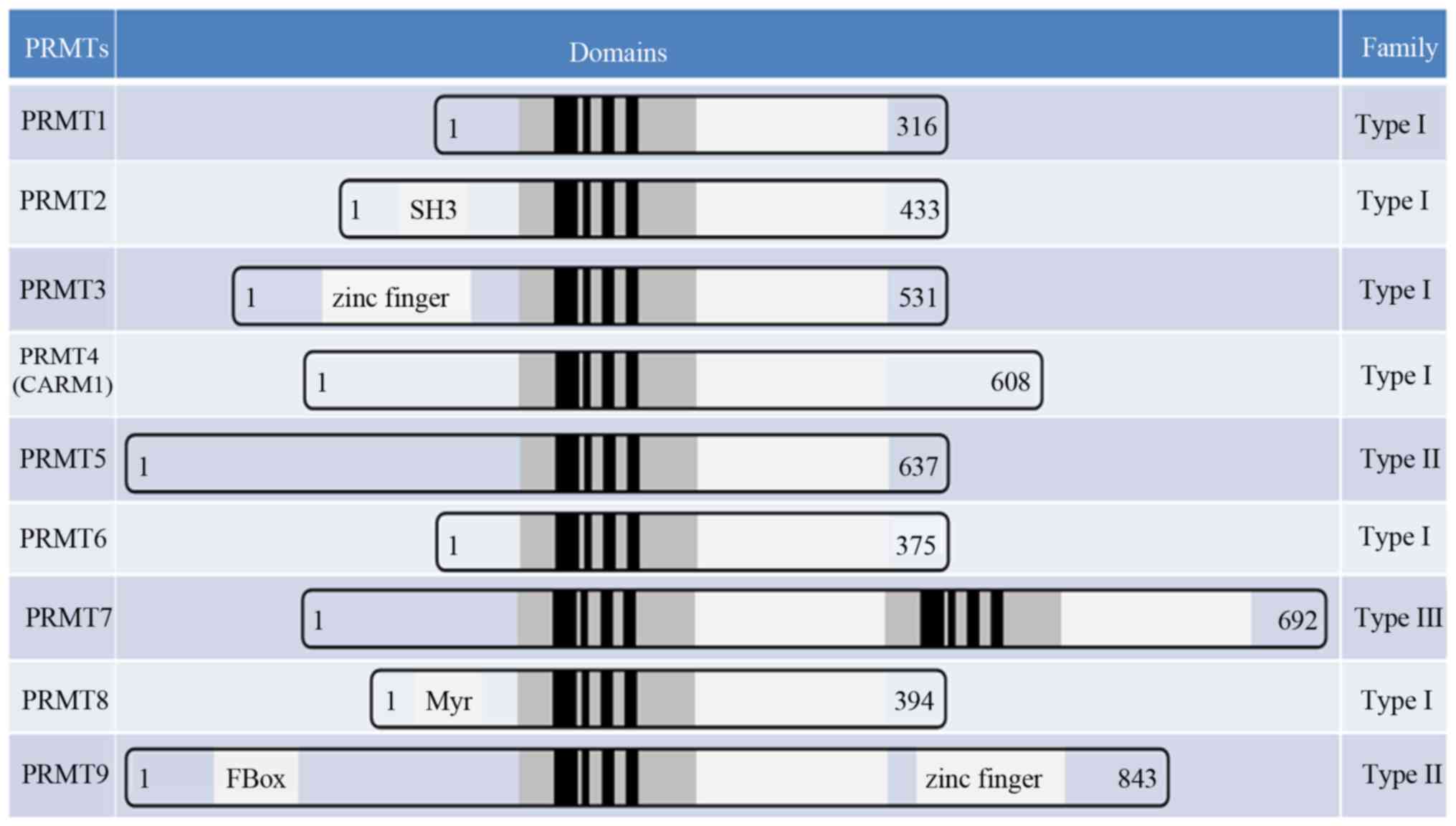
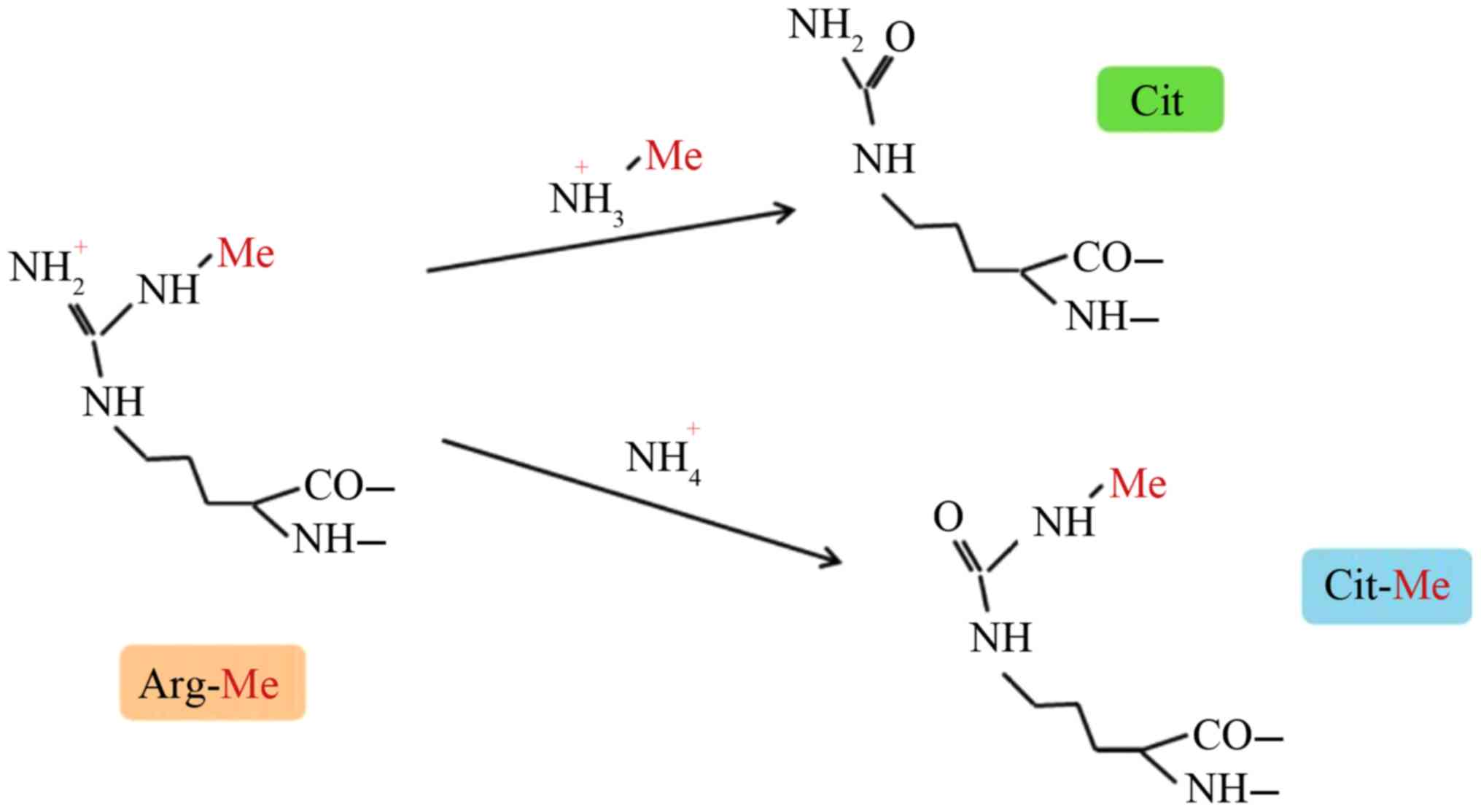
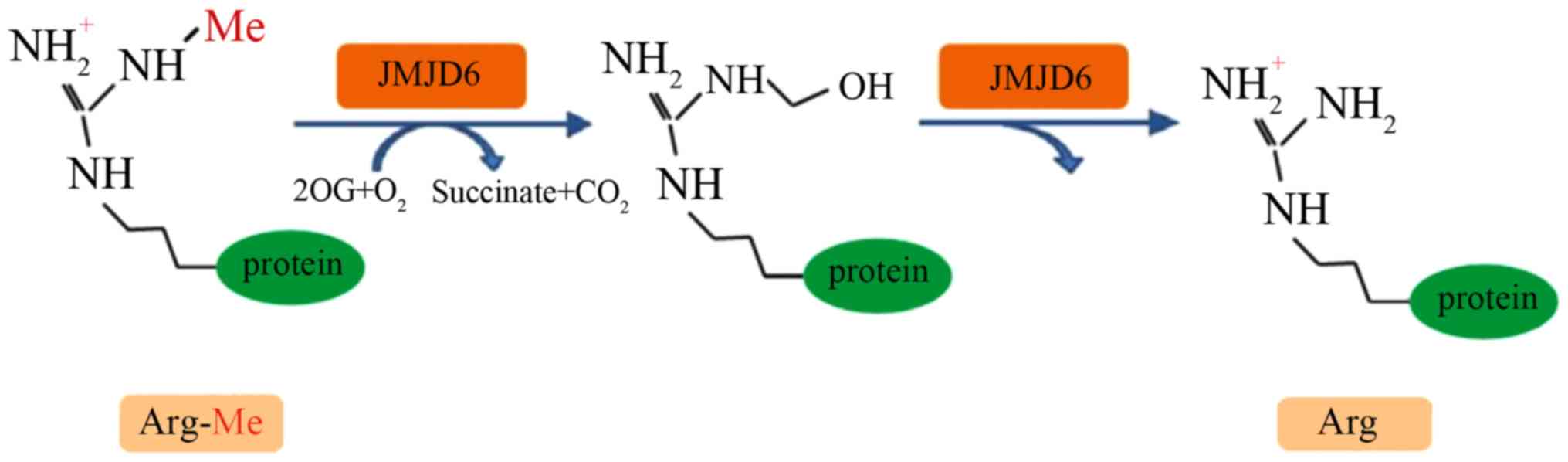
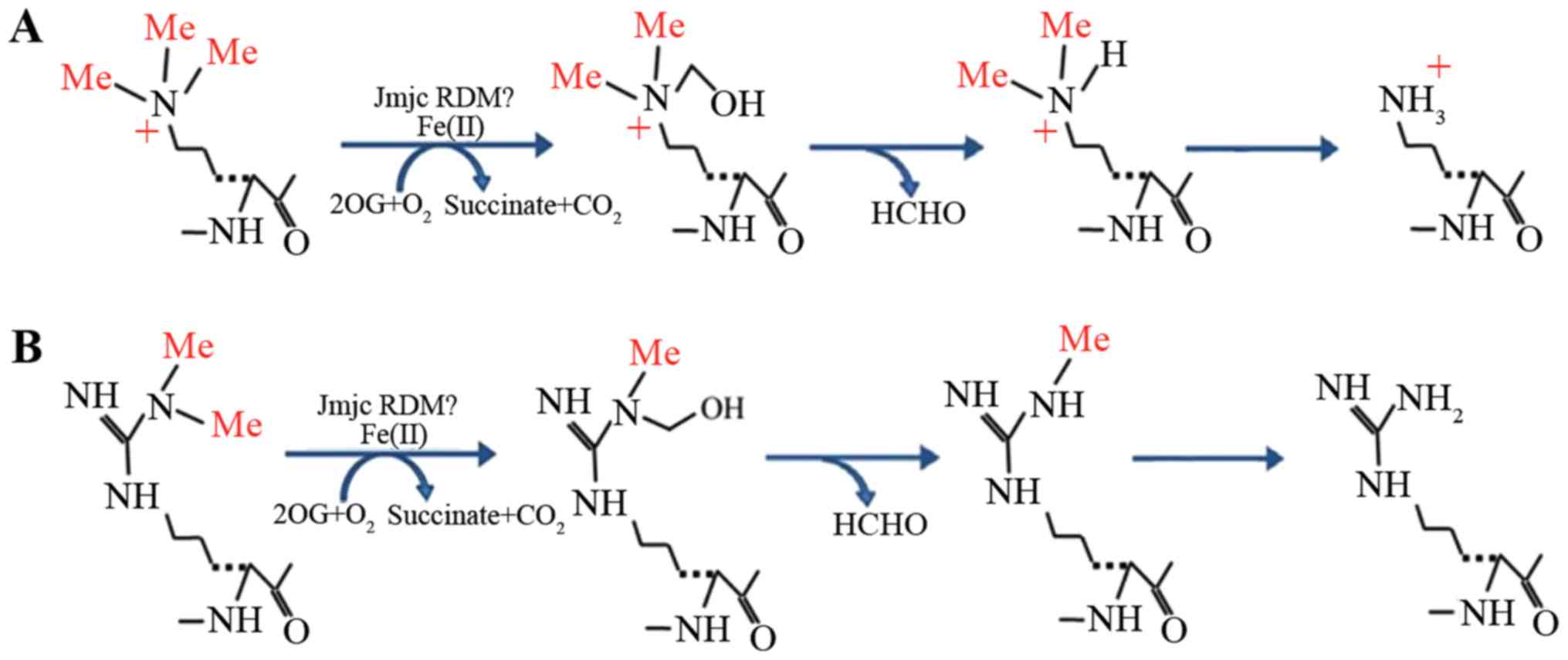
|
1
|
Bedford MT and Richard S: Arginine
methylation an emerging regulator of protein function. Mol Cell.
18:263–272. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rothbart SB and Strahl BD: Interpreting
the language of histone and DNA modifications. Biochim Biophys
Acta. 1839:627–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boisvert FM, Chénard CA and Richard S:
Protein interfaces in signaling regulated by arginine methylation.
Sci STKE. 2005:re22005.PubMed/NCBI
|
|
4
|
Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang
S and Bedford MT: TDRD3 is an effector molecule for
arginine-methylated histone marks. Mol Cell. 40:1016–1023. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gao Y, Zhao Y, Zhang J, Lu Y, Liu X, Geng
P, Huang B, Zhang Y and Lu J: The dual function of PRMT1 in
modulating epithelial-mesenchymal transition and cellular
senescence in breast cancer cells through regulation of ZEB1. Sci
Rep. 6:198742016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pal S, Baiocchi RA, Byrd JC, Grever MR,
Jacob ST and Sif S: Low levels of miR-92b/96 induce PRMT5
translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO
J. 26:3558–3569. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karkhanis V, Wang L, Tae S, Hu YJ,
Imbalzano AN and Sif S: Protein arginine methyltransferase 7
regulates cellular response to DNA damage by methylating promoter
histones H2A and H4 of the polymerase δ catalytic subunit gene,
POLD1. J Biol Chem. 287:29801–29814. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Frietze S, Lupien M, Silver PA and Brown
M: CARM1 regulates estrogen-stimulated breast cancer growth through
up-regulation of E2F1. Cancer Res. 68:301–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pal S, Vishwanath SN, Erdjument-Bromage H,
Tempst P and Sif S: Human SWI/SNF-associated PRMT5 methylates
histone H3 arginine 8 and negatively regulates expression of ST7
and NM23 tumor suppressor genes. Mol Cell Biol. 24:9630–9645. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mosammaparast N and Shi Y: Reversal of
histone methylation: Biochemical and molecular mechanisms of
histone demethylases. Annu Rev Biochem. 79:155–179. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bedford MT and Clarke SG: Protein arginine
methylation in mammals: Who, what, and why. Mol Cell. 33:1–13.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gary JD and Clarke S: RNA and protein
interactions modulated by protein arginine methylation. Prog
Nucleic Acid Res Mol Biol. 61:65–131. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bedford MT: Arginine methylation at a
glance. J Cell Sci. 120:4243–4246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee J and Bedford MT: PABP1 identified as
an arginine methyltransferase substrate using high-density protein
arrays. EMBO Rep. 3:268–273. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu MC: The Role of Protein Arginine
Methylation in mRNP Dynamics. Mol Biol Int. 2011:163827–163836.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nishioka K and Reinberg D: Methods and
tips for the purification of human histone methyltransferases.
Methods. 31:49–58. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gonsalvez GB, Tian L, Ospina JK, Boisvert
FM, Lamond AI and Matera AG: Two distinct arginine
methyltransferases are required for biogenesis of Sm-class
ribonucleoproteins. J Cell Biol. 178:733–740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei H, Mundade R, Lange KC and Lu T:
Protein arginine methylation of non-histone proteins and its role
in diseases. Cell Cycle. 13:32–41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsai WC, Gayatri S, Reineke LC, Sbardella
G, Bedford MT and Lloyd RE: Arginine Demethylation of G3BP1
Promotes Stress Granule Assembly. J Biol Chem. 291:22671–22685.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen EC, Henry MF, Weiss VH, Valentini SR,
Silver PA and Lee MS: Arginine methylation facilitates the nuclear
export of hnRNP proteins. Genes Dev. 12:679–691. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goulet I, Gauvin G, Boisvenue S and Côté
J: Alternative splicing yields protein arginine methyltransferase 1
isoforms with distinct activity, substrate specificity, and
subcellular localization. J Biol Chem. 282:33009–33021. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zou L, Zhang H, Du C, Liu X, Zhu S, Zhang
W, Li Z, Gao C, Zhao X, Mei M, et al: Correlation of SRSF1 and
PRMT1 expression with clinical status of pediatric acute
lymphoblastic leukemia. J Hematol Oncol. 5:422012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Elakoum R, Gauchotte G, Oussalah A,
Wissler MP, Clément-Duchêne C, Vignaud JM, Guéant JL and Namour F:
CARM1 and PRMT1 are dysregulated in lung cancer without
hierarchical features. Biochimie. 97:210–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shia WJ, Okumura AJ, Yan M, Sarkeshik A,
Lo MC, Matsuura S, Komeno Y, Zhao X, Nimer SD, Yates JR III, et al:
PRMT1 interacts with AML1-ETO to promote its transcriptional
activation and progenitor cell proliferative potential. Blood.
119:4953–4962. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mathioudaki K, Papadokostopoulou A,
Scorilas A, Xynopoulos D, Agnanti N and Talieri M: The PRMT1 gene
expression pattern in colon cancer. Br J Cancer. 99:2094–2099.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mathioudaki K, Scorilas A, Ardavanis A,
Lymberi P, Tsiambas E, Devetzi M, Apostolaki A and Talieri M:
Clinical evaluation of PRMT1 gene expression in breast cancer.
Tumour Biol. 32:575–582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mathioudakis N and Salvatori R:
Adult-onset growth hormone deficiency: Causes, complications and
treatment options. Curr Opin Endocrinol Diabetes Obes. 15:352–358.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Papadokostopoulou A, Mathioudaki K,
Scorilas A, Xynopoulos D, Ardavanis A, Kouroumalis E and Talieri M:
Colon cancer and protein arginine methyltransferase 1 gene
expression. Anticancer Res. 29:1361–1366. 2009.PubMed/NCBI
|
|
29
|
Zhong J, Cao RX, Zu XY, Hong T, Yang J,
Liu L, Xiao XH, Ding WJ, Zhao Q, Liu JH, et al: Identification and
characterization of novel spliced variants of PRMT2 in breast
carcinoma. FEBS J. 279:316–335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhong J, Cao RX, Hong T, Yang J, Zu XY,
Xiao XH, Liu JH and Wen GB: Identification and expression analysis
of a novel transcript of the human PRMT2 gene resulted from
alternative polyadenylation in breast cancer. Gene. 487:1–9. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takahashi Y, Iwai M, Kawai T, Arakawa A,
Ito T, Sakurai-Yageta M, Ito A, Goto A, Saito M, Kasumi F, et al:
Aberrant expression of tumor suppressors CADM1 and 4.1B in invasive
lesions of primary breast cancer. Breast Cancer. 19:242–252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singh V, Miranda TB, Jiang W, Frankel A,
Roemer ME, Robb VA, Gutmann DH, Herschman HR, Clarke S and Newsham
IF: DAL-1/4.1B tumor suppressor interacts with protein arginine
N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate
substrates in vitro and in vivo. Oncogene.
23:7761–7771. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hong H, Kao C, Jeng MH, Eble JN, Koch MO,
Gardner TA, Zhang S, Li L, Pan CX, Hu Z, et al: Aberrant expression
of CARM1, a transcriptional coactivator of androgen receptor, in
the development of prostate carcinoma and androgen-independent
status. Cancer. 101:83–89. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Majumder S, Liu Y, Ford OH III, Mohler JL
and Whang YE: Involvement of arginine methyltransferase CARM1 in
androgen receptor function and prostate cancer cell viability.
Prostate. 66:1292–1301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim YR, Lee BK, Park RY, Nguyen NT, Bae
JA, Kwon DD and Jung C: Differential CARM1 expression in prostate
and colorectal cancers. BMC Cancer. 10:1972010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Habashy HO, Rakha EA, Ellis IO and Powe
DG: The oestrogen receptor coactivator CARM1 has an oncogenic
effect and is associated with poor prognosis in breast cancer.
Breast Cancer Res Treat. 140:307–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang L, Pal S and Sif S: Protein arginine
methyltransferase 5 suppresses the transcription of the RB family
of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol.
28:6262–6277. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bao X, Zhao S, Liu T, Liu Y, Liu Y and
Yang X: Overexpression of PRMT5 promotes tumor cell growth and is
associated with poor disease prognosis in epithelial ovarian
cancer. J Histochem Cytochem. 61:206–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Powers MA, Fay MM, Factor RE, Welm AL and
Ullman KS: Protein arginine methyltransferase 5 accelerates tumor
growth by arginine methylation of the tumor suppressor programmed
cell death 4. Cancer Res. 71:5579–5587. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoshimatsu M, Toyokawa G, Hayami S, Unoki
M, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, et
al: Dysregulation of PRMT1 and PRMT6, Type I arginine
methyltransferases, is involved in various types of human cancers.
Int J Cancer. 128:562–573. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Thomassen M, Tan Q and Kruse TA: Gene
expression meta-analysis identifies chromosomal regions and
candidate genes involved in breast cancer metastasis. Breast Cancer
Res Treat. 113:239–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yao R, Jiang H, Ma Y, Wang L, Wang L, Du
J, Hou P, Gao Y, Zhao L, Wang G, et al: PRMT7 induces
epithelial-to-mesenchymal transition and promotes metastasis in
breast cancer. Cancer Res. 74:5656–5667. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baldwin RM, Haghandish N, Daneshmand M,
Amin S, Paris G, Falls TJ, Bell JC, Islam S and Côté J: Protein
arginine methyltransferase 7 promotes breast cancer cell invasion
through the induction of MMP9 expression. Oncotarget. 6:3013–3032.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tee WW, Pardo M, Theunissen TW, Yu L,
Choudhary JS, Hajkova P and Surani MA: Prmt5 is essential for early
mouse development and acts in the cytoplasm to maintain ES cell
pluripotency. Genes Dev. 24:2772–2777. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pawlak MR, Scherer CA, Chen J, Roshon MJ
and Ruley HE: Arginine N-methyltransferase 1 is required for early
postimplantation mouse development, but cells deficient in the
enzyme are viable. Mol Cell Biol. 20:4859–4869. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu Z, Chen T, Hébert J, Li E and Richard
S: A mouse PRMT1 null allele defines an essential role for arginine
methylation in genome maintenance and cell proliferation. Mol Cell
Biol. 29:2982–2996. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Blanc RS and Richard S: Arginine
Methylation: The Coming of Age. Mol Cell. 65:8–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of JmjC domain-containing proteins. Nature.
439:811–816. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lee DY, Teyssier C, Strahl BD and Stallcup
MR: Role of protein methylation in regulation of transcription.
Endocr Rev. 26:147–170. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wysocka J, Allis CD and Coonrod S: Histone
arginine methylation and its dynamic regulation. Front Biosci.
11:344–355. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ng SS, Yue WW, Oppermann U and Klose RJ:
Dynamic protein methylation in chromatin biology. Cell Mol Life
Sci. 66:407–422. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chang B, Chen Y, Zhao Y and Bruick RK:
JMJD6 is a histone arginine demethylase. Science. 318:444–447.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li S, Ali S, Duan X, Liu S, Du J, Liu C,
Dai H, Zhou M, Zhou L, Yang L, et al: JMJD1B Demethylates H4R3me2s
and H3K9me2 to Facilitate Gene Expression for Development of
Hematopoietic Stem and Progenitor Cells. Cell Rep. 23:389–403.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hino S, Kohrogi K and Nakao M: Histone
demethylase LSD1 controls the phenotypic plasticity of cancer
cells. Cancer Sci. 107:1187–1192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shen HJ, Xu WQ and Lan F: Histone lysine
demethylases in mammalian embryonic development. Exp Mol Med.
49:e3252017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Vossenaar ER, Zendman AJ, van Venrooij WJ
and Pruijn GJ: PAD, a growing family of citrullinating enzymes:
Genes, features and involvement in disease. BioEssays.
25:1106–1118. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nakashima K, Hagiwara T and Yamada M:
Nuclear localization of peptidylarginine deiminase V and histone
deimination in granulocytes. J Biol Chem. 277:49562–49568. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hagiwara T, Nakashima K, Hirano H, Senshu
T and Yamada M: Deimination of arginine residues in
nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem
Biophys Res Commun. 290:979–983. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Holbert MA and Marmorstein R: Structure
and activity of enzymes that remove histone modifications. Curr
Opin Struct Biol. 15:673–680. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cuthbert GL, Daujat S, Snowden AW,
Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD,
Tempst P, Bannister AJ, et al: Histone deimination antagonizes
arginine methylation. Cell. 118:545–553. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang Y, Wysocka J, Sayegh J, Lee YH,
Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y,
et al: Human PAD4 regulates histone arginine methylation levels via
demethylimination. Science. 306:279–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Fadok VA, Bratton DL, Rose DM, Pearson A,
Ezekewitz RA and Henson PM: A receptor for
phosphatidylserine-specific clearance of apoptotic cells. Nature.
405:85–90. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Poulard C, Rambaud J, Hussein N, Corbo L
and Le Romancer M: JMJD6 regulates ERα methylation on arginine.
PLoS One. 9:e879822014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Poulard C, Rambaud J, Lavergne E,
Jacquemetton J, Renoir JM, Trédan O, Chabaud S, Treilleux I, Corbo
L and Le Romancer M: Role of JMJD6 in Breast Tumourigenesis. PLoS
One. 10:e01261812015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lawrence P, Conderino JS and Rieder E:
Redistribution of demethylated RNA helicase A during foot-and-mouth
disease virus infection: Role of Jumonji C-domain containing
protein 6 in RHA demethylation. Virology. 452-453:1–11. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gao WW, Xiao RQ, Peng BL, Xu HT, Shen HF,
Huang MF, Shi TT, Yi J, Zhang WJ, Wu XN, et al: Arginine
methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene
activation. Proc Natl Acad Sci USA. 112:3327–3336. 2015. View Article : Google Scholar
|
|
69
|
Tsai WC, Reineke LC, Jain A, Jung SY and
Lloyd RE: Histone arginine demethylase JMJD6 is linked to stress
granule assembly through demethylation of the stress
granule-nucleating protein G3BP1. J Biol Chem. 292:18886–18896.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee
M, Jung KJ, Doyle MR, Amasino RM and Noh YS: Divergent roles of a
pair of homologous jumonji/zinc-finger-class transcription factor
proteins in the regulation of Arabidopsis flowering time. Plant
Cell. 16:2601–2613. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen X, Hu Y and Zhou DX: Epigenetic gene
regulation by plant Jumonji group of histone demethylase. Biochim
Biophys Acta. 1809:421–426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cho JN, Ryu JY, Jeong YM, Park J, Song JJ,
Amasino RM, Noh B and Noh YS: Control of seed germination by
light-induced histone arginine demethylation activity. Dev Cell.
22:736–748. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Webby CJ, Wolf A, Gromak N, Dreger M,
Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR
III, et al: Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a
protein associated with RNA splicing. Science. 325:90–93. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Heim A, Grimm C, Müller U, Häußler S,
Mackeen MM, Merl J, Hauck SM, Kessler BM, Schofield CJ, Wolf A, et
al: Jumonji domain containing protein 6 (Jmjd6) modulates splicing
and specifically interacts with arginine-serine-rich (RS) domains
of SR- and SR-like proteins. Nucleic Acids Res. 42:7833–7850. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Böttger A, Islam MS, Chowdhury R,
Schofield CJ and Wolf A: The oxygenase Jmjd6--a case study in
conflicting assignments. Biochem J. 468:191–202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Walport LJ, Hopkinson RJ, Chowdhury R,
Schiller R, Ge W, Kawamura A and Schofield CJ: Arginine
demethylation is catalysed by a subset of JmjC histone lysine
demethylases. Nat Commun. 7:119742016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Markolovic S, Wilkins SE and Schofield CJ:
Protein Hydroxylation Catalyzed by 2-Oxoglutarate-dependent
Oxygenases. J Biol Chem. 290:20712–20722. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hopkinson RJ, Walport LJ, Münzel M, Rose
NR, Smart TJ, Kawamura A, Claridge TD and Schofield CJ: Is JmjC
oxygenase catalysis limited to demethylation? Angew Chem Int Ed
Engl. 52:7709–7713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Williams ST, Walport LJ, Hopkinson RJ,
Madden SK, Chowdhury R, Schofield CJ and Kawamura A: Studies on the
catalytic domains of multiple JmjC oxygenases using peptide
substrates. Epigenetics. 9:1596–1603. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Klose RJ, Kallin EM and Zhang Y:
JmjC-domain-containing proteins and histone demethylation. Nat Rev
Genet. 7:715–727. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Klose RJ, Gardner KE, Liang G,
Erdjument-Bromage H, Tempst P and Zhang Y: Demethylation of histone
H3K36 and H3K9 by Rph1: A vestige of an H3K9 methylation system in
Saccharomyces cerevisiae? Mol Cell Biol. 27:3951–3961. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kim H, Kim D, Choi SA, Kim CR, Oh SK, Pyo
KE, Kim J, Lee SH, Yoon JB, Zhang Y, et al: KDM3A histone
demethylase functions as an essential factor for activation of
JAK2-STAT3 signaling pathway. Proc Natl Acad Sci USA.
115:11766–11771. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Tsukada Y and Zhang Y: Purification of
histone demethylases from HeLa cells. Methods. 40:318–326. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Larsen SC, Sylvestersen KB, Mund A, Lyon
D, Mullari M, Madsen MV, Daniel JA, Jensen LJ and Nielsen ML:
Proteome-wide analysis of arginine monomethylation reveals
widespread occurrence in human cells. Sci Signal. 9:rs92016.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yang Y, Yin X, Yang H and Xu Y: Histone
demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer
cell growth through promoting proteasomal degradation of OGT. Mol
Cell. 58:47–59. 2015. View Article : Google Scholar : PubMed/NCBI
|