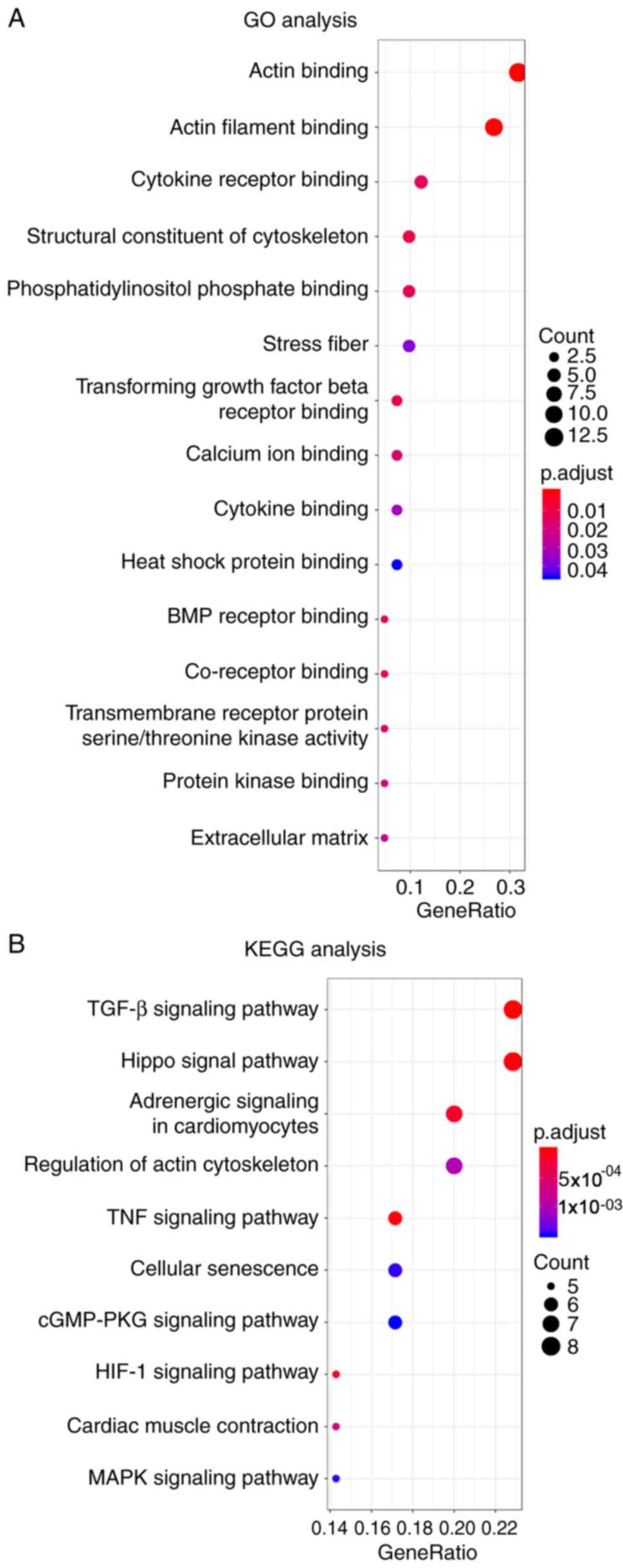
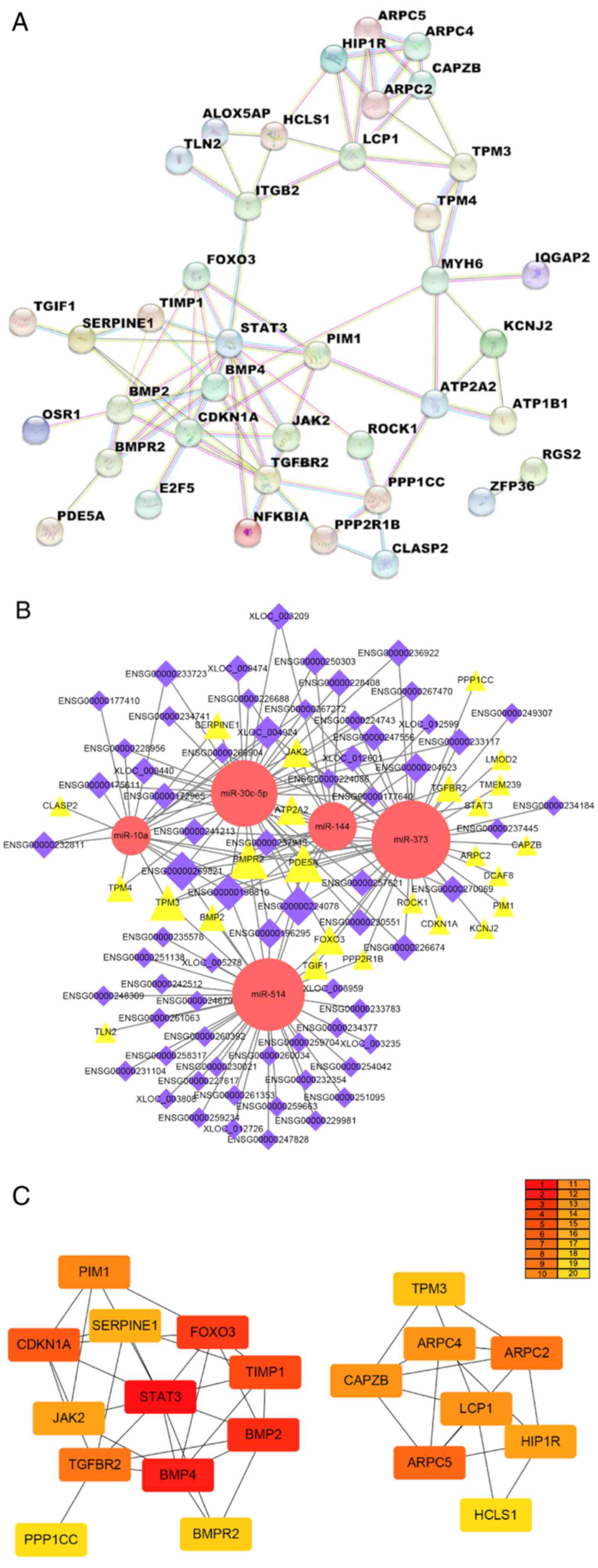
|
1
|
Maron BJ, Gardin JM, Flack JM, Gidding SS,
Kurosaki TT and Bild DE: Prevalence of hypertrophic cardiomyopathy
in a general population of young adults. Echocardiographic analysis
of 4111 subjects in the CARDIA study. Coronary artery risk
development in (Young) adults. Circulation. 92:785–789. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maron BJ, Maron MS and Semsarian C:
Genetics of hypertrophic cardiomyopathy after 20 years: Clinical
perspectives. J Am Coll Cardiol. 60:705–715. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Semsarian C, Ingles J, Maron MS and Maron
BJ: New perspectives on the prevalence of hypertrophic
cardiomyopathy. J Am Coll Cardiol. 65:1249–1254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Helms AS and Day SM: Other side of the
coin: The missing heritability in hypertrophic cardiomyopathy. Eur
Heart J. 38:3469–3471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song L, Su M, Wang SY, Zou Y, Wang X, Wang
Y, Cui H, Zhao P, Hui R and Wang J: MiR-451 is decreased in
hypertrophic cardiomyopathy and regulates autophagy by targeting
TSC1. J Cell Mol Med. 18:2266–2274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Papadakis M, Hadley G, Xilouri M, Hoyte
LC, Nagel S, McMenamin MM, Tsaknakis G, Watt SM, Drakesmith CW,
Chen R, et al: Tsc1 (hamartin) confers neuroprotection against
ischemia by inducing autophagy. Nat Med. 19:351–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li P, Piao Y, Shon HS and Ryu KH:
Comparing the normalization methods for the differential analysis
of Illumina high-throughput RNA-Seq data. BMC Bioinformatics.
16:3472015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Parker HS, Bravo HC and Leek JT: Removing
batch effects for prediction problems with frozen surrogate
variable analysis. PeerJ. 2:e5612014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Benjamini Y and Hochberg Y: Controlling
the False Discovery Rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
14
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-LncBase: Experimentally verified and computationally
predicted microRNA targets on long non-coding RNAs. Nucleic Acids
Res. 41:D239–D245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Volders PJ, Anckaert J, Verheggen K,
Nuytens J, Martens L, Mestdagh P and Vandesompele J: LNCipedia 5:
Towards a reference set of human long non-coding RNAs. Nucleic
Acids Res. 47:D135–D139. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xing C, Cai ZZ, Gong J, Zhou J, Xu JJ and
Guo F: Identification of potential biomarkers involved in gastric
cancer through integrated analysis of non-coding RNA associated
competing endogenous RNAs network. Clin Lab. 64:1661–1669. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. Aug 12–2015.(Epub ahead of print). doi: 10.7554/eLife.05005.
View Article : Google Scholar
|
|
21
|
Xiong DD, Dang YW, Lin P, Wen DY, He RQ,
Luo DZ, Feng ZB and Chen G: A circRNA-miRNA-mRNA network
identification for exploring underlying pathogenesis and therapy
strategy of hepatocellular carcinoma. J Transl Med. 16:2202018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
The Gene Ontology Consortium: The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rappaport N, Twik M, Plaschkes I, Nudel R,
Iny Stein T, Levitt J, Gershoni M, Morrey CP, Safran M and Lancet
D: MalaCards: An amalgamated human disease compendium with diverse
clinical and genetic annotation and structured search. Nucleic
Acids Res. 45(D1): D877–D887. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu GM, Wong L and Chua HN: Complex
discovery from weighted PPI networks. Bioinformatics. 25:1891–1897.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: A
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36:D901–D906. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Law V, Knox C, Djoumbou Y, Jewison T, Guo
AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al:
DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids
Res. 42:D1091–D1097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ebrahimie E, Fruzangohar M, Moussavi Nik
SH and Newman M: Gene ontology-based analysis of zebrafish omics
data using the web tool comparative gene ontology. Zebrafish.
14:492–494. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fang L, Ellims AH, Beale AL, Taylor AJ,
Murphy A and Dart AM: Systemic inflammation is associated with
myocardial fibrosis, diastolic dysfunction, and cardiac hypertrophy
in patients with hypertrophic cardiomyopathy. Am J Transl Res.
9:5063–5073. 2017.PubMed/NCBI
|
|
35
|
Zen K, Irie H, Doue T, Takamiya M, Yamano
T, Sawada T, Azuma A and Matsubara H: Analysis of circulating
apoptosis mediators and proinflammatory cytokines in patients with
idiopathic hypertrophic cardiomyopathy: Comparison between
nonobstructive and dilated-phase hypertrophic cardiomyopathy. Int
Heart J. 46:231–244. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mirtschink P and Krek W: Hypoxia-driven
glycolytic and fructolytic metabolic programs: Pivotal to
hypertrophic heart disease. Biochim Biophys Acta. 1863:1822–1828.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Semenza GL: Signal transduction to
hypoxia-inducible factor 1. Biochem Pharmacol. 64:993–998. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sherrid MV: Drug therapy for hypertrophic
cardiomypathy: Physiology and practice. Curr Cardiol Rev. 12:52–65.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ammirati E, Contri R, Coppini R, Cecchi F,
Frigerio M and Olivotto I: Pharmacological treatment of
hypertrophic cardiomyopathy: Current practice and novel
perspectives. Eur J Heart Fail. 18:1106–1118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Burns C, Bagnall RD, Lam L, Semsarian C
and Ingles J: Multiple gene variants in hypertrophic cardiomyopathy
in the era of next-generation sequencing. Circ Cardiovasc Genet.
10(pii): e0016662017.PubMed/NCBI
|
|
41
|
Ntelios D, Meditskou S, Efthimiadis G,
Pitsis A, Nikolakaki E, Girtovitis F, Parcharidou D, Zegkos T,
Kouidou S, Karvounis H and Tzimagiorgis G: Elevated plasma levels
of miR-29a are associated with hemolysis in patients with
hypertrophic cardiomyopathy. Clin Chim Acta. 471:321–326. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Salman OF, El-Rayess HM, Abi Khalil C,
Nemer G and Refaat MM: Inherited cardiomyopathies and the role of
mutations in non-coding regions of the genome. Front Cardiovasc
Med. 5:772018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang L, Ellims A, Moore XL, White DA,
Taylor AJ, Chin-Dusting J and Dart AM: Circulating microRNAs as
biomarkers for diffuse myocardial fibrosis in patients with
hypertrophic cardiomyopathy. J Transl Med. 13:3142015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xue J, Zhou D, Poulsen O, Hartley I,
Imamura T, Xie EX and Haddad GG: Exploring miRNA-mRNA regulatory
network in cardiac pathology in Na+/H+
exchanger isoform 1 transgenic mice. Physiol Genomics. 50:846–861.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yamada S, Hsiao YW, Chang SL, Lin YJ, Lo
LW, Chung FP, Chiang SJ, Hu YF, Tuan TC, Chao TF, et al:
Circulating microRNAs in arrhythmogenic right ventricular
cardiomyopathy with ventricular arrhythmia. Europace 20 (FI1).
F37–F45. 2018. View Article : Google Scholar
|
|
46
|
Lee JD, Lee TH, Huang YC, Chang YJ, Chang
CH, Hsu HL, Lin YH, Wu CY, Lee M, Huang YC, et al: ALOX5AP genetic
variants and risk of atherothrombotic stroke in the Taiwanese
population. J Clin Neurosci. 18:1634–1638. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Helgadottir A, Manolescu A, Thorleifsson
G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ,
Gudmundsson G, Grant SF, Thorgeirsson G, et al: The gene encoding
5-lipoxygenase activating protein confers risk of myocardial
infarction and stroke. Nat Genet. 36:233–239. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lai HM, Li XM, Yang YN, Ma YT, Xu R, Pan
S, Zhai H, Liu F, Chen BD and Zhao Q: Genetic variation in NFKB1
and NFKBIA and susceptibility to coronary artery disease in a
Chinese Uygur population. PLoS One. 10:e01291442015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Han P, Li W, Lin CH, Yang J, Shang C,
Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al: A long noncoding
RNA protects the heart from pathological hypertrophy. Nature.
514:102–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang W, Li Y, He F and Wu H: Microarray
profiling of long non-coding RNA (lncRNA) associated with
hypertrophic cardiomyopathy. BMC Cardiovasc Disord. 15:622015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sherrid MV, Shetty A, Winson G, Kim B,
Musat D, Alviar CL, Homel P, Balaram SK and Swistel DG: Treatment
of obstructive hypertrophic cardiomyopathy symptoms and gradient
resistant to first-line therapy with β-blockade or verapamil. Circ
Heart Fail. 6:694–702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Östman-Smith I: Beta-Blockers in pediatric
hypertrophic cardiomyopathies. Rev Recent Clin Trials. 9:82–85.
2014. View Article : Google Scholar : PubMed/NCBI
|