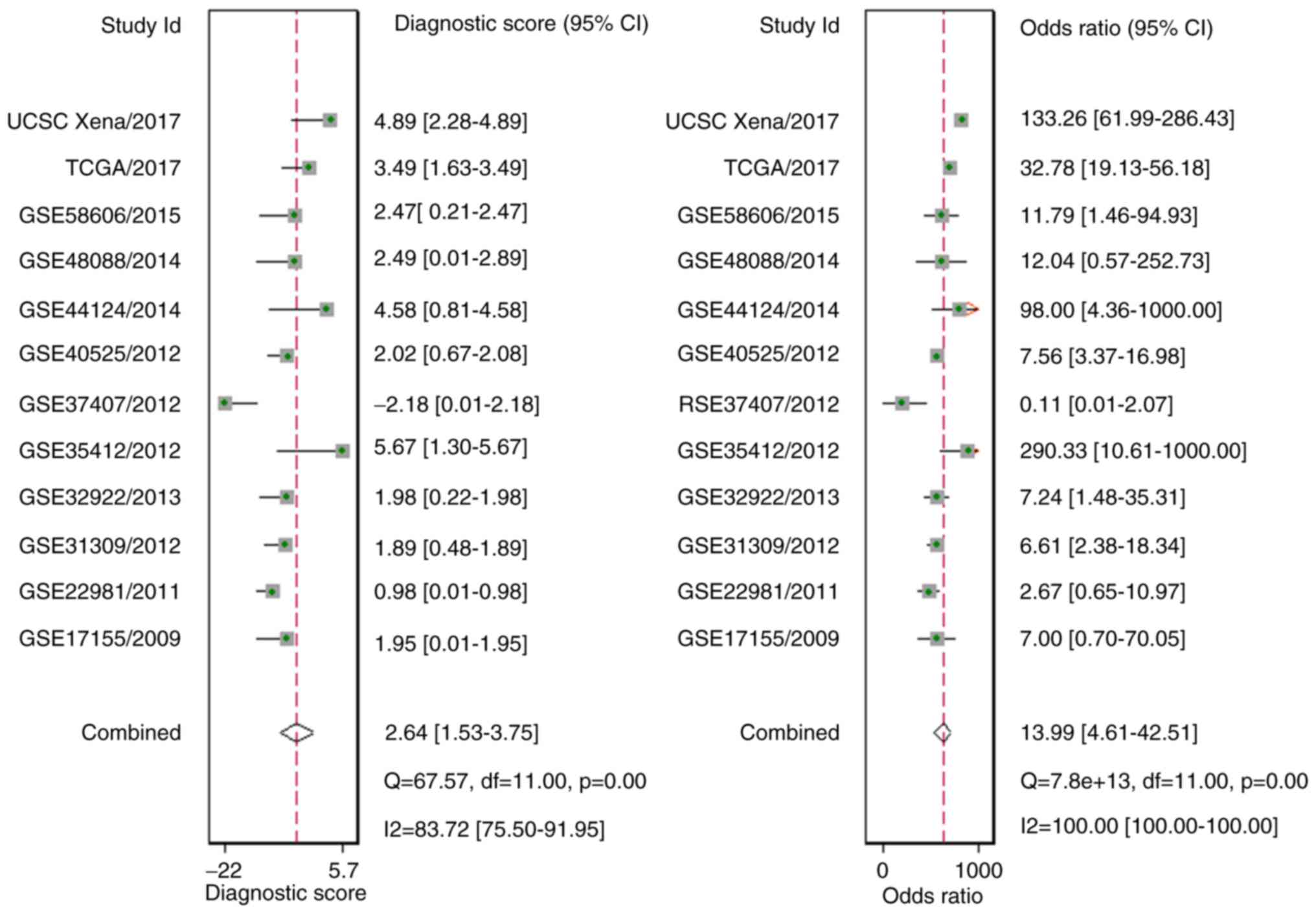
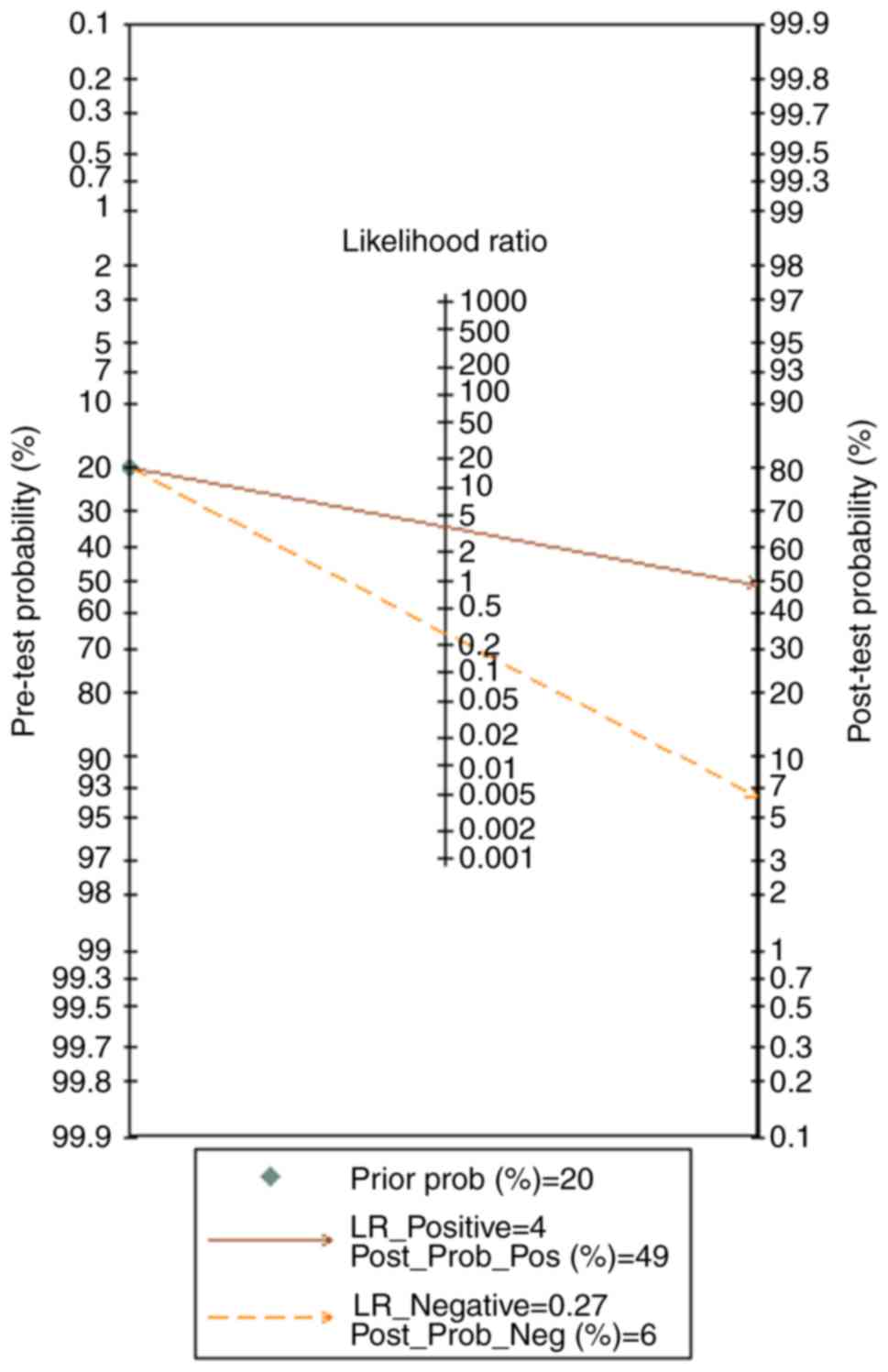
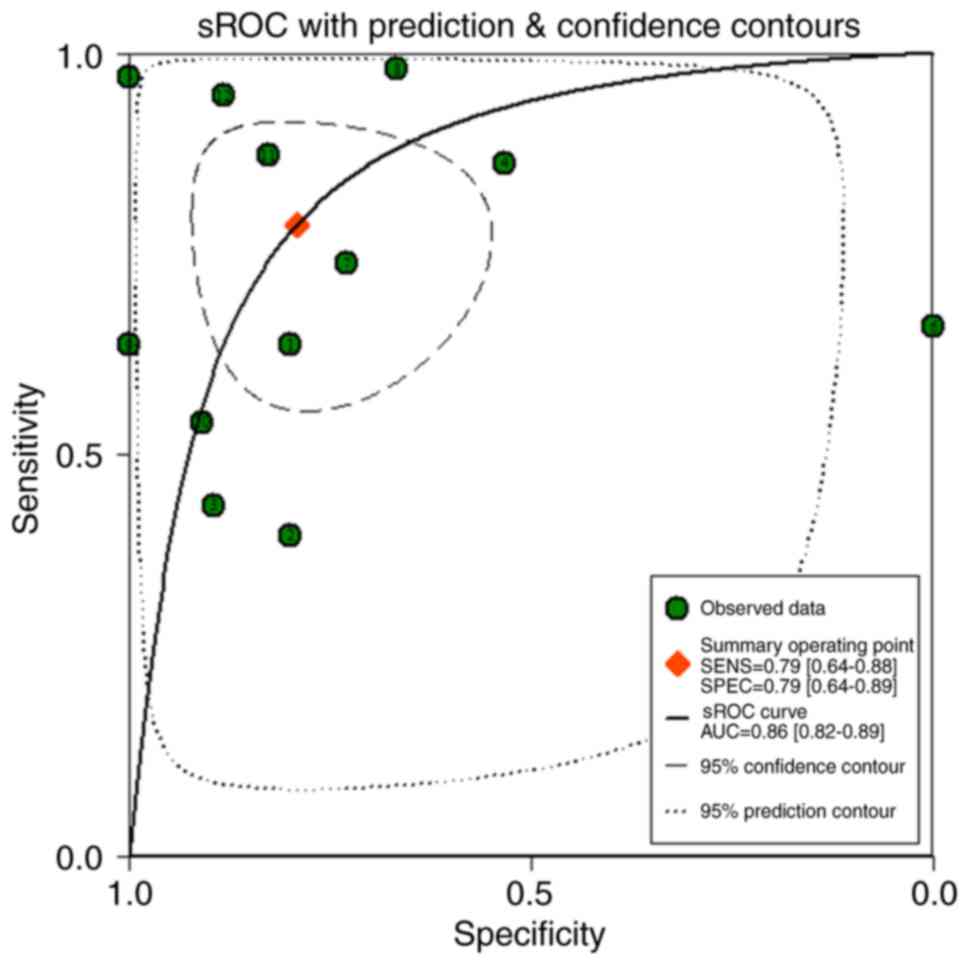
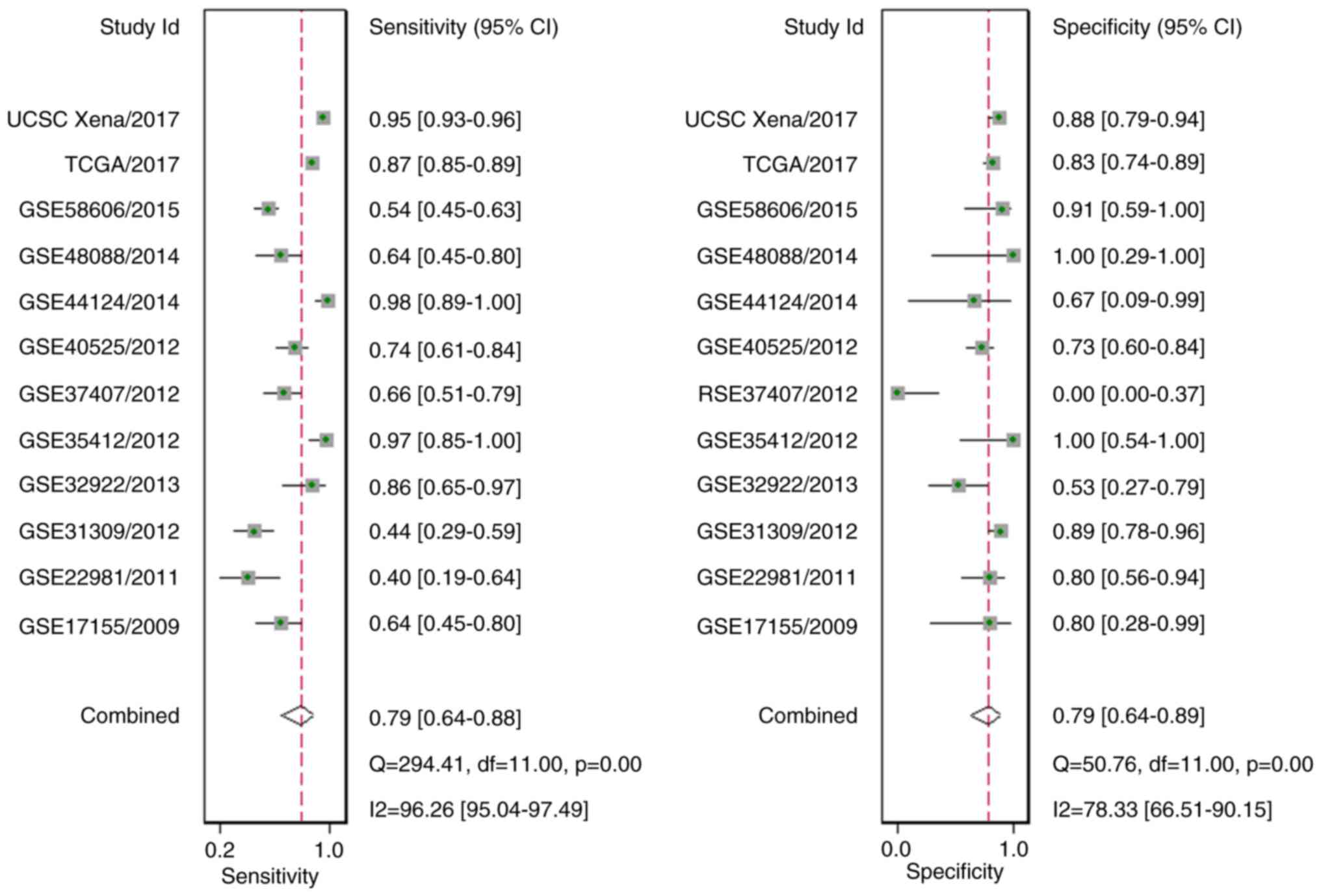
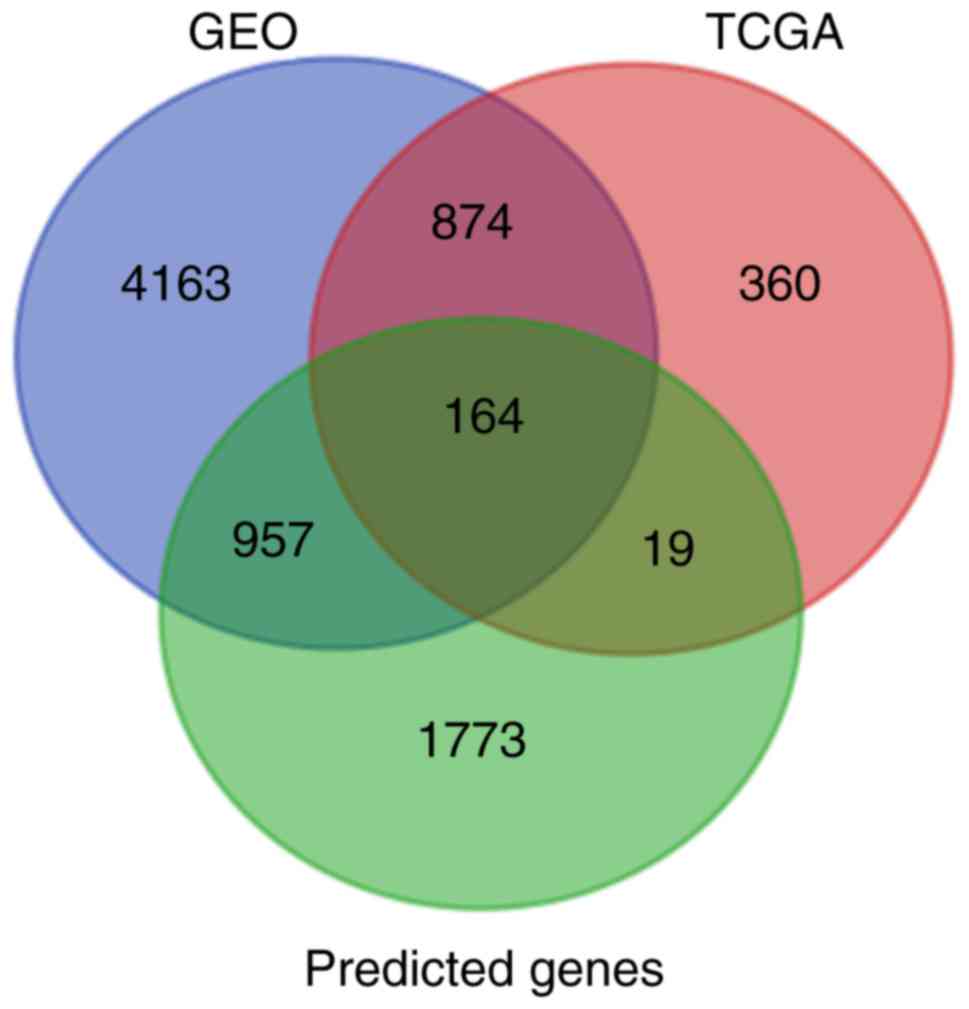
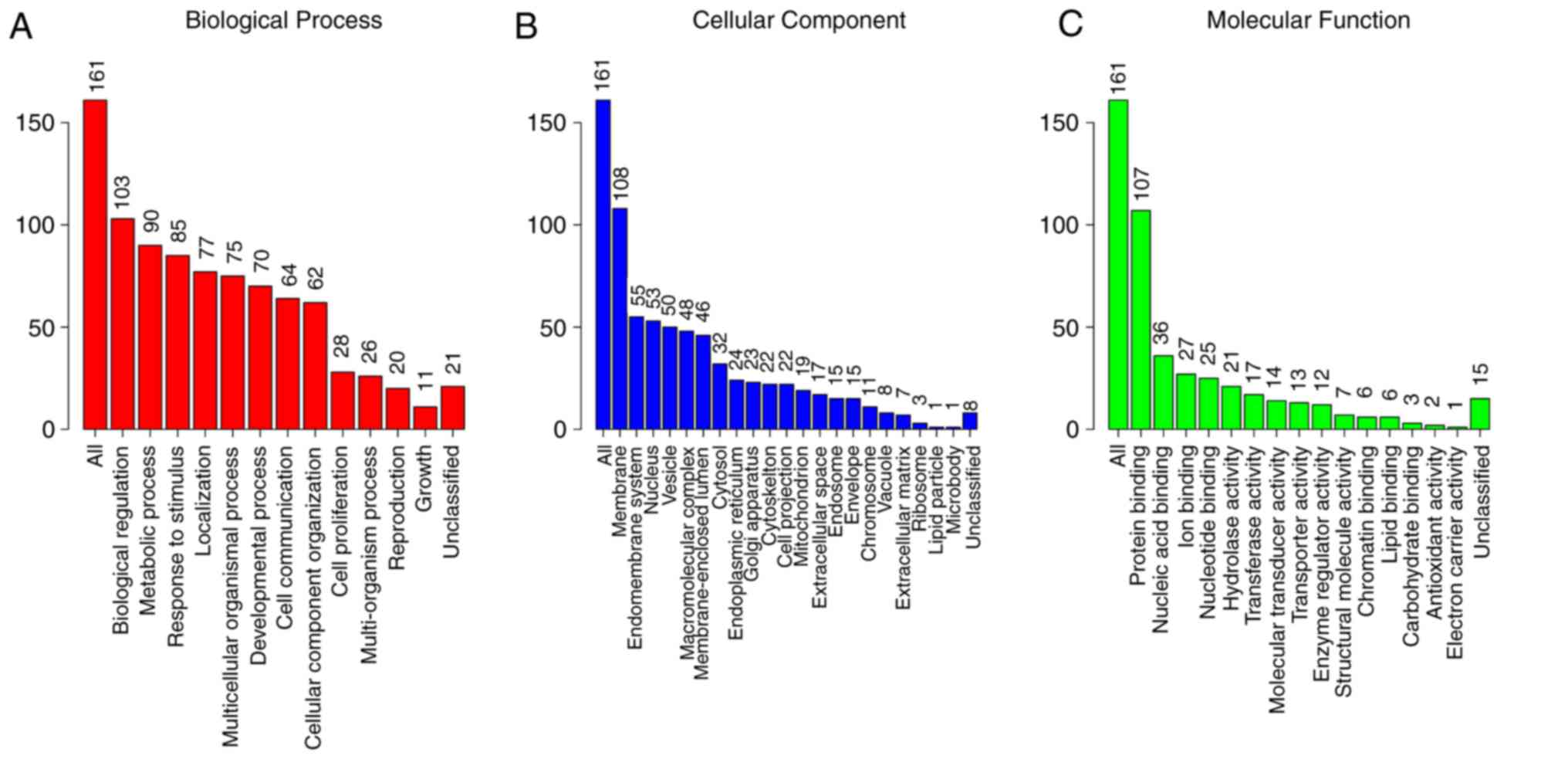
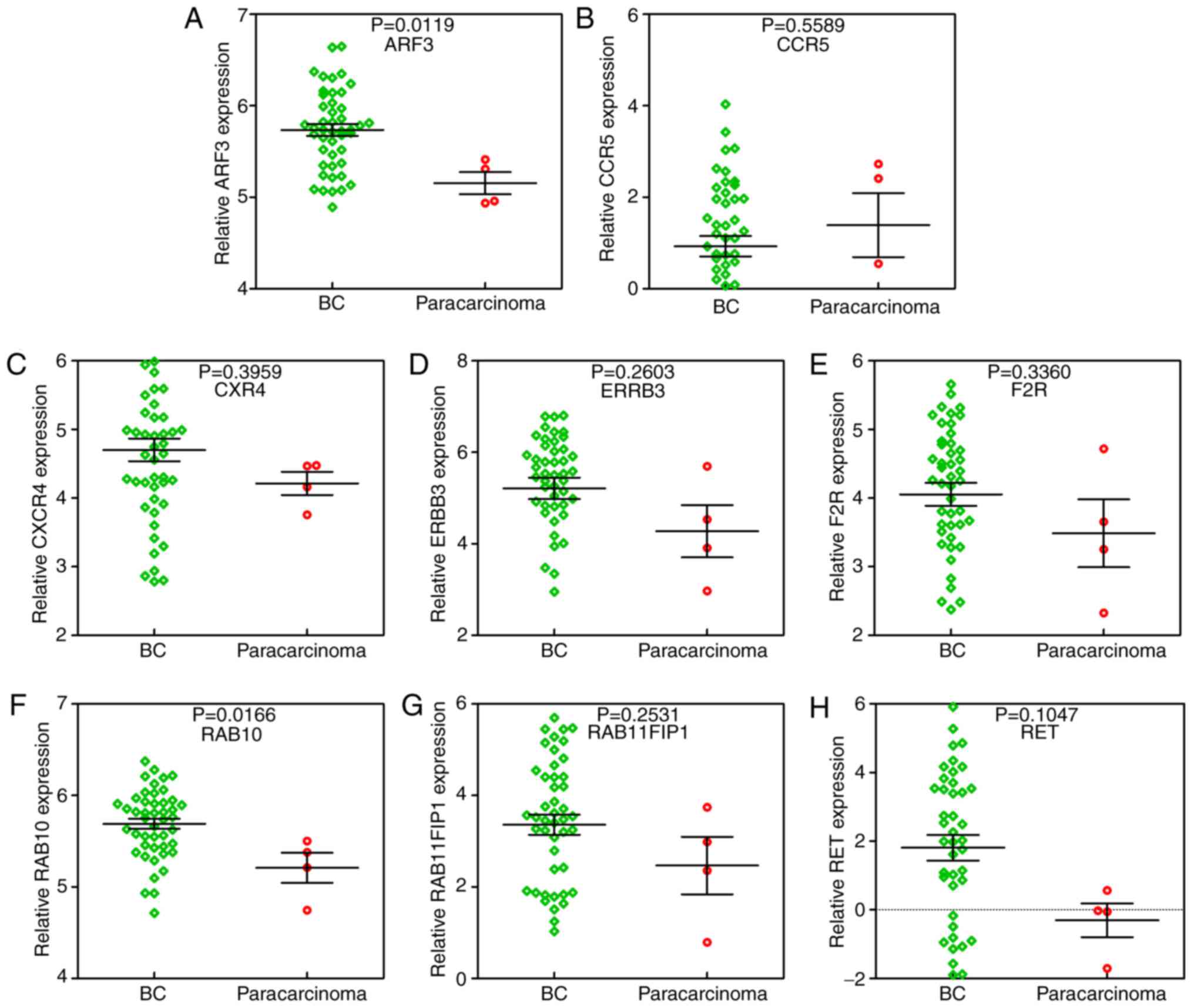
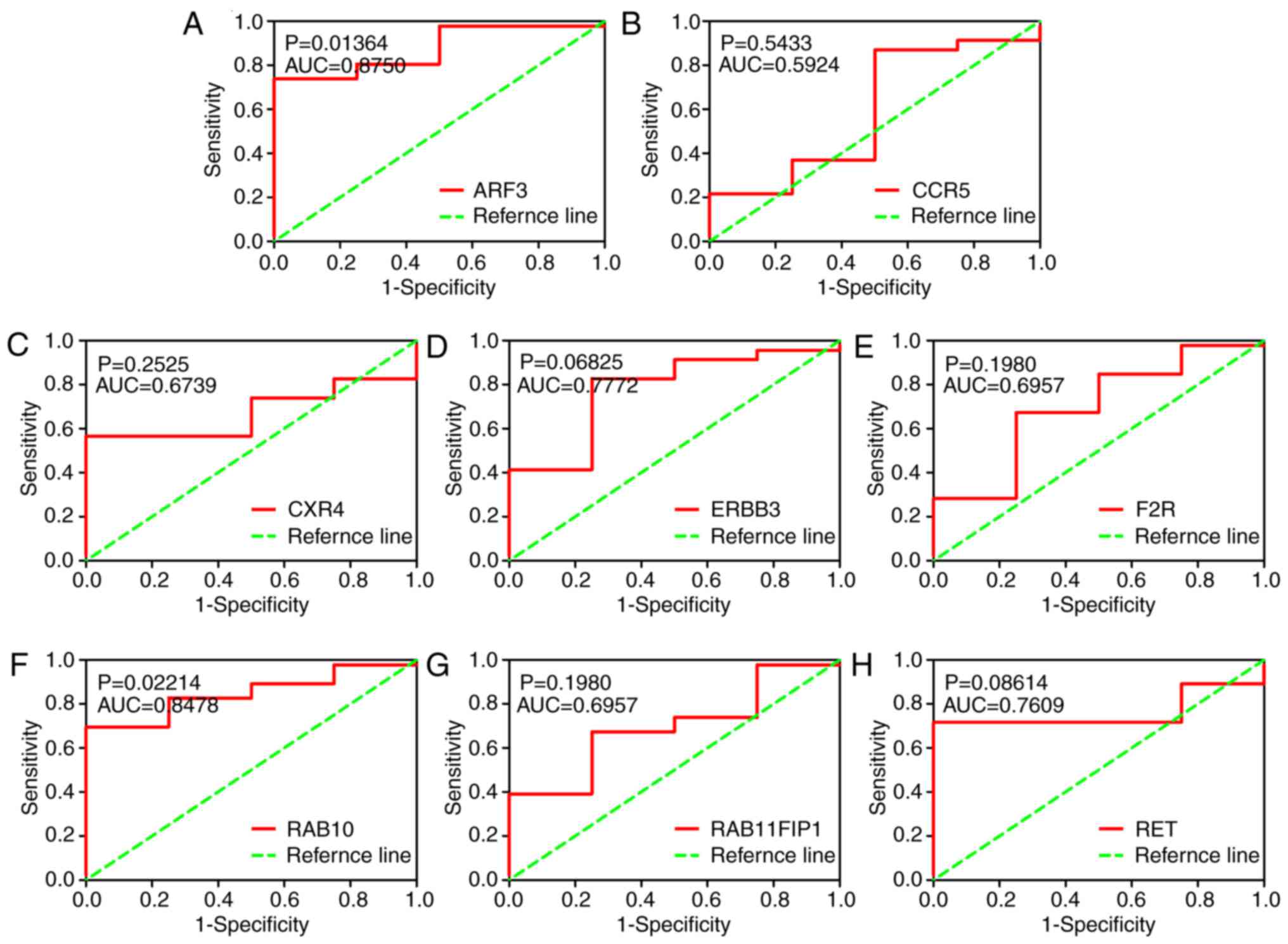
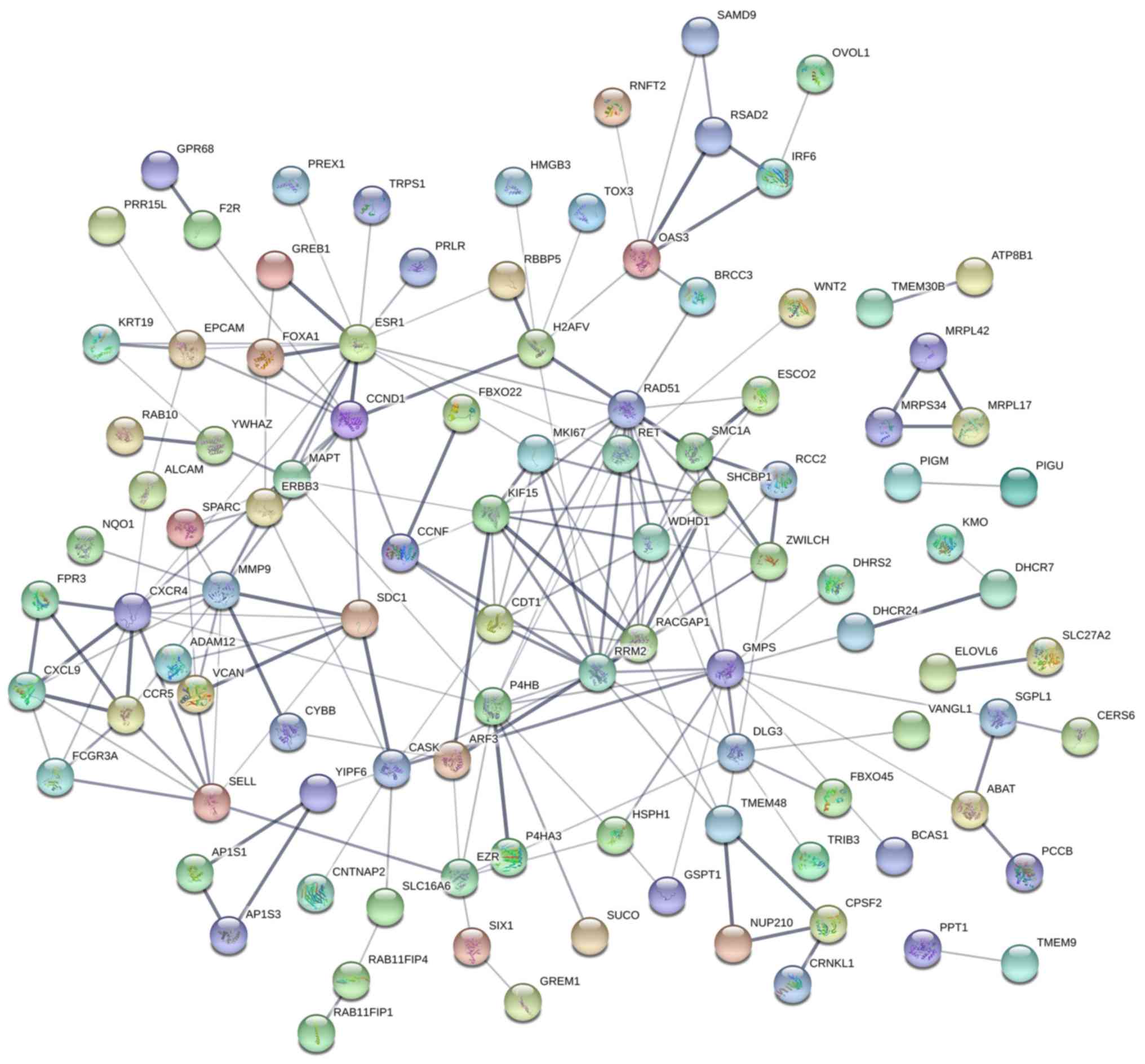
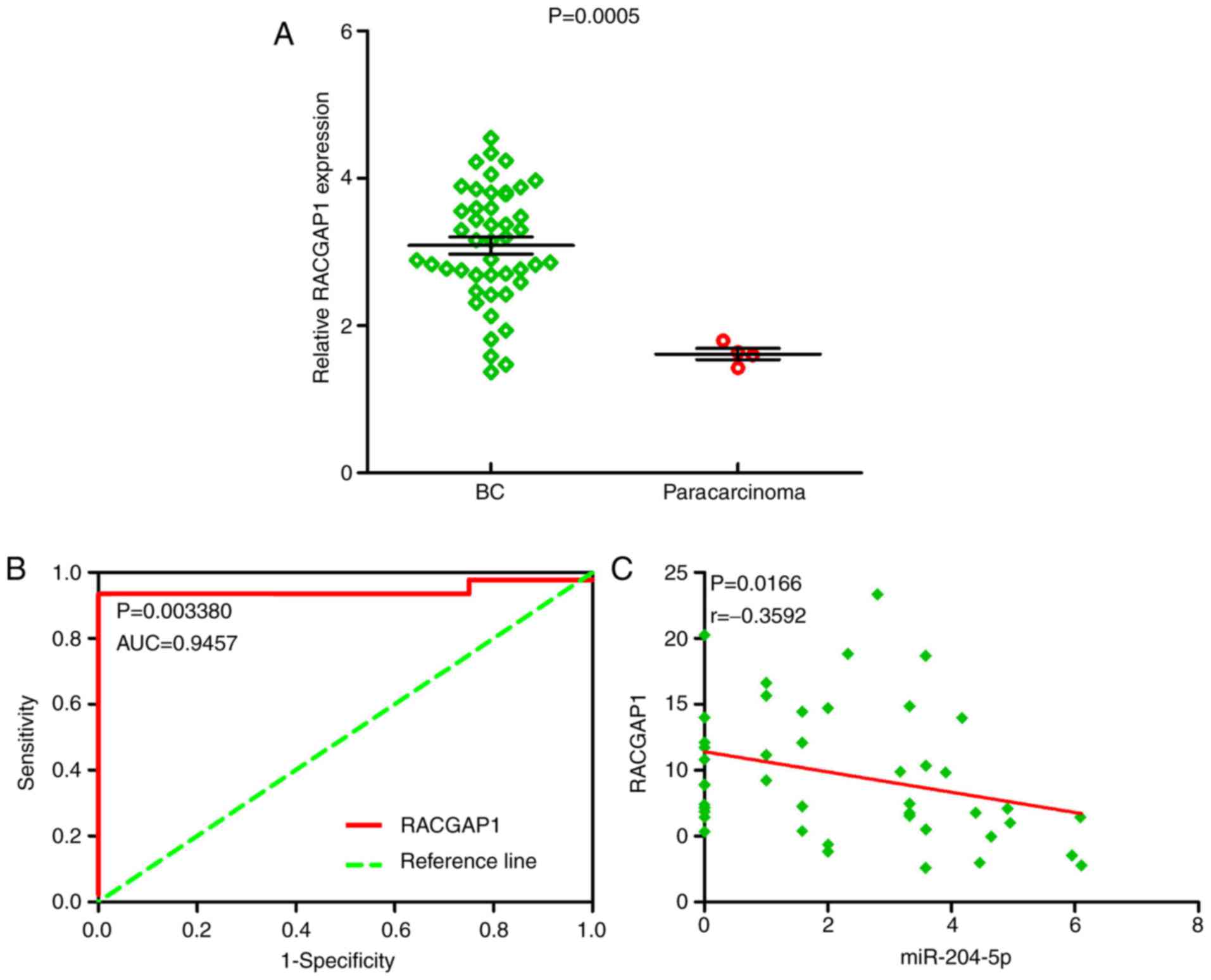
|
1
|
Sun W, Jiang YZ, Liu YR, Ma D and Shao ZM:
Nomograms to estimate long-term overall survival and breast
cancer-specific survival of patients with luminal breast cancer.
Oncotarget. 7:20496–20506. 2016.PubMed/NCBI
|
|
2
|
Samson M, Porter N, Orekoya O, Hebert JR,
Adams SA, Bennett CL and Steck SE: Progestin and breast cancer
risk: A systematic review. Breast Cancer Res Treat. 155:3–12. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chavez-MacGregor M, Clarke CA,
Lichtensztajn DY and Giordano SH: Delayed initiation of adjuvant
chemotherapy among patients with breast cancer. JAMA Oncol.
2:322–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Biglia N, D'Alonzo M, Sgro LG, Tomasi Cont
N, Bounous V and Robba E: Breast cancer treatment in mutation
carriers: Surgical treatment. Minerva Ginecol. 68:548–556.
2016.PubMed/NCBI
|
|
5
|
Monroy-Cisneros K, Esparza-Romero J,
Valencia ME, Guevara-Torres AG, Méndez-Estrada RO, Anduro-Corona I
and Astiazarán-García H: Antineoplastic treatment effect on bone
mineral density in Mexican breast cancer patients. BMC Cancer.
16:8602016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Frasier LL, Holden S, Holden T, Schumacher
JR, Leverson G, Anderson B, Greenberg CC and Neuman HB: Temporal
trends in postmastectomy radiation therapy and breast
reconstruction associated with changes in national comprehensive
cancer network guidelines. JAMA Oncol. 2:95–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu L, Peng S, Huang Q, Liu Y, Jiang H, Li
X and Wang J: Expression status of cyclaseassociated protein 2 as a
prognostic marker for human breast cancer. Oncol Rep. 36:1981–1988.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang X, Li X and Xie X, Ye F, Chen B,
Song C, Tang H and Xie X: High expressions of LDHA and AMPK as
prognostic biomarkers for breast cancer. Breast. 30:39–46. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Wu J, Yuan J, Zhu X, Wu H and Li
M: Midline2 is overexpressed and a prognostic indicator in human
breast cancer and promotes breast cancer cell proliferation in
vitro and in vivo. Front Med. 10:41–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iravani O, Yip GW, Thike AA, Chua PJ, Jane
Scully O, Tan PH and Bay BH: Prognostic significance of Claudin 12
in estrogen receptor-negative breast cancer. J Clin Pathol.
69:878–883. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Q, Zhao GS, Yuan XL, Li XH, Yang Z,
Cui YF, Guan QL, Sun XY, Shen W, Xu TA and Wang QS: Tumor necrosis
factor alpha-238G/A polymorphism and risk of breast cancer: An
update by meta-analysis. Medicine (Baltimore). 96:e74422017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baretta Z, Mocellin S, Goldin E, Olopade
OI and Huo D: Effect of BRCA germline mutations on breast cancer
prognosis: A systematic review and meta-analysis. Medicine
(Baltimore). 95:e49752016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y, Song Y and Zhu X: MicroRNA-181a
regulates apoptosis and autophagy process in Parkinson's disease by
inhibiting p38 mitogen-activated protein kinase (MAPK)/c-Jun
N-terminal kinases (JNK) signaling pathways. Med Sci Monit.
23:1597–1606. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Erbes T, Hirschfeld M, Rücker G, Jaeger M,
Boas J, Iborra S, Mayer S, Gitsch G and Stickeler E: Feasibility of
urinary microRNA detection in breast cancer patients and its
potential as an innovative non-invasive biomarker. BMC Cancer.
15:1932015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mesci A, Huang X, Taeb S, Jahangiri S, Kim
Y, Fokas E, Bruce J, Leong HS and Liu SK: Targeting of CCBE1 by
miR-330-3p in human breast cancer promotes metastasis. Br J Cancer.
116:1350–1357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song L, Zhang W, Chang Z, Pan Y, Zong H,
Fan Q and Wang L: miR-4417 targets tripartite motif-containing 35
(TRIM35) and regulates pyruvate kinase muscle 2 (PKM2)
phosphorylation to promote proliferation and suppress apoptosis in
hepatocellular carcinoma cells. Med Sci Monit. 23:1741–1750. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Q, He M, Guan S, Ma M, Wu H, Yu Z,
Jiang L, Wang Y, Zong X, Jin F and Wei M: MicroRNA-100 suppresses
the migration and invasion of breast cancer cells by targeting
FZD-8 and inhibiting Wnt/β-catenin signaling pathway. Tumour Biol.
37:5001–5011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Wang YW, Xing AY, Xiang S, Shi DB,
Liu L, Li YX and Gao P: Suppression of SPIN1-mediated PI3K-Akt
pathway by miR-489 increases chemosensitivity in breast cancer. J
Pathol. 239:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Lin S, Tseng KF, Han K, Wang Y,
Gan ZH, Min DL and Hu HY: Selumetinib suppresses cell
proliferation, migration and trigger apoptosis, G1 arrest in
triple-negative breast cancer cells. BMC Cancer. 16:8182016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia M, Li H, Wang JJ, Zeng HJ and Wang SH:
MiR-99a suppress proliferation, migration and invasion through
regulating insulin-like growth factor 1 receptor in breast cancer.
Eur Rev Med Pharmacol Sci. 20:1755–1763. 2016.PubMed/NCBI
|
|
22
|
Zhan Y, Liang X, Li L, Wang B, Ding F, Li
Y, Wang X, Zhan Q and Liu Z: MicroRNA-548j functions as a
metastasis promoter in human breast cancer by targeting Tensin1.
Mol Oncol. 10:838–849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mohammadi-Yeganeh S, Paryan M, Arefian E,
Vasei M, Ghanbarian H, Mahdian R, Karimipoor M and Soleimani M:
MicroRNA-340 inhibits the migration, invasion, and metastasis of
breast cancer cells by targeting Wnt pathway. Tumour Biol.
37:8993–9000. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo YH, Tang W, Zhang X, Tan Z, Guo WL,
Zhao N, Pang SM, Dang YW, Rong MH and Cao J: Promising significance
of the association of miR-204-5p expression with
clinicopathological features of hepatocellular carcinoma. Medicine
(Baltimore). 96:e75452017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao W, Wu Y, He X, Zhang C, Zhu M, Chen B,
Liu Q, Qu X, Li W, Wen S and Wang B: MicroRNA-204-5p inhibits
invasion and metastasis of laryngeal squamous cell carcinoma by
suppressing forkhead box C1. J Cancer. 8:2356–2368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J,
Ruan H, Ma S and Xu B: miR-204-5p acts as a tumor suppressor by
targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in
malignant melanoma. Onco Targets Ther. 10:1237–1246. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Li F and Zhou X: miR-204-5p
regulates cell proliferation and metastasis through inhibiting
CXCR4 expression in OSCC. Biomed Pharmacother. 82:202–207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu L, Wang J, Li X, Ma J, Shi C, Zhu H,
Xi Q, Zhang J, Zhao X and Gu M: MiR-204-5p suppresses cell
proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma.
Biochem Biophys Res Commun. 457:621–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li T, Pan H and Li R: The dual regulatory
role of miR-204 in cancer. Tumour Biol. 37:11667–11677. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Li G, Luo Q, Xie J and Gan C:
Integrated TCGA analysis implicates lncRNA CTB-193M12.5 as a
prognostic factor in lung adenocarcinoma. Cancer Cell Int.
18:272018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao K, Li Z and Tian H: Twenty-gene-based
prognostic model predicts lung adenocarcinoma survival. Onco
Targets Ther. 11:3415–3424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Q, Zhao G, Yang Z, Liu X and Xie P:
Downregulation of microRNA1243p suppresses the mTOR signaling
pathway by targeting DDIT4 in males with major depressive disorder.
Int J Mol Med. 41:493–500. 2018.PubMed/NCBI
|
|
34
|
Shang J, Wang F, Chen P, Wang X, Ding F,
Liu S and Zhao Q: Co-expression network analysis identified COL8A1
is associated with the progression and prognosis in human colon
adenocarcinoma. Dig Dis Sci. 63:1219–1228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Su L, Wang C, Zheng C, Wei H and Song X: A
meta-analysis of public microarray data identifies biological
regulatory networks in Parkinson's disease. BMC Med Genomics.
11:402018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu W, Li J and Wu B: Gene expression
profiling of the mouse gut: Effect of intestinal flora on
intestinal health. Mol Med Rep. 17:3667–3673. 2018.PubMed/NCBI
|
|
38
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun W, Qiu Z, Huang W and Cao M: Gene
expression profiles and proteinprotein interaction networks during
tongue carcinogenesis in the tumor microenvironment. Mol Med Rep.
17:165–171. 2018.PubMed/NCBI
|
|
40
|
Liu J, Li H, Sun L, Wang Z, Xing C and
Yuan Y: Aberrantly methylated-differentially expressed genes and
pathways in colorectal cancer. Cancer Cell Int. 17:752017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Y, Teng L, Liu W, Cao Y, Ding D, Wang
W, Chen H, Li C and An R: Identification of biological targets of
therapeutic intervention for clear cell renal cell carcinoma based
on bioinformatics approach. Cancer Cell Int. 16:162016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fassan M, Baffa R, Palazzo JP, Lloyd J,
Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM and
Rosenberg A: MicroRNA expression profiling of male breast cancer.
Breast Cancer Res. 11:R582009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao H, Shen J, Medico L, Wang D,
Ambrosone CB and Liu S: A pilot study of circulating miRNAs as
potential biomarkers of early stage breast cancer. PLoS One.
5:e137352010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7:e297702012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tanic M, Gómez-López G, Benítez J and
Martínez-Delgado B: Comparison between hereditary breast tumors and
normal breast tissue samples.
|
|
46
|
Romero-Cordoba S, Rodriguez-Cuevas S,
Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez
G, Bautista-Piña V, Arellano-Llamas R and Hidalgo-Miranda A:
Identification and pathway analysis of microRNAs with no previous
involvement in breast cancer. PLoS One. 7:e319042012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gravgaard KH, Lyng MB, Laenkholm AV,
Søkilde R, Nielsen BS, Litman T and Ditzel HJ: The miRNA-200 family
and miRNA-9 exhibit differential expression in primary versus
corresponding metastatic tissue in breast cancer. Breast Cancer Res
Treat. 134:207–217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Biagioni F, Bossel Ben-Moshe N, Fontemaggi
G, Canu V, Mori F, Antoniani B, Di Benedetto A, Santoro R, Germoni
S, De Angelis F, et al: miR-10b*, a master inhibitor of the cell
cycle, is down-regulated in human breast tumours. EMBO Mol Med.
4:1214–1229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Feliciano A, Castellvi J, Artero-Castro A,
Leal JA, Romagosa C, Hernández-Losa J, Peg V, Fabra A, Vidal F,
Kondoh H, et al: miR-125b acts as a tumor suppressor in breast
tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and
MEGF9. PLoS One. 8:e762472013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pena-Chilet M, Martínez MT, Pérez-Fidalgo
JA, Peiró-Chova L, Oltra SS, Tormo E, Alonso-Yuste E,
Martinez-Delgado B, Eroles P, Climent J, et al: MicroRNA profile in
very young women with breast cancer. BMC Cancer. 14:5292014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Matamala N, Vargas MT, Gonzalez-Campora R,
Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G,
Yanowsky K, Calvete-Candenas J, et al: Tumor microRNA expression
profiling identifies circulating microRNAs for early breast cancer
detection. Clin Chem. 61:1098–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang S, Gao L, Thakur A, Shi P, Liu F,
Feng J, Wang T, Liang Y, Liu JJ, Chen M and Ren H: miRNA-204
suppresses human non-small cell lung cancer by targeting ATF2.
Tumour Biol. 37:11177–11186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu Z, Long J, Du R, Ge C, Guo K and Xu Y:
miR-204 regulates the EMT by targeting snai1 to suppress the
invasion and migration of gastric cancer. Tumour Biol.
37:8327–8335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu G, Wang J, Chen G and Zhao X:
microRNA-204 modulates chemosensitivity and apoptosis of prostate
cancer cells by targeting zinc-finger E-box-binding homeobox 1
(ZEB1). Am J Transl Res. 9:3599–3610. 2017.PubMed/NCBI
|
|
55
|
Sun Y, Yu X and Bai Q: miR-204 inhibits
invasion and epithelial-mesenchymal transition by targeting FOXM1
in esophageal cancer. Int J Clin Exp Pathol. 8:12775–12783.
2015.PubMed/NCBI
|
|
56
|
Wang X, Qiu W, Zhang G, Xu S, Gao Q and
Yang Z: MicroRNA-204 targets JAK2 in breast cancer and induces cell
apoptosis through the STAT3/BCl-2/survivin pathway. Int J Clin Exp
Pathol. 8:5017–5025. 2015.PubMed/NCBI
|
|
57
|
Shen SQ, Huang LS, Xiao XL, Zhu XF, Xiong
DD, Cao XM, Wei KL, Chen G and Feng ZB: miR-204 regulates the
biological behavior of breast cancer MCF-7 cells by directly
targeting FOXA1. Oncol Rep. 38:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li W, Jin X, Zhang Q, Zhang G, Deng X and
Ma L: Decreased expression of miR-204 is associated with poor
prognosis in patients with breast cancer. Int J Clin Exp Pathol.
7:3287–3292. 2014.PubMed/NCBI
|
|
59
|
Ye ZH, Wen DY, Cai XY, Liang L, Wu PR, Qin
H, Yang H, He Y and Chen G: The protective value of miR-204-5p for
prognosis and its potential gene network in various malignancies: A
comprehensive exploration based on RNA-seq high-throughput data and
bioinformatics. Oncotarget. 8:104960–104980. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Flores-Peréz A, Marchat LA,
Rodriguez-Cuevas S, Bautista-Piña V, Hidalgo-Miranda A, Ocampo EA,
Martínez MS, Palma-Flores C, Fonseca-Sánchez MA, Astudillo-de la
Vega H, et al: Dual targeting of ANGPT1 and TGFBR2 genes by miR-204
controls angiogenesis in breast cancer. Sci Rep. 6:345042016.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zeng J, Wei M, Shi R, Cai C, Liu X, Li T
and Ma W: MiR-204-5p/Six1 feedback loop promotes
epithelial-mesenchymal transition in breast cancer. Tumour Biol.
37:2729–2735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu J and Li Y: Trichostatin A and
Tamoxifen inhibit breast cancer cell growth by miR-204 and ERα
reducing AKT/mTOR pathway. Biochem Biophys Res Commun. 467:242–247.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee H, Lee S, Bae H, Kang HS and Kim SJ:
Genome-wide identification of target genes for miR-204 and miR-211
identifies their proliferation stimulatory role in breast cancer
cells. Sci Rep. 6:252872016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang L, Tian H, Yuan J, Wu H, Wu J and Zhu
X: CONSORT: Sam68 Is directly regulated by MiR-204 and promotes the
self-renewal potential of breast cancer cells by activating the
Wnt/Beta-catenin signaling pathway. Medicine (Baltimore).
94:e22282015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Saigusa S, Tanaka K, Mohri Y, Ohi M,
Shimura T, Kitajima T, Kondo S, Okugawa Y, Toiyama Y, Inoue Y and
Kusunoki M: Clinical significance of RacGAP1 expression at the
invasive front of gastric cancer. Gastric Cancer. 18:84–92. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Imaoka H, Toiyama Y, Saigusa S, Kawamura
M, Kawamoto A, Okugawa Y, Hiro J, Tanaka K, Inoue Y, Mohri Y and
Kusunoki M: RacGAP1 expression, increasing tumor malignant
potential, as a predictive biomarker for lymph node metastasis and
poor prognosis in colorectal cancer. Carcinogenesis. 36:346–354.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lawson CD and Der CJ: Filling GAPs in our
knowledge: ARHGAP11A and RACGAP1 act as oncogenes in basal-like
breast cancers. Small GTPases. 9:290–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sahin S, Işık Gönül İ, Çakır A, Seçkin S
and Uluoğlu Ö: Clinicopathological significance of the
proliferation markers Ki67, RacGAP1, and topoisomerase 2 alpha in
breast cancer. Int J Surg Pathol. 24:607–613. 2016. View Article : Google Scholar : PubMed/NCBI
|