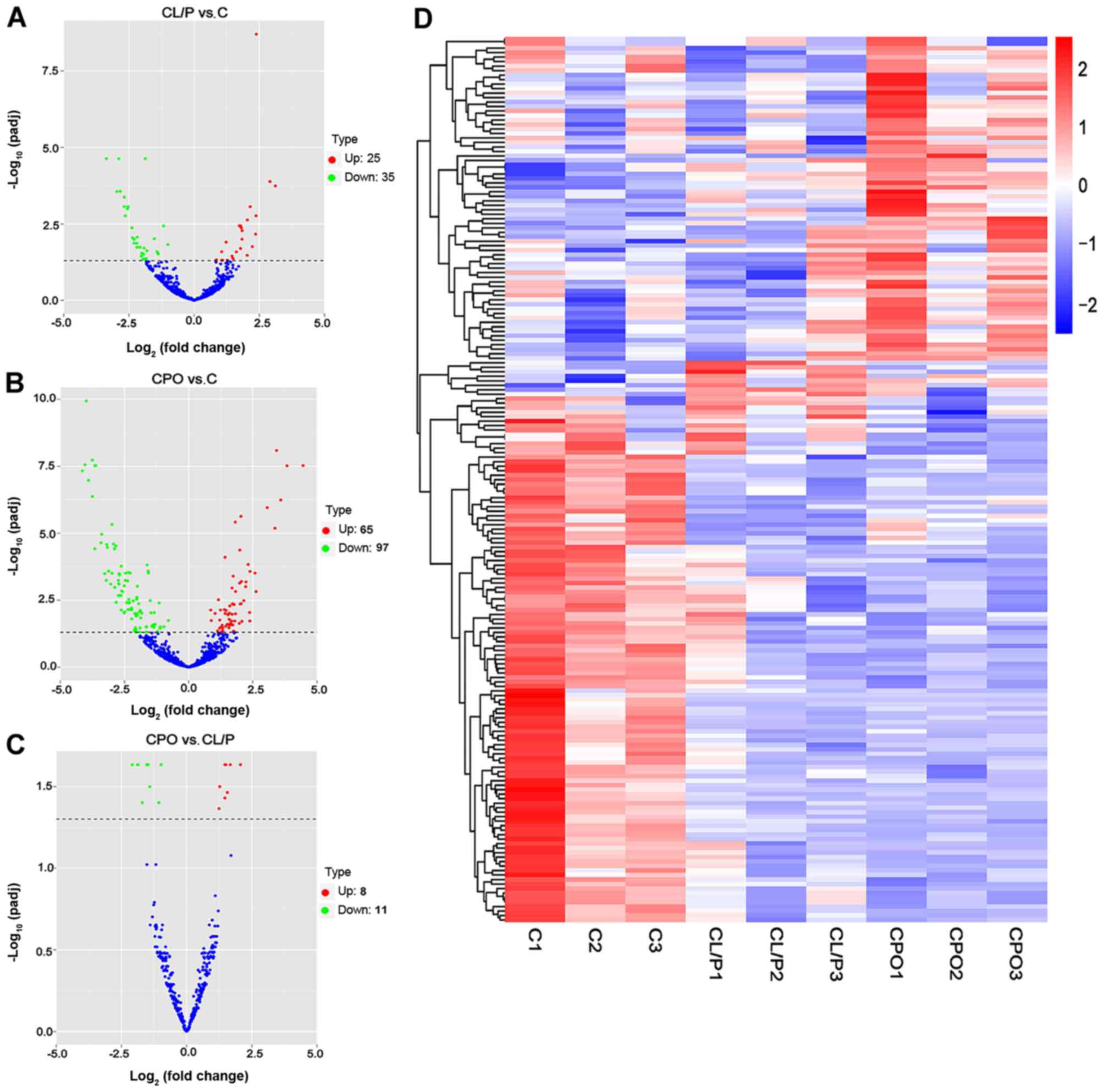
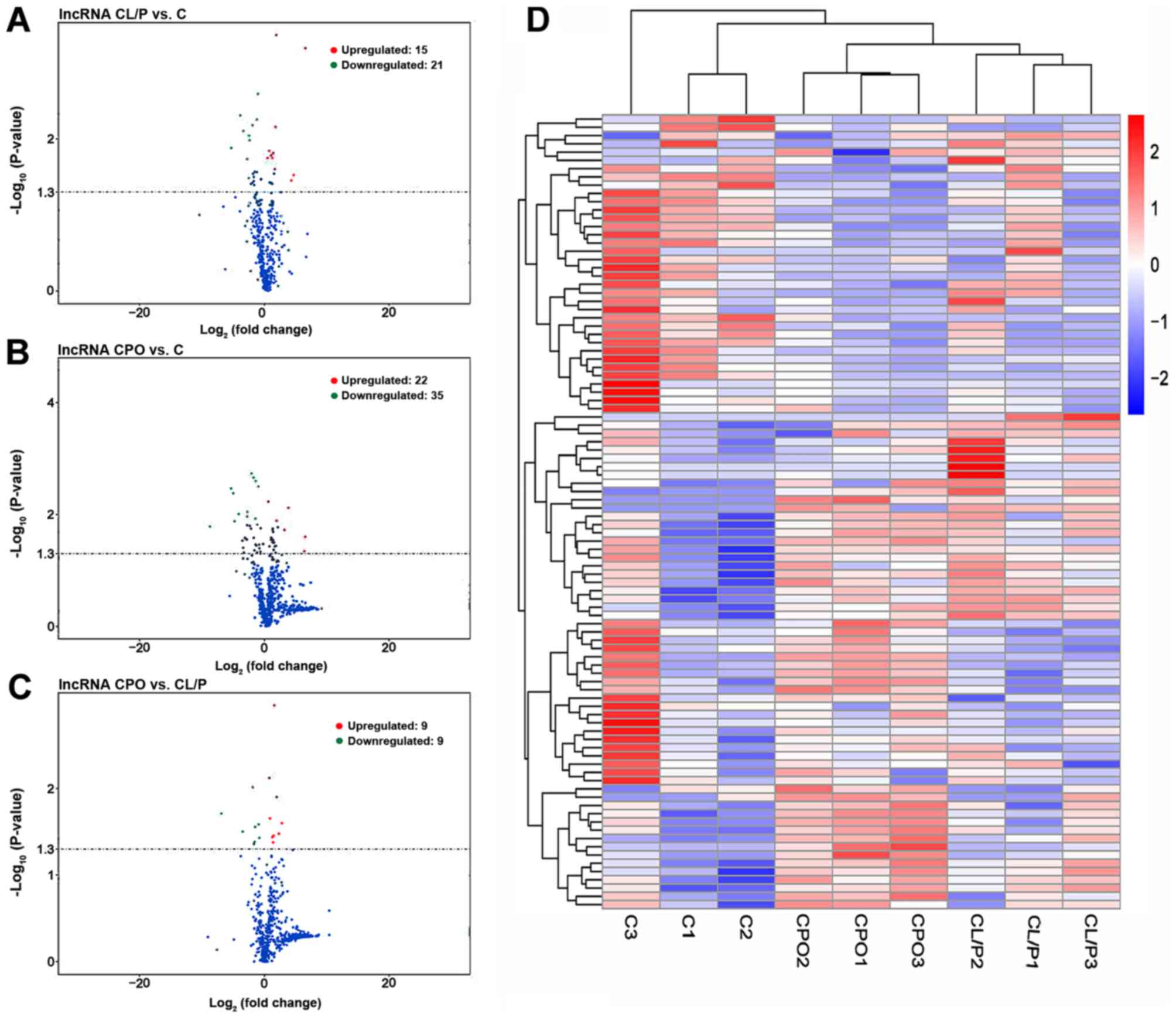
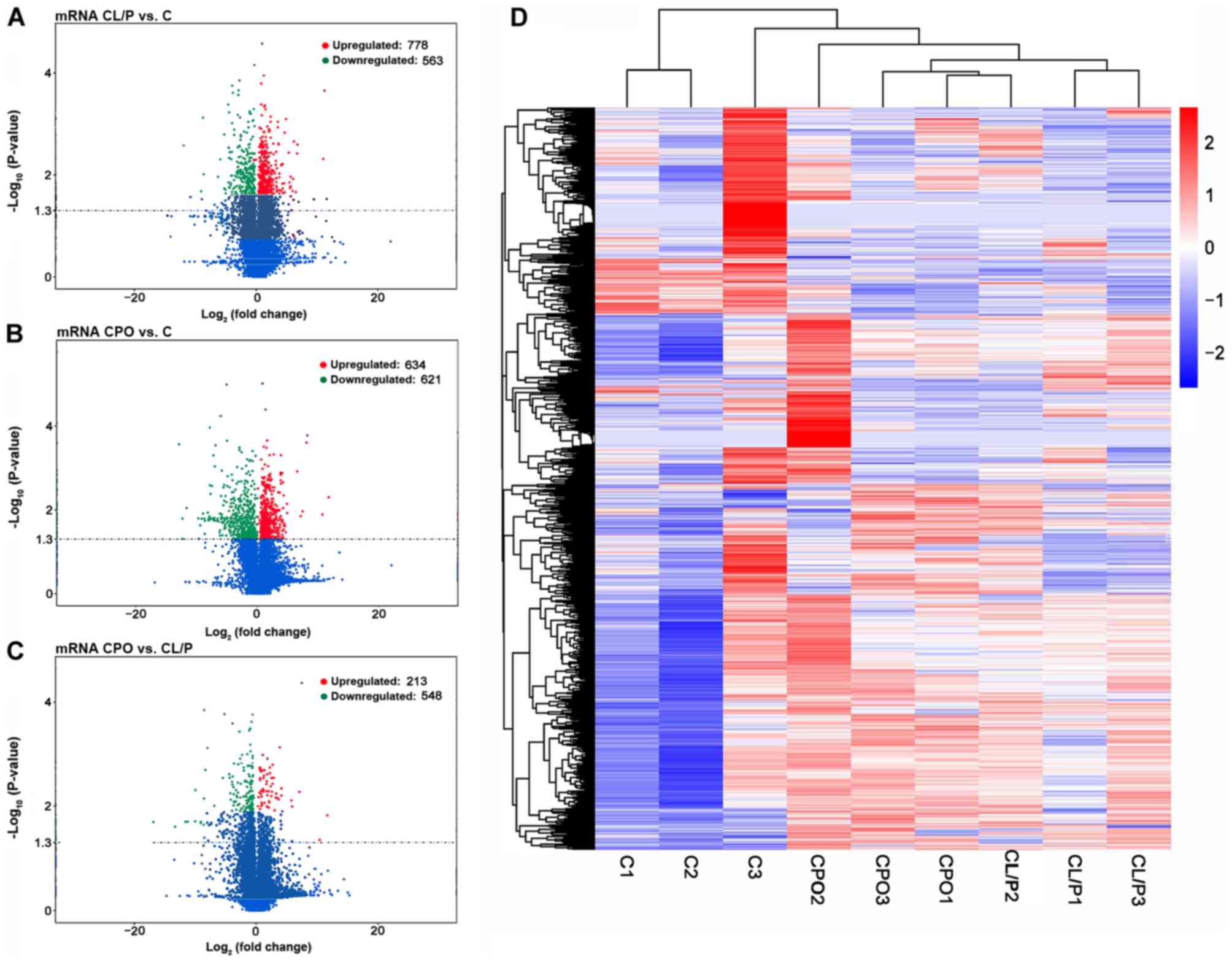
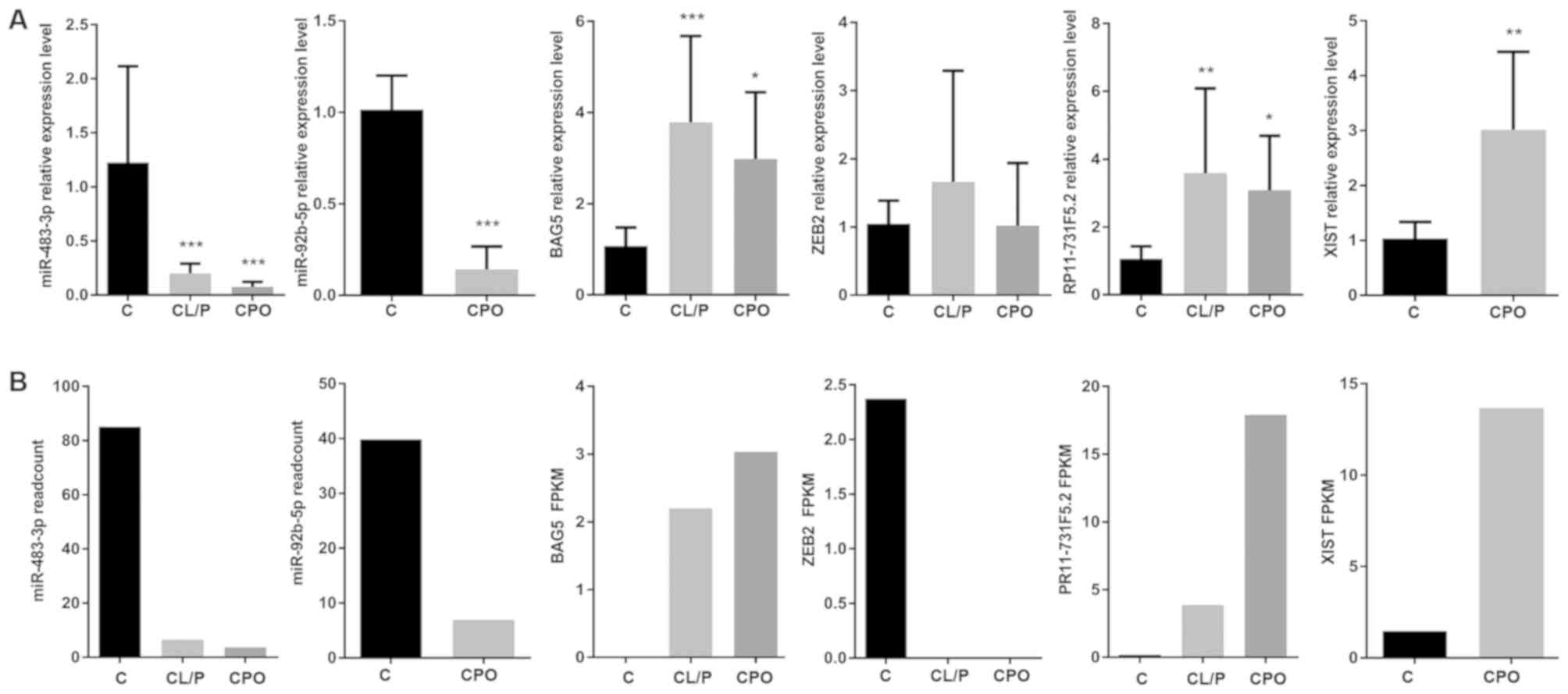
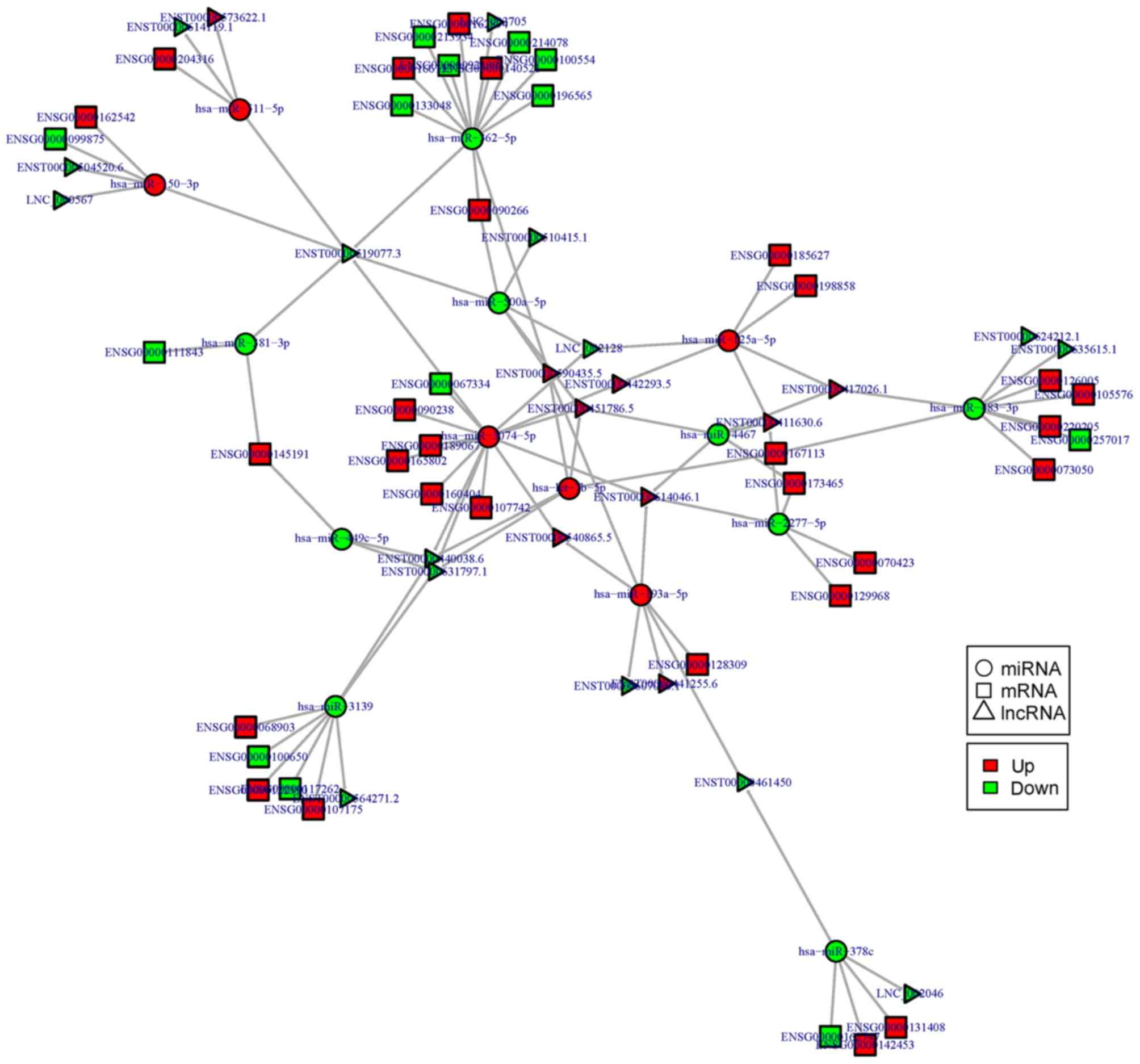
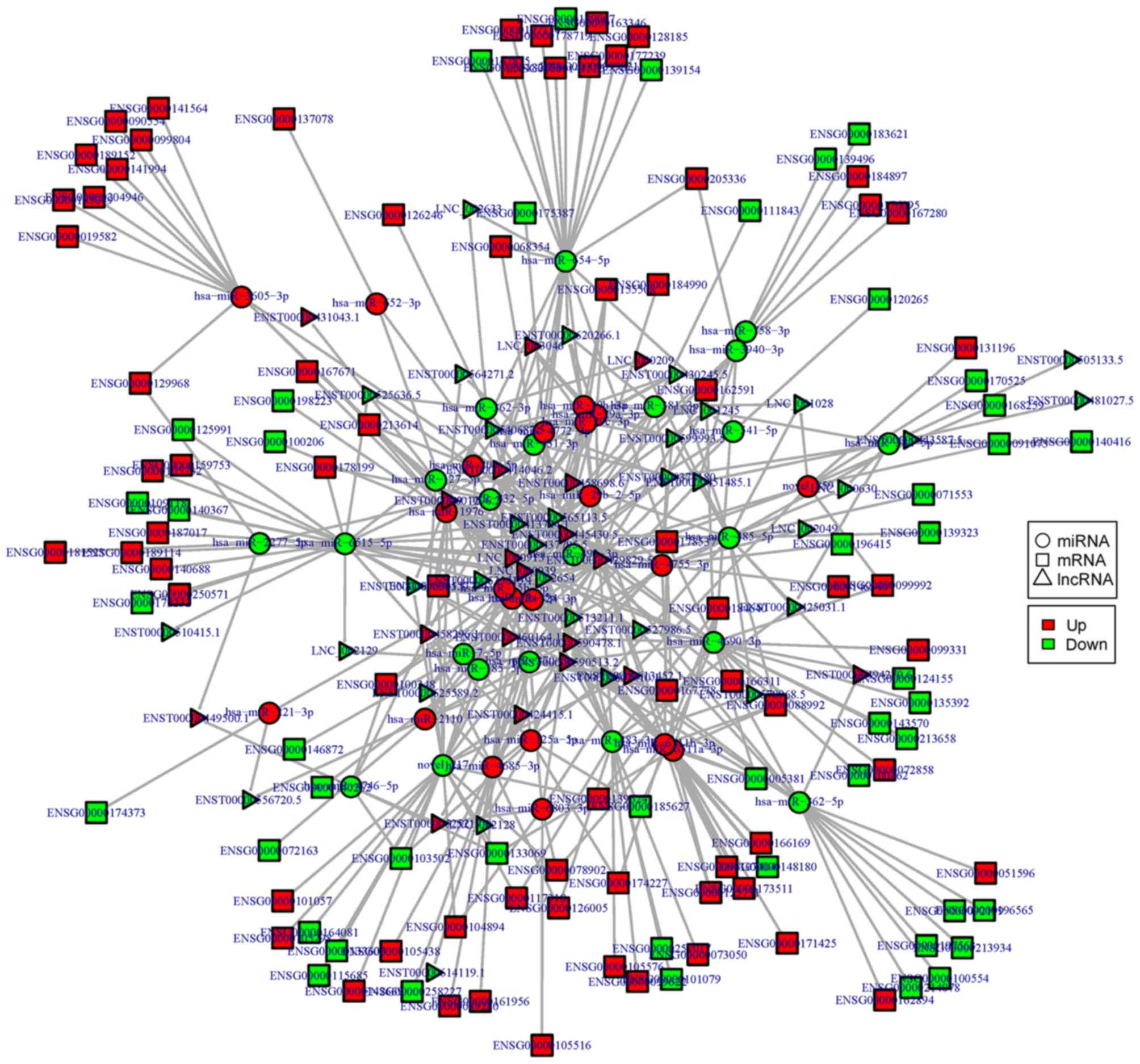
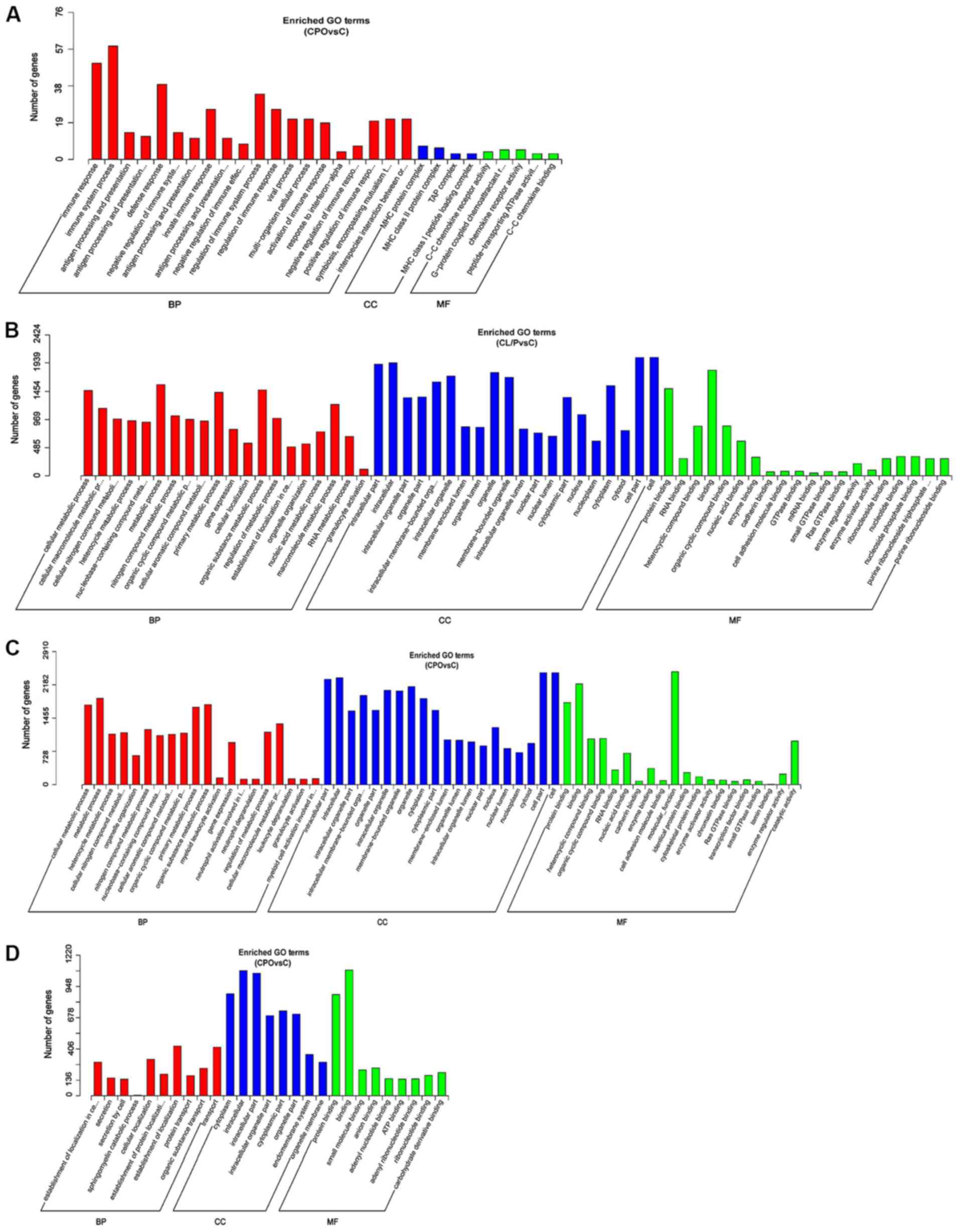
|
1
|
Mossey PA, Little J, Munger RG, Dixon MJ
and Shaw WC: Cleft lip and palate. Lancet. 374:1773–1785. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leslie EJ and Marizita ML: Genetics of
cleft lip and cleft palate. Am J Med Genet C Semin Med Genet 163C.
246–258. 2013. View Article : Google Scholar
|
|
3
|
Ladd-Acosta C and Beaty TH: Integrating
RNA expression identifies candidate gene for orofacial clefts. J
Dent Res. 97:31–32. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chiquet BT, Hashmi SS, Henry R, Burt A,
Mulliken JB, Stal S, Bray M, Blanton SH and Hecht JT: Genomic
screening identifies novel linkages and provides further evidence
for a role of MYH9 in nonsyndromic cleft lip and palate. Eur J Hum
Genet. 17:195–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chiquet BT, Yuan Q, Swindell EC, Maili L,
Plant R, Dyke J, Boyer R, Teichgraeber JF, Greives MR, Mulliken JB,
et al: Knockdown of Crispld2 in zebrafish identifies a novel
network for nonsyndromic cleft lip with or without cleft palate
candidate genes. Eur J Hum Genet. 26:1441–1450. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ludwig KU, Mangold E, Herms S, Nowak S,
Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E, et al:
Genome-wide meta-analyses of nonsyndromic cleft lip with or without
cleft palate identify six new risk loci. Nat Genet. 44:968–971.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Beaty TH, Murray JC, Marazita ML, Munger
RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin, et
al: A genome-wide association study of cleft lip with and without
cleft palate identifies risk variants near MAFB and ABCA4. Nat
Genet. 42:525–529. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C,
Meng L, Wang W, Song Y, Cheng Y, et al: Genome-wide analyses of
non-syndromic cleft lip with palate identify 14 novel loci and
genetic heterogeneity. Nat Commun. 8:143642017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schoen C, Aschrafi A, Thonissen M,
Poelmans G, Von den Hoff JW and Carels CEL: MicroRNAs in
palatogenesis and cleft palate. Front Physiol. 8:1652017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moreno-Moya JM, Vilella F and Simón C:
MicroRNA: Key gene expression regulators. Fertil Steril.
101:1516–1523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou J, Xiong Q, Chen H, Yang C and Fan Y:
Identification of the spinal expression profile of Non-coding RNAs
involved in neuropathic pain following spared nerve injury by
sequence analysis. Front Mol Neurosci. 10:912017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang YK and Yu JC: Circulating microRNAs
and long non-coding RNAs in gastric cancer diagnosis: An update and
review. World J Gastroenterol. 21:9863–9886. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao B, Lu M, Wang D, Li H and He X:
Genome-wide identification of long noncoding RNAs in human
intervertebral disc degeneration by RNA sequencing. Biomed Res Int.
2016:36848752016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mukhopadhyay P, Brock G, Pihur V, Webb C,
Pisano MM and Greene RM: Developmental microRNA expression
profiling of murine embryonic orofacial tissue. Birth Defects Res A
Clin Mol Teratol. 88:511–534. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding HL, Hooper JE, Batzel P, Eames BF,
Postlethwait JH, Artinger KB and Clouthier DE: MicroRNA profiling
during craniofacial development: Potential roles for Mir23b and
Mir133b. Front Physiol. 7:2812016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dey BK, Mueller AC and Dutta A: Long
non-coding RNAs as emerging regulators of differentiation,
development, and disease. Transcription. 5:e9440142014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ozturk F, Li Y, Zhu X, Guda C and Naπwshad
A: Systematic analysis of palatal transcriptome to identify cleft
palate genes within TGFβ3-knockout mice alleles: RNA-Seq analysis
of TGFβ3 Mice. BMC Genomics. 14:1132013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao L, Yin J and Wu W: Long non-coding RNA
H19-mediated mouse cleft palate induced by
2,3,7,8-tetrachlorodibenzo- p-dioxin. Exp Ther Med. 11:2355–2360.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao L, Liu Y, Wen Y and Wu W: LncRNA
H19-mediated mouse cleft palate induced by all-trans retinoic acid.
Hum Exp Toxicol. 36:395–401. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C,
Liu Y, Chen R and Zhao Y: Utilizing sequence intrinsic composition
to classify protein-coding and long non-coding transcripts. Nucleic
Acids Res. 41:e1662013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ,
Wei L and Gao G: CPC: Assess the protein-coding potential of
transcripts using sequence features and support vector machine.
Nucleic Acids Res 35 (Web Server Issue). W345–W349. 2007.
View Article : Google Scholar
|
|
24
|
Mistry J, Bateman A and Finn RD:
Predicting active site residue annotations in the Pfam database.
BMC Bioinformatics. 8:2982007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin MF, Jungreis I and Kellis M: PhyloCSF:
A comparative genomics method to distinguish protein coding and
non-coding regions. Bioinformatics. 27:i275–i282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10:R252009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wen M, Shen Y, Shi S and Tang T: miREvo:
An integrative microRNA evolutionary analysis platform for
next-generation sequencing experiments. BMC Bioinformatics.
13:1402012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11:R142010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mao X, Cai T, Olyarchuk JG and Wei L:
Automated genome annotation and pathway identification using the
KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics.
21:3787–3793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ponting CP, OliPver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huan T, Joehanes R, Schurmann C, Schramm
K, Pilling LC, Peters MJ, Mägi R, DeMeo D, O'Connor GT, Ferrucci L,
et al: A whole-blood transcriptome meta-analysis identifies gene
expression signatures of cigarette smoking. Hum Mol Genet.
25:4611–4623. 2016.PubMed/NCBI
|
|
38
|
Hardy JJ, Mooney SR, Pearson AN, McGuire
D, Correa DJ, Simon RP and Meller R: Assessing the accuracy of
blood RNA profiles to identify patients with post-concussion
syndrome: A pilot study in a military patient population. PLoS One.
12:e01831132017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Zhou S, Zhang Q, Feng S, Chen Y,
Zheng H, Wang X, Zhao W, Zhang T, Zhou Y, et al: Proteomic analysis
of RBP4/vitamin A in children with cleft lip and/or palate. J Dent
Res. 93:547–552. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Zou J, Li Q, Chen L, Gao Y, Yan H,
Zhou B and Li J: Assessment of differentially expressed plasma
microRNAs in nonsyndromic cleft palate and nonsyndromic cleft lip
with cleft palate. Oncotarget. 7:86266–86279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zou J, Li J, Li J, Ji C, Li Q and Guo X:
Expression profile of plasma microRNAs in nonsyndromic cleft lip
and their clinical significance as biomarkers. Biomed Pharmacother.
82:459–466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Iamaroon A, Wallon UM, Overall CM and
Diewert VM: Expression of 72-kDa gelatinase (matrix
metalloproteinase-2) in the developing mouse craniofacial complex.
Arch Oral Biol. 41:1109–1119. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Morris-Wiman J, Du Y and Brinkley L:
Occurrence and temporal variation in matrix metalloproteinases and
their inhibitors during murine secondary palatal morphogenesis. J
Craniofac Genet Dev Biol. 19:201–212. 1999.PubMed/NCBI
|
|
44
|
Morris-Wiman J, Burch H and Basco E:
Temporospatial distribution of matrix metalloproteinase and tissue
inhibitors of matrix metalloproteinases during murine secondary
palate morphogenesi. Anat Embryol (Berl). 202:129–141. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Letra A, da Silva RA, Menezes R, de Souza
AP, de Almeida AL, Sogayar MC and Granjeiro JM: Studies with MMP9
gene promoter polymorphism and nonsyndromic cleft lip and palate.
Am J Med Genet A 143A. 89–91. 2007. View Article : Google Scholar
|
|
46
|
Schoen C, Glennon JC, Abghari S, Bloemen
M, Aschrafi A, Carels CEL and Von den Hoff JW: Differential
microRNA expression in cultured palatal fibroblasts from infants
with cleft palate and controls. Eur J Orthod. 40:90–96. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu T, Chen Y, Du Y, Tao J, Zhou Z and Yang
Z: Serum exosomal MiR-92b-5p as a potential biomarker for acute
heart failure caused by dilated cardiomyopathy. Cell Physiol
Biochem. 46:1939–1950. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wu T, Chen Y, Du Y, Tao J, Li W, Zhou Z
and Yang Z: Circulating exosomal miR-92b-5p is a promising
diagnostic biomarker of heart failure with reduced ejection
fraction patients hospitalized for acute heart failure. J Thorac
Dis. 10:6211–6220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao C, Zhao F, Feng H, Xu S and Qin G:
MicroRNA-92b inhibits epithelial-mesenchymal transition-induced
migration and invasion by targeting Smad3 in nasopharyngeal cancer.
Oncotarget. 8:91603–91613. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhuang LK, Yang YT, Ma X, Han B, Wang ZS,
Zhao QY, Wu LQ and Qu ZQ: MicroRNA-92b promotes hepatocellular
carcinoma progression by targeting Smad7 and is mediated by long
non-coding RNA XIST. Cell Death Dis. 7:e22032016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao S, Moreno M, Eliason S, Cao H, Li X,
Yu W, Bidlack FB, Margolis HC, Baldini A and Amendt BA: TBX1
protein interactions and microRNA-96-5p regulation controls cell
proliferation during craniofacial and dental development:
Implications for 22q11.2 deletion syndrome. Hum Mol Genet.
24:2330–2348. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Baroni T, Bellucci C, Lilli C, Pezzetti F,
Carinci F, Lumare E, Palmieri A, Stabellini G and Bodo M: Human
cleft lip and palate fibroblasts and normal nicotine-treated
fibroblasts show altered in vitro expressions of genes related to
molecular signaling pathways and extracellular matrix metabolism. J
Cell Physiol. 222:748–756. 2010.PubMed/NCBI
|
|
53
|
Park JW, Cai J, McIntosh I, Jabs EW,
Fallin MD, Ingersoll R, Hetmanski JB, Vekemans M, Attie-Bitach T,
Lovett M, et al: High throughput SNP and expression analyses of
candidate genes for non-syndromic oral clefts. J Med Genet.
43:598–608. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yamamoto H, Ito K, Kawai M, Murakami Y,
Bessho K and Ito Y: Runx3 expression during mouse tongue and palate
development. Anat Rec A Discov Mol Cell Evol Biol. 288:695–699.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nawshad A, Medici D and Liu CC: TGFbeta3
inhibits E-cadherin gene expression in palate medial-edge
epithelial cells through a Smad2-Smad4-LEF1 transcription complex.
J Cell Sci. 120:1646–1653. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nawshad A, LaGamba D and Hay ED:
Transforming growth factor beta (TGFbeta) signalling in palatal
growth, apoptosis and epithelial mesenchymal transformation (EMT).
Arch Oral Biol. 49:675–689. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cui XM, Shiomi N, Chen J, Saito T,
Yamamoto T, Ito Y, Bringas P, Chai Y and Shuler CF: Overexpression
of Smad2 in Tgf-beta3-null mutant mice rescues cleft palate. Dev
Biol. 278:193–202. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shin JO, Lee JM, Cho KW, Kwak S, Kwon HJ,
Lee MJ, Cho SW, Kim KS and Jung HS: MiR-200b is involved in Tgf-β
signaling to regulate mammalian palate development. Histochem Cell
Biol. 137:67–78. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shin JO, Nakagawa E, Kim EJ, Cho KW, Lee
JM, Cho SW and Jung HS: miR-200b regulates cell migration via Zeb
family during mouse palate development. Histochem Cell Biol.
137:459–470. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mutz KO, Heilkenbrinker A, Lönne M, Walter
JG and Stahl F: Transcriptome analysis using next-generation
sequencing. Curr Opin Biotechnol. 24:22–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Mantione KJ, Kream RM, Kuzelova H, Ptacek
R, Raboch J, Samuel JM and Stefano GB: Comparing bioinformatic gene
expression profiling methods: Microarray and RNA-Seq. Med Sci Monit
Basic Res. 20:138–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lowe R, Shirley N, Bleackley M, Dolan S
and Shafee T: Transcriptomics technologies. PLoS Comput Biol.
13:e10054572017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Warner DR, Mukhopadhyay P, Brock G, Webb
CL, Michele Pisano M and Greene RM: MicroRNA expression profiling
of the developing murine upper lip. Dev Growth Differ. 56:434–447.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Charoenchaikorn K, Yokomizo T, Rice DP,
Honjo T, Matsuzaki K, Shintaku Y, Imai Y, Wakamatsu A, Takahashi S,
Ito Y, et al: Runx1 is involved in the fusion of the primary and
the secondary palatal shelves. Dev Biol. 326:392–402. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu Y, El-Naggar S, Darling DS, Higashi Y
and Dean DC: Zeb1 links epithelial-mesenchymal transition and
cellular senescence. Development. 135:579–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lai K, Jia S, Yu S, Luo J and He Y:
Genome-wide analysis of aberrantly expressed lncRNAs and miRNAs
with associated co-expression and ceRNA networks in β-thalassemia
and hereditary persistence of fetal hemoglobin. Oncotarget.
5:49931–49943. 2017.
|
|
68
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Dai J, Yu H, Si J, Fang B and Shen SG:
Irf6-related gene regulatory network involved in palate and lip
development. J Craniofac Surg. 26:1600–1605. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Choi JW, Park HW, Kwon YJ and Park BY:
Role of apoptosis in retinoic acid-induced cleft palate. J
Craniofac Surg. 22:1567–1571. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chenevix-Trench G, Jones K, Green AC,
Duffy DL and Martin NG: Cleft lip with or without cleft palate:
Associations with transforming growth factor alpha and retinoic
acid receptor loci. Am J Hum Genet. 51:1377–1385. 1992.PubMed/NCBI
|
|
72
|
Zhu X, Ozturk F, Pandey S, Guda CB and
Nawshad A: Implications of TGFβ on transcriptome and cellular
biofunctions of palatal mesenchyme. Front Physiol. 3:852012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Christensen K, Juel K, Herskind AM and
Murray JC: Long term follow up study of survival associated with
cleft lip and palate at birth. BMJ. 328:14052004. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
De Oliveira PSN, Coutinho LL, Tizioto PC,
Cesar ASM, de Oliveira GB, Diniz WJDS, De Lima AO, Reecy JM, Mourão
GB, Zerlotini A and Regitano LCA: An integrative transcriptome
analysis indicates regulatory mRNA-miRNA networks for residual feed
intake in Nelore cattle. Sci Rep. 8:170722018. View Article : Google Scholar : PubMed/NCBI
|