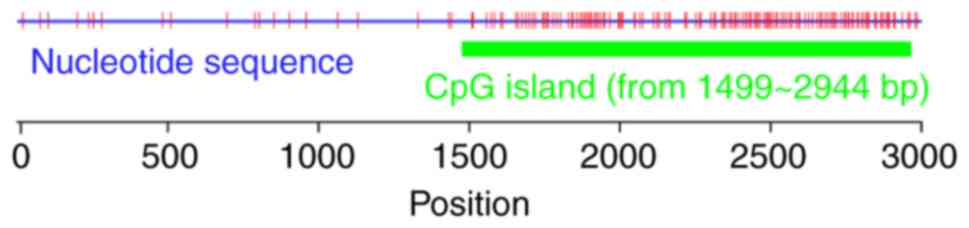
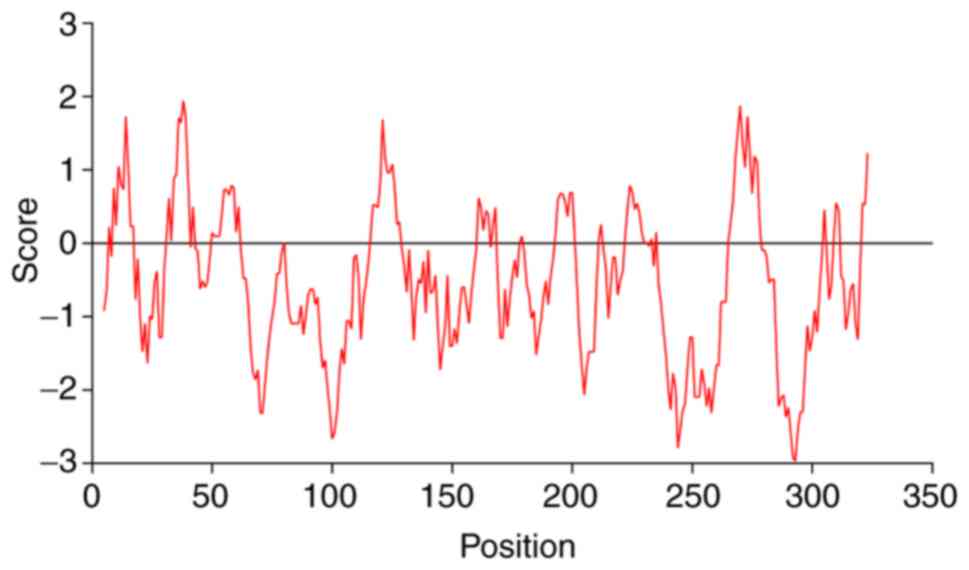
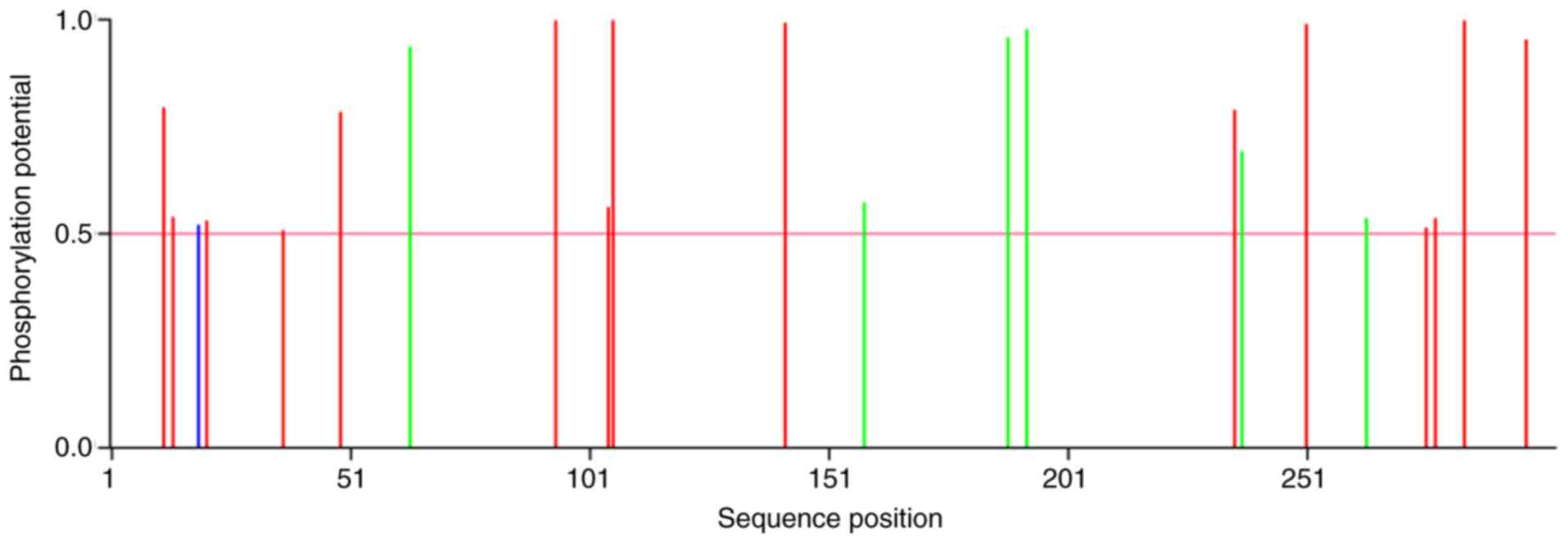
|
1
|
Williams M, Lyu MS, Yang YL, Lin EP,
Dunbrack R, Birren B, Cunningham J and Hunter K: Ier5, a novel
member of the slow-kinetics immediate-early genes. Genomics.
55:327–334. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takaya T, Kasatani K, Noguchi S and Nikawa
J: Functional analyses of immediate early gene ETR101 expressed in
yeast. Biosci Biotechnol Biochem. 73:1653–1660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Savitz J, Frank MB, Victor T, Bebak M,
Marino JH, Bellgowan PS, McKinney BA, Bodurka J, Kent Teague T and
Drevets WC: Inflammation and neurological disease-related genes are
differentially expressed in depressed patients with mood disorders
and correlate with morphometric and functional imaging
abnormalities. Brain Behav Immun. 31:161–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ishikawa Y and Sakurai H: Heat-induced
expression of the immediate-early gene IER5 and its involvement in
the proliferation of heat-shocked cells. FEBS J. 282:332–340. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ishikawa Y, Kawabata S and Sakurai H: HSF1
transcriptional activity is modulated by IER5 and PP2A/B55. FEBS
Lett. 589:1150–1155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asano Y, Kawase T, Okabe A, Tsutsumi S,
Ichikawa H, Tatebe S, Kitabayashi I, Tashiro F, Namiki H, Kondo T,
et al: IER5 generates a novel hypo-phosphorylated active form of
HSF1 and contributes to tumorigenesis. Sci Rep. 6:191742016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li XN, Ji C, Zhou PK and Wu YM:
Establishments of IER5 silence and overexpression cervical cancer
SiHa cell lines and analysis of radiosensitivity. Int J Clin Exp
Pathol. 9:6671–6682. 2016.
|
|
8
|
Kawabata S, Ishita Y, Ishikawa Y and
Sakurai H: Immediate-early response 5 (IER5) interacts with protein
phosphatase 2A and regulates the phosphorylation of ribosomal
protein S6 kinase and heat shock factor 1. FEBS Lett.
589:3679–3685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakamura S, Nagata Y, Tan L, Takemura T,
Shibata K, Fujie M, Fujisawa S, Tanaka Y, Toda M, Makita R, et al:
Transcriptional repression of Cdc25B by IER5 inhibits the
proliferation of leukemic progenitor cells through NF-YB and p300
in acute myeloid leukemia. PLoS One. 6:e280112011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Tian M, Zhao H, He Y, Li F, Li X,
Yu X, Ding K, Zhou P and Wu Y: IER5 as a promising predictive
marker promotes irradiation-induced apoptosis in cervical cancer
tissues from patients undergoing chemoradiotherapy. Oncotarget.
8:36438–36448. 2017.PubMed/NCBI
|
|
11
|
Shi HM, Ding KK, Zhou PK, Guo DM, Chen D,
Li YS, Zhao CL, Zhao CC and Zhang X: Radiation-induced expression
of IER5 is dose-dependent and not associated with the clinical
outcomes of radiotherapy in cervical cancer. Oncol Lett.
11:1309–1314. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang C, Yang M, Feng Z, Liu X, Yin L, Zhou
P and Ding K: Radiation modulated the interaction of IER5 protein
and CDC25B promoter DNA in primary hepatocellular carcinoma. Int J
Clin Exp Pathol. 9:2888–2895. 2016.
|
|
13
|
Yang C, Wang Y, Hao C, Yuan Z, Liu X, Yang
F, Jiang H, Jiang X, Zhou P and Ding K: IER5 promotes irradiation-
and cisplatin-induced apoptosis in human hepatocellular carcinoma
cells. Am J Transl Res. 8:1789–1798. 2016.PubMed/NCBI
|
|
14
|
Ding KK, Shang ZF, Hao C, Xu QZ, Shen JJ,
Yang CJ, Xie YH, Qiao C, Wang Y, Xu LL and Zhou PK: Induced
expression of the IER5 gene by gamma-ray irradiation and its
involvement in cell cycle checkpoint control and survival. Radiat
Environ Biophys. 48:205–213. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang C, Yin L, Zhou P, Liu X, Yang M, Yang
F, Jiang H and Ding K: Transcriptional regulation of IER5 in
response to radiation in HepG2. Cancer Gene Ther. 23:61–65. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu XP, Wu YM, Liu Y, Tian M, Wang JD, Ding
KK, Ma T and Zhou PK: IER5 is involved in DNA double-strand breaks
repair in association with PAPR1 in Hela cells. Int J Med Sci.
14:1292–1300. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ohtsubo K and Marth JD: Glycosylation in
Cellular mechanisms of health and disease. Cell. 126:855–867. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Singh V, Ram M, Kumar R, Prasad R, Roy BK
and Singh KK: Phosphorylation: Implications in cancer. Protein J.
36:1–6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jones DT: Protein secondary structure
prediction based on position-specific scoring matrices. J Mol Biol.
292:195–202. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J and Zhang Y: I-TASSER server: New
development for protein structure and function predictions. Nucleic
Acids Res. 43:W174–W181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y: I-TASSER: Fully automated protein
structure prediction in CASP8. Proteins. 77 (Suppl 9):S100–S113.
2009. View Article : Google Scholar
|
|
22
|
Hashimshony T, Zhang J, Keshet I, Bustin M
and Cedar H: The role of DNA methylation in setting up chromatin
structure during development. Nat Genet. 34:187–192. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balada E, Ordi-Ros J, Serrano-Acedo S,
Martinez-Lostao L, Rosa-Leyva M and Vilardell-Tarrés M: Transcript
levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T
cells from patients with systemic lupus erythematosus. Immunology.
124:339–347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Illingworth RS and Bird AP: CpG islands-‘a
rough guide’. FEBS Lett. 583:1713–1720. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eckhardt F, Lewin J, Cortese R, Rakyan VK,
Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al:
DNA methylation profiling of human chromosomes 6, 20 and 22. Nat
Genet. 38:1378–1385. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Palmer KJ, Konkel JE and Stephens DJ:
PCTAIRE protein kinases interact directly with the COPII complex
and modulate secretory cargo transport. J Cell Sci. 118:3839–3847.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bard F, Mazelin L, Péchoux-Longin C,
Malhotra V and Jurdic P: Src regulates golgi structure and KDEL
receptor-dependent retrograde transport to the endoplasmic
reticulum. J Biol Chem. 278:46601–46606. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wintjens R and Rooman M: Structural
classification of HTH DNA-binding domains and protein-DNA
interaction modes. J Mol Biol. 262:294–313. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aravind L and Koonin EV: DNA-binding
proteins and evolution of transcription regulation in the archaea.
Nucleic Acids Res. 27:4658–4670. 1999. View Article : Google Scholar : PubMed/NCBI
|