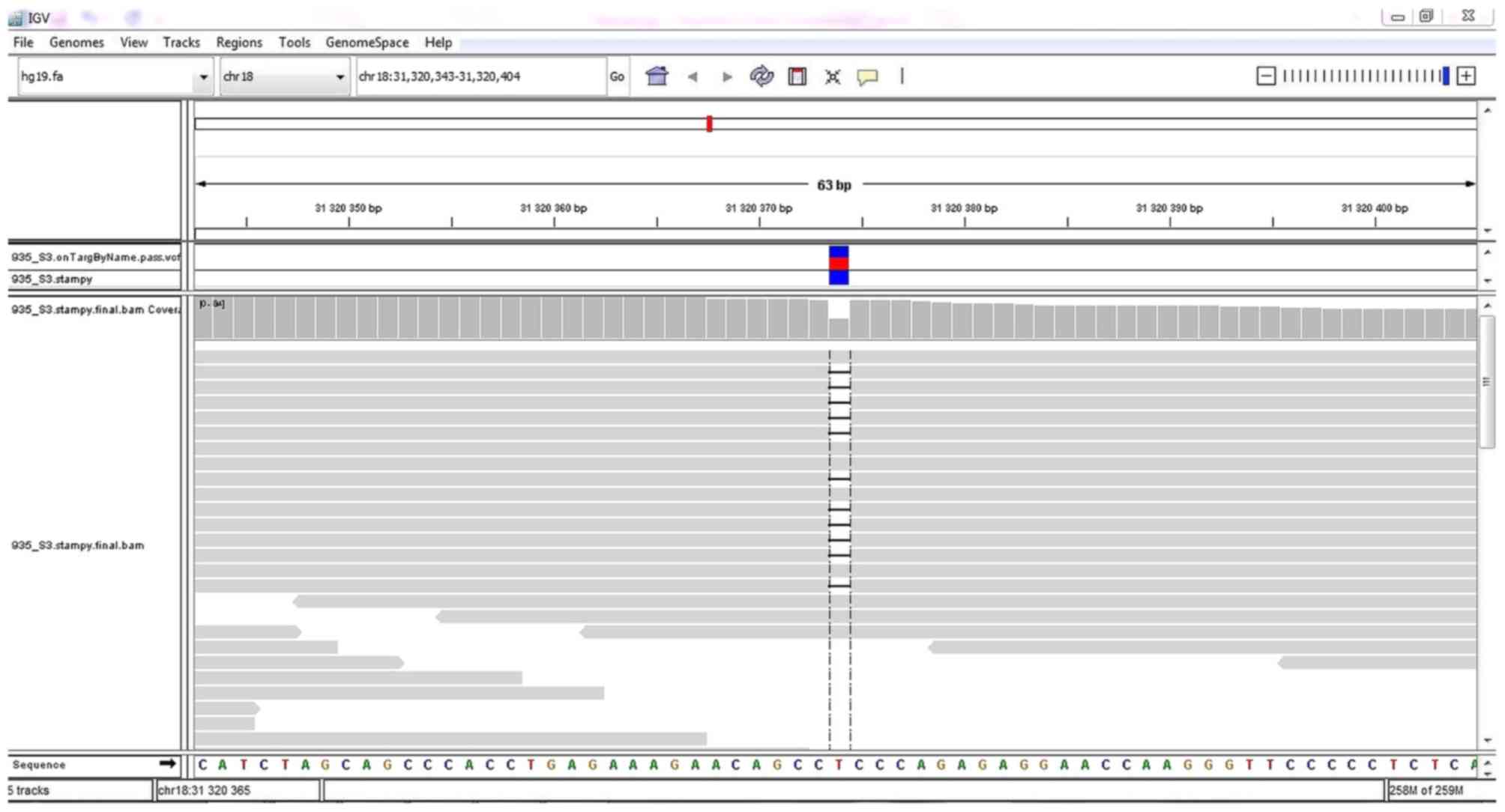
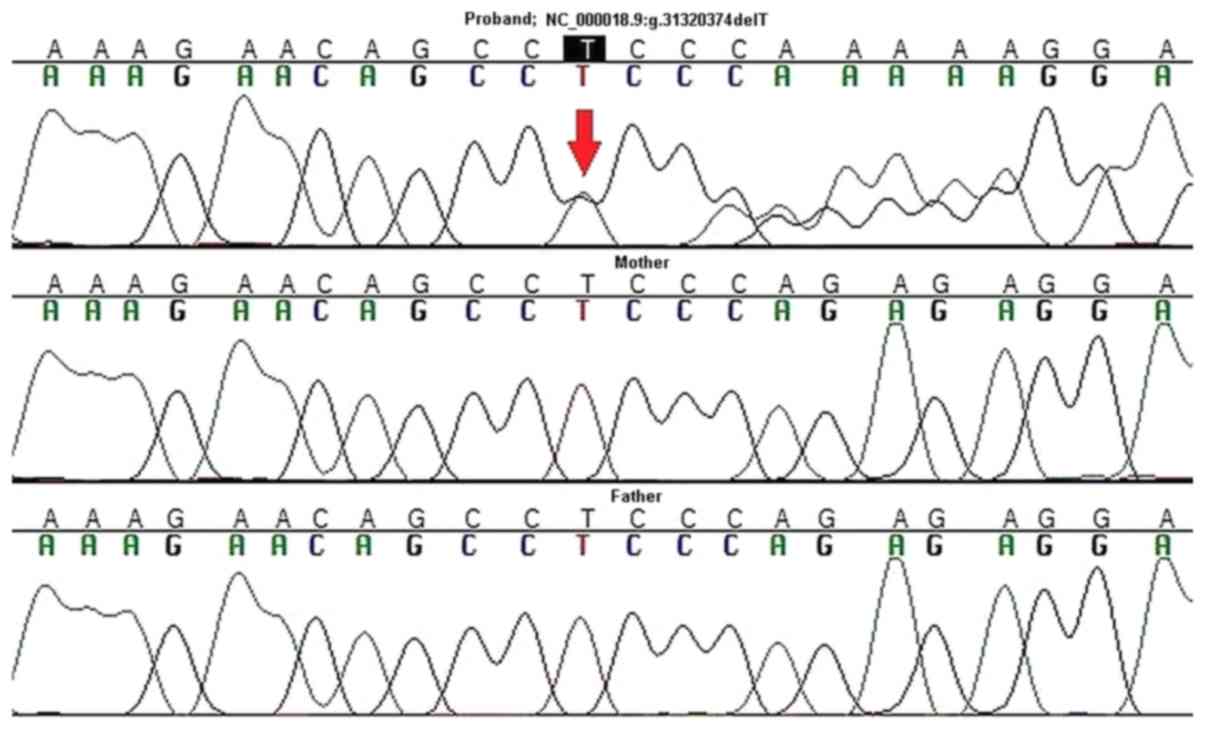
|
1
|
Kochinke K, Zweier C, Nijhof B, Fenckova
M, Cizek P, Honti F, Keerthikumar S, Oortveld MA, Kleefstra T,
Kramer JM, et al: Systematic phenomics analysis deconvolutes genes
mutated in intellectual disability into biologically coherent
modules. Am J Hum Genet. 98:149–164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Redin C, Gérard B, Lauer J, Herenger Y,
Muller J, Quartier A, Masurel-Paulet A, Willems M, Lesca G,
El-Chehadeh S, et al: Efficient strategy for the molecular
diagnosis of intellectual disability using targeted high-throughput
sequencing. J Med Genet. 51:724–736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rauch A, Wieczorek D, Graf E, Wieland T,
Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di
Donato N, et al: Range of genetic mutations associated with severe
non-syndromic sporadic intellectual disability: An exome sequencing
study. Lancet. 380:1674–1682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Srivastava A, Ritesh KC, Tsan YC, Liao R,
Su F, Cao X, Hannibal MC, Keegan CE, Chinnaiyan AM, Martin DM and
Bielas SL: De novo dominant ASXL3 mutations alter H2A
deubiquitination and transcription in Bainbridge-Ropers syndrome.
Hum Mol Genet. 25:597–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bainbridge MN, Hu H, Muzny DM, Musante L,
Lupski JR, Graham BH, Chen W, Gripp KW, Jenny K, Wienker TF, et al:
De novo truncating mutations in ASXL3 are associated with a novel
clinical phenotype with similarities to Bohring-Opitz syndrome.
Genome Med. 5:112013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kuechler A, Czeschik JC, Graf E, Grasshoff
U, Hüffmeier U, Busa T, Beck-Woedl S, Faivre L, Rivière JB, Bader
I, et al: Bainbridge-Ropers syndrome caused by loss-of-function
variants in ASXL3: A recognizable condition. Eur J Hum Genet.
25:183–191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vojta V and Peters A: Das Vojta Prinzip.
3rd edition. Springer; Heidelberg: 2007,
|
|
8
|
Jung MW, Landenberger M, Jung T,
Lindenthal T and Philippi H: Vojta therapy and neurodevelopmental
treatment in children with infantile postural asymmetry: A
randomized controlled trial. J Phys Ther Sci. 29:301–306. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silva M, de Leeuw N, Mann K, Schuring-Blom
H, Morgan S, Giardino D, Rack K and Hastings R: European guidelines
for constitutional cytogenomic analysis. Eur J Hum Genet. 27:1–16.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Howe B, Umrigar A and Tsien F: Chromosome
preparation from cultured cells. J Vis Exp. e502032014.PubMed/NCBI
|
|
11
|
Roy S and Motsinger Reif A: Evaluation of
calling algorithms for array-CGH. Front Genet. 4:2172013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davis MP, van Dongen S, Abreu-Goodger C,
Bartonicek N and Enright AJ: Kraken: A set of tools for quality
control and analysis of high-throughput sequence data. Methods.
63:41–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet.journal.
17:10–12. 2011. View Article : Google Scholar
|
|
14
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Speir ML, Zweig AS, Rosenbloom KR, Raney
BJ, Paten B, Nejad P, Lee BT, Learned K, Karolchik D, Hinrichs AS,
et al: The UCSC Genome Browser database: 2016 update. Nucleic Acids
Res. 44:D717–D725. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lunter G and Goodson M: Stampy: A
statistical algorithm for sensitive and fast mapping of Illumina
sequence reads. Genome Res. 21:936–939. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; 1000 Genome
Project Data Processing Subgroup, : The Sequence Alignment/Map
format and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M, et al: The Genome Analysis Toolkit: A MapReduce framework for
analyzing next-generation DNA sequencing data. Genome Res.
20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cingolani P, Patel VM, Coon M, Nguyen T,
Land SJ, Ruden DM and Lu X: Using Drosophila melanogaster as a
model for genotoxic chemical mutational studies with a new program,
SnpSift. Front Genet. 3:352012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Landrum MJ, Lee JM, Benson M, Brown G,
Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al
ClinVar, : Public archive of interpretations of clinically relevant
variants. Nucleic Acids Res. 44:D862–D868. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cingolani P, Platts A, Wang le L, Coon M,
Nguyen T, Wang L, Land SJ, Lu X and Ruden DM: A program for
annotating and predicting the effects of single nucleotide
polymorphisms, SnpEff: SNPs in the genome of Drosophila
melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Danecek P, Auton A, Abecasis G, Albers CA,
Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST,
et al: The variant call format and VCFtools. Bioinformatics.
27:2156–2158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Obenchain V, Lawrence M, Carey V, Gogarten
S, Shannon P and Morgan M: VariantAnnotation: A Bioconductor
package for exploration and annotation of genetic variants.
Bioinformatics. 30:2076–2078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Robinson JT, Thorvaldsdóttir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thorvaldsdóttir H, Robinson JT and Mesirov
JP: Integrative Genomics Viewer (IGV): High-performance genomics
data visualization and exploration. Brief Bioinform. 14:178–192.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American College
of Medical Genetics and Genomics and the Association for Molecular
Pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hamosh A, Scott AF, Amberger JS, Bocchini
CA and McKusick VA: Online Mendelian Inheritance in Man (OMIM), a
knowledgebase of human genes and genetic disorders. Nucleic Acids
Res. 33:D514–D517. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
UniProt Consortium: The universal protein
resource (UniProt). Nucleic Acids Res. 36:D190–D195.
2008.PubMed/NCBI
|
|
32
|
Lek M, Karczewski KJ, Minikel EV, Samocha
KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ,
Cummings BB, et al: Analysis of protein-coding genetic variation in
60,706 humans. Nature. 536:285–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
1000 Genomes Project Consortium, ; Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Choi Y, Sims GE, Murphy S, Miller JR and
Chan AP: Predicting the functional effect of amino acid
substitutions and indels. PLoS One. 7:e466882012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kopanos C, Tsiolkas V, Kouris A, Chapple
CE, Albarca Aguilera M, Meyer R and Massouras A: VarSome: The human
genomic variant search engine. Bioinformatics. 2018.
|
|
38
|
Untergasser A, Cutcutache I, Koressaar T,
Ye J, Faircloth BC, Remm M and Rozen SG: Primer3-new capabilities
and interfaces. Nucleic Acids Res. 40:e1152012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koressaar T and Remm M: Enhancements and
modifications of primer design program Primer3. Bioinformatics.
23:1289–1291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ye J, Coulouris G, Zaretskaya I,
Cutcutache I, Rozen S and Madden TL: Primer-BLAST: A tool to design
target-specific primers for polymerase chain reaction. BMC
Bioinformatics. 13:1342012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Katoh M and Katoh M: Identification and
characterization of ASXL3 gene in silico. Int J Oncol.
24:1617–1622. 2004.PubMed/NCBI
|
|
42
|
Katoh M: Functional and cancer genomics of
ASXL family members. Br J Cancer. 109:299–306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sinclair DA, Milne TA, Hodgson JW,
Shellard J, Salinas CA, Kyba M, Randazzo F and Brock HW: The
Additional sex combs gene of Drosophila encodes a chromatin protein
that binds to shared and unique Polycomb group sites on polytene
chromosomes. Development. 125:1207–1216. 1998.PubMed/NCBI
|
|
44
|
Gaytán de Ayala Alonso A, Gutiérrez L,
Fritsch C, Papp B, Beuchle D and Müller J: A genetic screen
identifies novel polycomb group genes in Drosophila. Genetics.
176:2099–2108. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Balasubramanian M, Willoughby J, Fry AE,
Weber A, Firth HV, Deshpande C, Berg JN, Chandler K, Metcalfe KA,
Lam W, et al: Delineating the phenotypic spectrum of
Bainbridge-Ropers syndrome: 12 new patients with de novo,
heterozygous, loss-of-function mutations in ASXL3 and review of
published literature. J Med Genet. 54:537–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bohring A, Silengo M, Lerone M, Superneau
DW, Spaich C, Braddock SR, Poss A and Opitz JM: Severe end of Opitz
trigonocephaly (C) syndrome or new syndrome? Am J Med Genet.
85:438–446. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shashi V, Pena LD, Kim K, Burton B, Hempel
M, Schoch K, Walkiewicz M, McLaughlin HM, Cho M, Stong N, et al: De
novo truncating variants in ASXL2 are associated with a unique and
recognizable clinical phenotype. Am J Hum Genet. 99:991–999. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chinen Y, Nakamura S, Ganaha A, Hayashi S,
Inazawa J, Yanagi K, Nakanishi K, Kaname T and Naritomi K: Mild
prominence of Sylvian fissure in a Bainbridge-Ropers syndrome
patient with a novel frameshift variant in ASXL3. Clin Case Rep.
6:330–336. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Giri D, Rigden D, Didi M, Peak M, McNamara
P and Senniappan S: Novel compound heterozygous ASXL3
mutation causing Bainbridge-Ropers like syndrome and primary IGF1
deficiency. Int J Pediatr Endocrinol. 2017:82017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
De Rubeis S, He X, Goldberg AP, Poultney
CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al:
Synaptic, transcriptional and chromatin genes disrupted in autism.
Nature. 515:209–215. 2014. View Article : Google Scholar : PubMed/NCBI
|