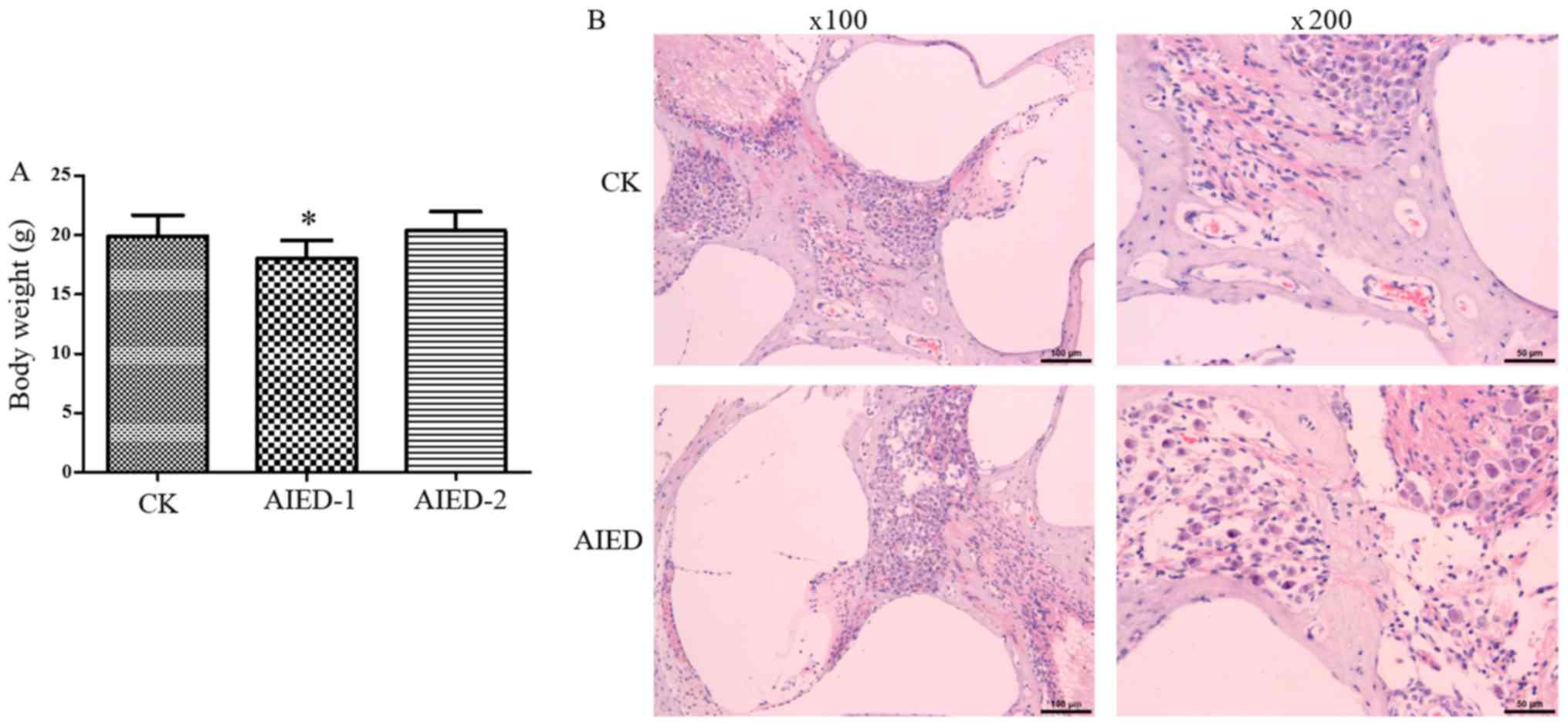
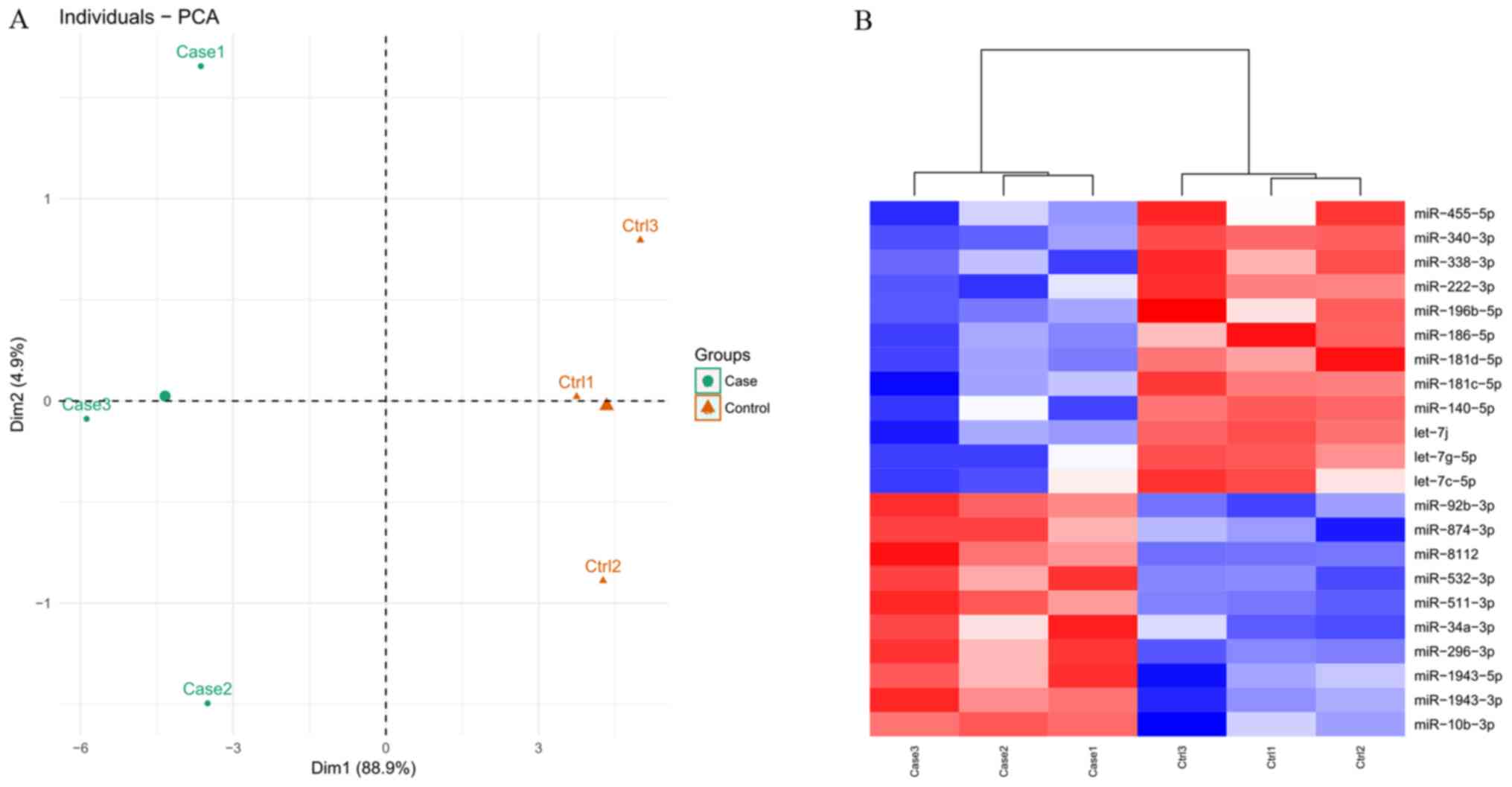
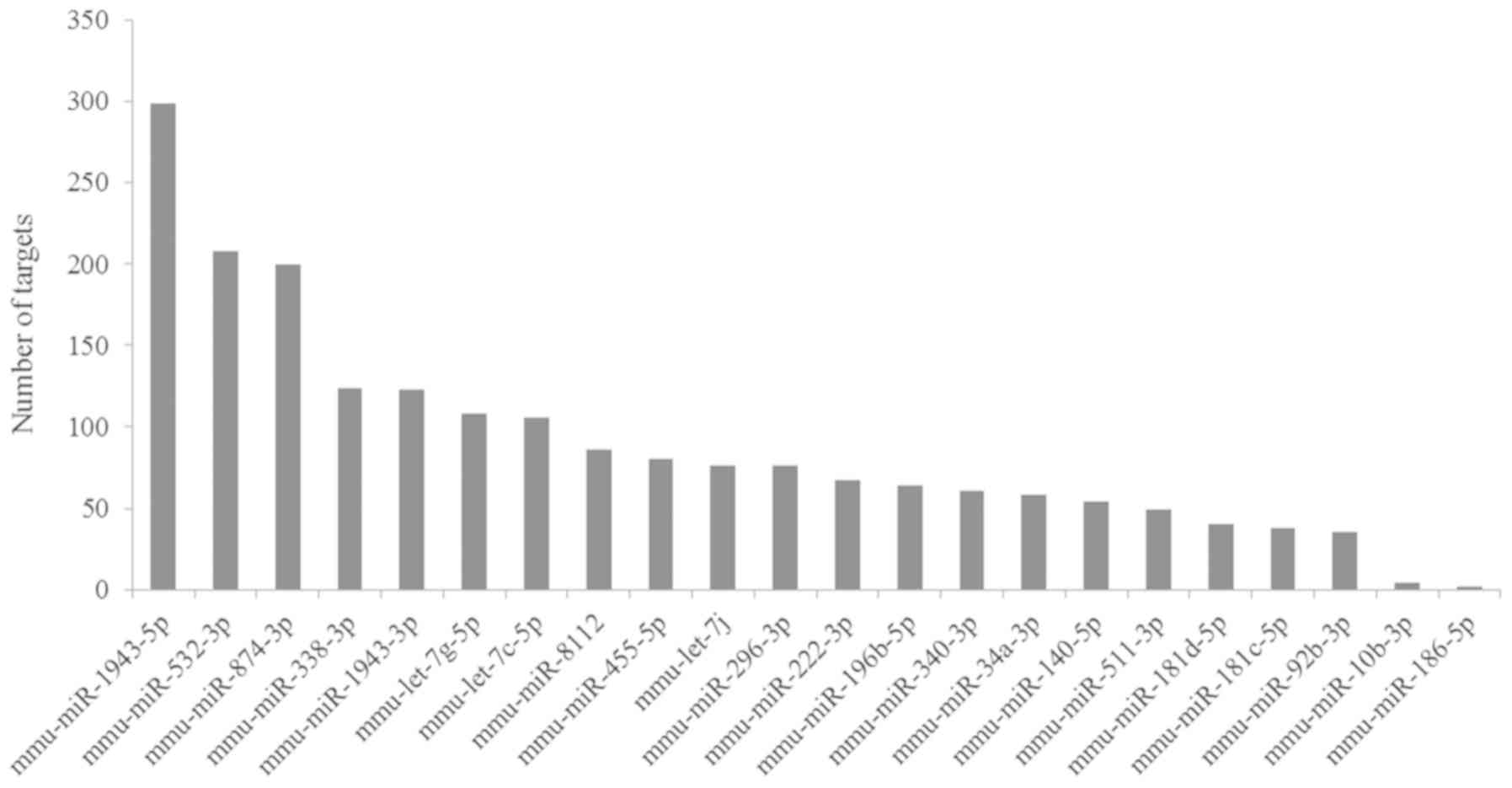
|
1
|
Matsuoka AJ and Harris JP: Autoimmune
inner ear disease: A retrospective review of forty-seven patients.
Audiol Neurotol. 18:228–239. 2013. View Article : Google Scholar
|
|
2
|
Chen J, Liang J, Ou J and Cai W: Mental
health in adults with sudden sensorineural hearing loss: An
assessment of depressive symptoms and its correlates. J Psychosom
Res. 75:72–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carlsson PI, Hall M, Lind KJ and Danermark
B: Quality of life, psychosocial consequences, and audiological
rehabilitation after sudden sensorineural hearing loss. Int J
Audiol. 50:139–144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goodall AF and Siddiq MA: Current
understanding of the pathogenesis of autoimmune inner ear disease:
A review. Clin Otolaryngol. 40:412–419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gopen Q, Keithley EM and Harris JP:
Mechanisms underlying autoimmune inner ear disease. Drug Discov
Today Dis Mech. 3:137–142. 2006. View Article : Google Scholar
|
|
6
|
Satoh H, Firestein GS, Billings PB, Harris
JP and Keithley EM: Proinflammatory cytokine expression in the
endolymphatic sac during inner ear inflammation. J Assoc Res
Otolaryngol. 4:139–147. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Svrakic M, Pathak S, Goldofsky E, Hoffman
R, Chandrasekhar SS, Sperling N, Alexiades G, Ashbach M and
Vambutas A: Diagnostic and prognostic utility of measuring tumor
necrosis factor in the peripheral circulation of patients with
immune-mediated sensorineural hearing loss. Arch Otolaryngol Head
Neck Surg. 138:1052–1058. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pathak S, Hatam LJ, Bonagura V and
Vambutas A: Innate immune recognition of molds and homology to the
inner ear protein, cochlin, in patients with autoimmune inner ear
disease. J Clin Immunol. 33:1204–1215. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee JM, Kim JY, Bok J, Kim KS, Choi JY and
Kim SH: Identification of evidence for autoimmune pathology of
bilateral sudden sensorineural hearing loss using proteomic
analysis. Clin Immunol. 183:24–35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lorenz RR, Solares CA, Williams P, Sikora
J, Pelfrey CM, Hughes GB and Tuohy VK: Interferon-γ production to
inner ear antigens by T cells from patients with autoimmune
sensorineural hearing loss. J Neuroimmunol. 130:173–178. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen JQ, Papp G, Szodoray P and Zeher M:
The role of microRNAs in the pathogenesis of autoimmune diseases.
Autoimmun Rev. 15:1171–1180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishida W, Fukuda K, Higuchi T, Kajisako M,
Sakamoto S and Fukushima A: Dynamic changes of microRNAs in the eye
during the development of experimental autoimmune uveoretinitis.
Invest Ophthalmol Vis Sci. 52:611–617. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fang X, Sun D, Wang Z, Yu Z, Liu W, Pu Y,
Wang D, Huang A, Liu M, Xiang Z, et al: miR-30a positively
regulates the inflammatory response of microglia in experimental
autoimmune encephalomyelitis. Neurosci Bull. 33:603–615. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rudnicki A, Shivatzki S, Beyer LA, Takada
Y, Raphael Y and Avraham KB: MicroRNA-224 regulates Pentraxin 3, a
component of the humoral arm of innate immunity, in inner ear
inflammation. Hum Mol Genet. 23:3138–3146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xin Q, Li J, Dang J, Bian X, Shan S, Yuan
J, Qian Y, Liu Z, Liu G, Yuan Q, et al: miR-155 deficiency
ameliorates autoimmune inflammation of systemic lupus erythematosus
by targeting S1pr1 in Faslpr/lpr mice. J Immunol. 194:5437–5445.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
National Research Council (US) Institute
for Laboratory Animal Research, . Guide for the Care and Use of
Laboratory Animals. National Academies Press (US); Washingto, DC:
1996
|
|
17
|
Watanabe K, Inai S, Jinnouchi K, Baba S
and Yagi T: Expression of caspase-activated deoxyribonuclease (CAD)
and caspase 3 (CPP32) in the cochlea of cisplatin (CDDP)-treated
guinea pigs. Auris Nasus Larynx. 30:219–225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Silva-Gomes R, Marcq E, Trigo G, Gonçalves
CM, Longatto-Filho A, Castro AG, Pedrosa J and Fraga AG:
Spontaneous healing of Mycobacterium ulcerans lesions in the
guinea pig model. PLoS Negl Trop Dis. 9:e00042652015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poonawala T, Levay-Young BK, Hebbel RP and
Gupta K: Opioids heal ischemic wounds in the rat. Wound Repair
Regen. 13:165–174. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet J.
17:10–12. 2011. View Article : Google Scholar
|
|
21
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Haeussler M, Zweig AS, Tyner C, Speir ML,
Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN,
et al: The UCSC Genome Browser database: 2019 update. Nucleic Acids
Res. 47(D1): D853–D858. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42(D1): D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lorenz R, Bernhart SH, Höner Zu
Siederdissen C, Tafer H, Flamm C, Stadler PF and Hofacker IL:
ViennaRNA Package 2.0. Algorithms Mol Biol. 6:262011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bonnet E, Wuyts J, Rouzé P and Van de Peer
Y: Evidence that microRNA precursors, unlike other non-coding RNAs,
have lower folding free energies than random sequences.
Bioinformatics. 20:2911–2917. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kassambara A and Mundt F: factoextra:
Extract and Visualize the Results of Multivariate Data Analyses. R
package version 1.0.3, 2017. https://rpkgs.datanovia.com/factoextra/index.html
|
|
27
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carlson M: org.Mm.eg.db: Genome wide
annotation for Mouse. R package version 3.7.0, 2018. https://bioconductor.org/packages/release/data/annotation/html/org.Mm.eg.db.html
|
|
32
|
Carlson M.; KEGG.db, : A set of annotation
maps for KEGG. R package version 3.2.3, 2016. https://bioconductor.org/packages/release/data/annotation/html/KEGG.db.html
|
|
33
|
Ding L, Liu J, Shen HX, Pan LP, Liu QD,
Zhang HD, Han L, Shuai LG, Ding EM, Zhao QN, et al: Analysis of
plasma microRNA expression profiles in male textile workers with
noise-induced hearing loss. Hear Res. 333:275–282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li YH, Yang Y, Yan YT, Xu LW, Ma HY, Shao
YX, Cao CJ, Wu X, Qi MJ, Wu YY, et al: Analysis of serum microRNA
expression in male workers with occupational noise-induced hearing
loss. Braz J Med Biol Res. 51:e64262018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen L, Al-Mossawi MH, Ridley A, Sekine T,
Hammitzsch A, de Wit J, Simone D, Shi H, Penkava F,
Kurowska-Stolarska M, et al: miR-10b-5p is a novel Th17 regulator
present in Th17 cells from ankylosing spondylitis. Ann Rheum Dis.
76:620–625. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Eryilmaz A, Dagli M, Karabulut H, Sivas
Acar F, Erkol Inal E and Gocer C: Evaluation of hearing loss in
patients with ankylosing spondylitis. J Laryngol Otol. 121:845–849.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kahveci OK, Demirdal US, Duran A, Altuntas
A, Kavuncu V and Okur E: Hearing and cochlear function of patients
with ankylosing spondylitis. Clin Rheumatol. 31:1103–1108. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sugahara K, Hashimoto M, Hirose Y,
Shimogori H and Yamashita H: Autoimmune inner ear disease
associated with ankylosing spondylitis. Egypt J Otolaryngol.
30:176–179. 2014. View Article : Google Scholar
|
|
39
|
Mojsilovic-Petrovic J, Callaghan D, Cui H,
Dean C, Stanimirovic DB and Zhang W: Hypoxia-inducible factor-1
(HIF-1) is involved in the regulation of hypoxia-stimulated
expression of monocyte chemoattractant protein-1 (MCP-1/CCL2) and
MCP-5 (Ccl12) in astrocytes. J Neuroinflammation. 4:122007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Webb A, Johnson A, Fortunato M, Platt A,
Crabbe T, Christie MI, Watt GF, Ward SG and Jopling LA: Evidence
for PI-3K-dependent migration of Th17-polarized cells in response
to CCR2 and CCR6 agonists. J Leukoc Biol. 84:1202–1212. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoon EL, Yeon JE, Ko E, Lee HJ, Je JH, Yoo
YJ, Kang SH, Suh SJ, Kim JH, Seo YS, et al: An explorative analysis
for the role of serum miR-10b-3p levels in predicting response to
sorafenib in patients with advanced hepatocellular carcinoma. J
Korean Med Sci. 32:212–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Zhang X, Zhao Z and Xu H:
Preliminary analysis of microRNAs expression profiling in MC3T3-E1
cells exposed to fluoride. Biol Trace Elem Res. 176:367–373. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Haimanot RT: Neurological complications of
endemic skeletal fluorosis, with special emphasis on
radiculo-myelopathy. Paraplegia. 28:244–251. 1990.PubMed/NCBI
|
|
44
|
Liu L, Chen Y, Qi J, Zhang Y, He Y, Ni W,
Li W, Zhang S, Sun S, Taketo MM, et al: Wnt activation protects
against neomycin-induced hair cell damage in the mouse cochlea.
Cell Death Dis. 7:e21362016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xia MY, Zhao XY, Huang QL, Sun HY, Sun C,
Yuan J, He C, Sun Y, Huang X, Kong W, et al: Activation of
Wnt/β-catenin signaling by lithium chloride attenuates
d-galactose-induced neurodegeneration in the auditory cortex of a
rat model of aging. FEBS Open Bio. 7:759–776. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ni W, Zeng S, Li W, Chen Y, Zhang S, Tang
M, Sun S, Chai R and Li H: Wnt activation followed by Notch
inhibition promotes mitotic hair cell regeneration in the postnatal
mouse cochlea. Oncotarget. 7:66754–66768. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Brennan E, Wang B, McClelland A, Mohan M,
Marai M, Beuscart O, Derouiche S, Gray S, Pickering R, Tikellis C,
et al: Protective effect of let-7 miRNA family in regulating
inflammation in diabetes-associated atherosclerosis. Diabetes.
66:2266–2277. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kumar M, Ahmad T, Sharma A, Mabalirajan U,
Kulshreshtha A, Agrawal A and Ghosh B: Let-7 microRNA-mediated
regulation of IL-13 and allergic airway inflammation. J Allergy
Clin Immunol. 128:1077–85.e1, 10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Block H, Ley K and Zarbock A: Severe
impairment of leukocyte recruitment in ppGalNAcT-1-deficient mice.
J Immunol. 188:5674–5681. 2012. View Article : Google Scholar : PubMed/NCBI
|