|
1
|
simpleBreastcancer.orgUS Breast Cancer Statistics.
http://www.breastcancer.org/symptoms/understand_bc/statisticsbreast
cancer org. February 18–2019
|
|
2
|
Linos E, Spanos D, Rosner BA, Linos K,
Hesketh T, Qu JD, Gao YT, Zheng W and Colditz GA: Effects of
reproductive and demographic changes on breast cancer incidence in
China: A modeling analysis. J Natl Cancer Inst. 100:1352–1360.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Higgins MJ and Baselga J: Targeted
therapies for breast cancer. J Clin Invest. 121:3797–3803. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Győrffy B, Hatzis C, Sanft T, Hofstatter
E, Aktas B and Pusztai L: Multigene prognostic tests in breast
cancer: Past, present, future. Breast Cancer Res. 17:112015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z,
Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al: A functional
genomic approach identifies FAL1 as an oncogenic long noncoding RNA
that associates with BMI1 and represses p21 expression in cancer.
Cancer Cell. 26:344–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng S, Zhang J, Su W, Bai S, Xiao L, Chen
X, Lin J, Reddy RM, Chang AC, Beer DG and Chen G: Overexpression of
LINC00152 correlates with poor patient survival and knockdown
impairs cell proliferation in lung cancer. Sci Rep. 7:29822017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sang H, Liu H, Xiong P and Zhu M: Long
non-coding RNA functions in lung cancer. Tumor Biol. 36:4027–4037.
2015. View Article : Google Scholar
|
|
11
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Lu T, Wang Q, Liu J and Jiao W:
Circular RNAs: Crucial regulators in the human body (Review). Oncol
Rep. 40:3119–3135. 2018.PubMed/NCBI
|
|
13
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo LL, Song CH, Wang P, Dai LP, Zhang JY
and Wang KJ: Competing endogenous RNA networks and gastric cancer.
World J Gastroenterol. 21:11680–11687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y
and Chen Y: The emerging function and mechanism of ceRNAs in
cancer. Trends Genet. 32:211–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng DL, Xiang YY, Ji LJ and Lu XJ:
Competing endogenous RNA interplay in cancer: Mechanism,
methodology, and perspectives. Tumor Biol. 36:479–488. 2015.
View Article : Google Scholar
|
|
19
|
Cline MS, Craft B, Swatloski T, Goldman M,
Ma S, Haussler D and Zhu J: Exploring TCGA pan-cancer data at the
UCSC cancer genomics browser. Sci Rep. 3:26522013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
R Core Team R, . A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: version 3.4.3. http://www.R-project.org/November 30–2017
|
|
21
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46(D1): D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:Aug 12–2015.(Epub ahead of print). doi:
10.7554/eLife.05005. View Article : Google Scholar
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen WJ, Tang RX, He RQ, Li DY, Liang L,
Zeng JH, Hu XH, Ma J, Li SK and Chen G: Clinical roles of the
aberrantly expressed lncRNAs in lung squamous cell carcinoma: A
study based on RNA-sequencing and microarray data mining.
Oncotarget. 8:61282–61304. 2017.PubMed/NCBI
|
|
26
|
Nik-Zainal S, Davies H, Staaf J,
Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB,
Martin S, Wedge DC, et al: Landscape of somatic mutations in 560
breast cancer whole-genome sequences. Nature. 534:47–54. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng
J, Xiao W, Yu G, Yao W, Zhou H, et al: lncRNA MALAT1 functions as a
competing endogenous RNA to regulate ZEB2 expression by sponging
miR-200s in clear cell kidney carcinoma. Oncotarget. 6:38005–38015.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Niu L, Jiang S, Zhai J, Wang P,
Kong F and Jin X: Comprehensive analysis of aberrantly expressed
profiles of lncRNAs and miRNAs with associated ceRNA network in
muscle-invasive bladder cancer. Oncotarget. 7:86174–86185. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li F, Huang C, Li Q and Wu X: Construction
and comprehensive analysis for dysregulated long non-coding RNA
(lncRNA)-associated competing endogenous RNA (ceRNA) network in
gastric cancer. Med Sci Monit. 24:37–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou S, Wang L, Yang Q, Liu H, Meng Q,
Jiang L, Wang S and Jiang W: Systematical analysis of lncRNA-mRNA
competing endogenous RNA network in breast cancer subtypes. Breast
Cancer Res Treat. 169:267–275. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D,
Yan X, Chen B, Yu L, Li J, et al: Identification of novel long
non-coding RNAs in triple-negative breast cancer. Oncotarget.
6:21730–21739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang F, Lv SX, Lv L, Liu YH, Dong SY, Yao
ZH, Dai XX, Zhang XH and Wang OC: Identification of lncRNA
FAM83H-AS1 as a novel prognostic marker in luminal subtype breast
cancer. Onco Targets Ther. 9:7039–7045. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheang MC, Chia SK, Voduc D, Gao D, Leung
S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, et al: Ki67
index, HER2 status, and prognosis of patients with luminal B breast
cancer. J Natl Cancer Inst. 101:736–750. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan N, Zhang G, Bie F, Ma M, Ma Y, Jiang
X, Wang Y and Hao X: Integrative analysis of lncRNAs and miRNAs
with coding RNAs associated with ceRNA crosstalk network in triple
negative breast cancer. Onco Targets Ther. 10:5883–5897. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang H, Fu Z, Dai C, Cao J, Liu X, Xu J,
Lv M, Gu Y, Zhang J, Hua X, et al: lncRNAs expression profiling in
normal ovary, benign ovarian cyst and malignant epithelial ovarian
cancer. Sci Rep. 6:389832016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Z, Yao Q, Zhao S, Wang Y, Li Y and Wang
Z: Comprehensive analysis of differential co-expression patterns
reveal transcriptional dysregulation mechanism and identify novel
prognostic lncRNAs in esophageal squamous cell carcinoma. Onco
Targets Ther. 10:3095–3105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Crippa M, Bestetti I, Perotti M,
Castronovo C, Tabano S, Picinelli C, Grassi G, Larizza L, Pincelli
AI and Finelli P: New case of trichorinophalangeal syndrome-like
phenotype with a de novo t(2;8)(p16.1;q23.3) translocation which
does not disrupt the TRPS1 gene. BMC Med Genet. 15:522014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang N, Tan HY, Chan YT, Guo W, Li S and
Feng Y: Identification of WT1 as determinant of heptatocellular
carcinoma and its inhibition by Chinese herbal medicine Salvia
chinensis benth and its active ingredient protocatechualdehyde.
Oncotarget. 8:105848–105859. 2017.PubMed/NCBI
|
|
40
|
Cai C, Huo Q, Wang X, Chen B and Yang Q:
SNHG16 contributes to breast cancer cell migration by competitively
binding miR-98 with E2F5. Biochem Biophys Res Commun. 485:272–278.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kong Q and Qiu M: Long noncoding RNA
SNHG15 promotes human breast cancer proliferation, migration and
invasion by sponging miR-211-3p. Biochem Biophys Res Commun.
495:1594–1600. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang W, Luo P, Guo W, Shi Y, Xu D, Zheng H
and Jia L: lncRNA SNHG20 knockdown suppresses the osteosarcoma
tumorigenesis through the mitochondrial apoptosis pathway by
miR-139/RUNX2 axis. Biochem Biophys Res Commun. 503:1927–1933.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C,
Xu M, Wu F and Mo YY: Negative regulation of lncRNA GAS5 by miR-21.
Cell Death Differ. 20:1558–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yuan X, Wang S, Liu M, Lu Z, Zhan Y, Wang
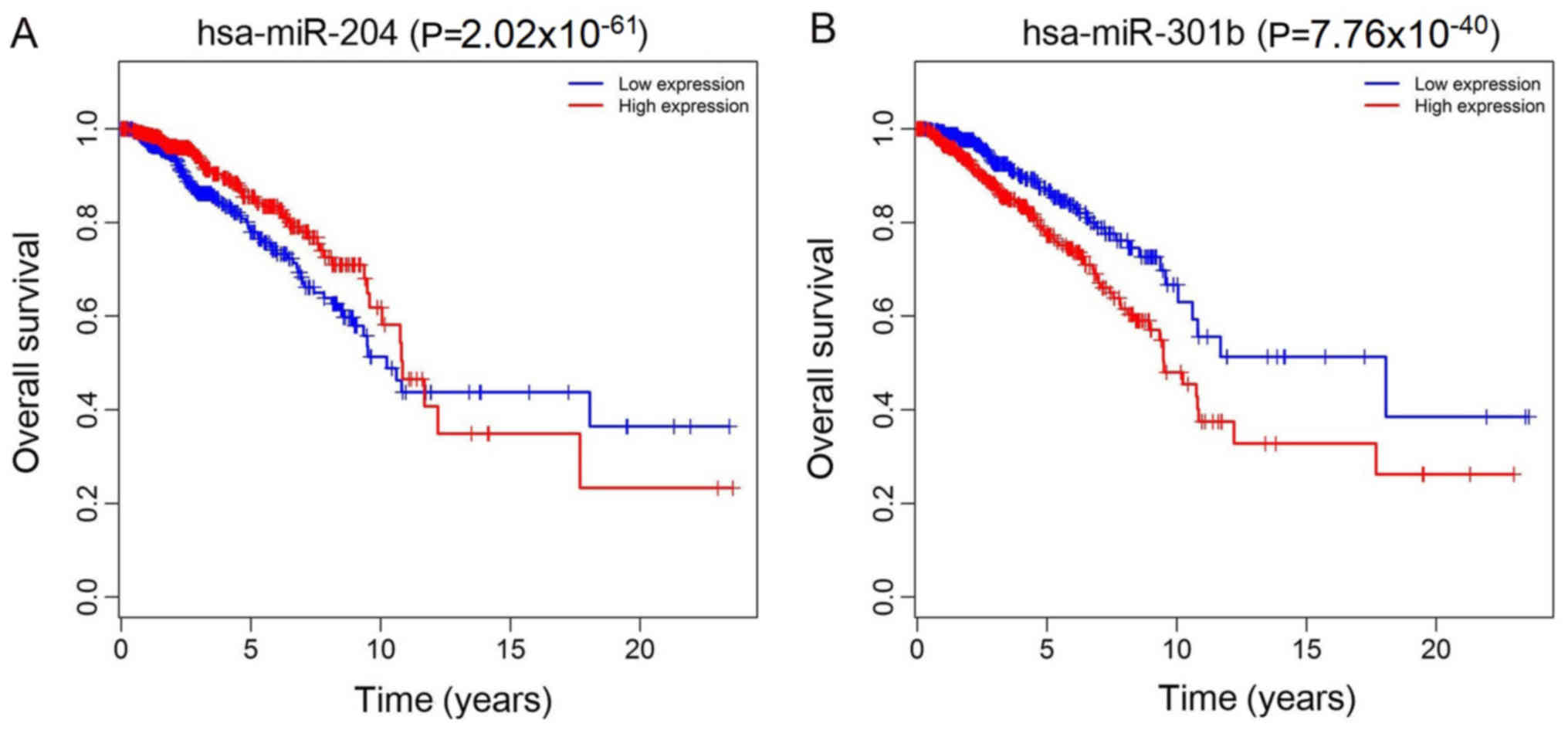
W and Xu AM: Histological and pathological assessment of miR-204
and SOX4 levels in gastric cancer patients. Biomed Res Int.
2017:68946752017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Todorova K, Metodiev MV, Metodieva G,
Mincheff M, Fernández N and Hayrabedyan S: Micro-RNA-204
participates in TMPRSS2/ERG regulation and androgen receptor
reprogramming in prostate cancer. Horm Cancer. 8:28–48. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Abmutalib NS, Othman SN, Mohamad Yusof A,
Abdullah Suhaimi SN, Muhammad R and Jamal R: Integrated microRNA,
gene expression and transcription factors signature in papillary
thyroid cancer with lymph node metastasis. Peer J. 4:e21192016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Geng Y, Deng L, Su D, Xiao J, Ge D, Bao Y
and Jing H: Identification of crucial microRNAs and genes in
hypoxia-induced human lung adenocarcinoma cells. Onco Targets Ther.
9:4605–4616. 2016. View Article : Google Scholar : PubMed/NCBI
|