|
1
|
Arnautovic K and Arnautovic A:
Extramedullary intradural spinal tumors: A review of modern
diagnostic and treatment options and a report of a series. Bosn J
Basic Med Sci. 9 (Suppl 1):S40–S45. 2009. View Article : Google Scholar :
|
|
2
|
Allen JC, Siffert J and Hukin J: Clinical
manifestations of childhood ependymoma: A multitude of syndromes.
Pediatr Neurosurg. 28:49–55. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thuppal S, Propp JM and McCarthy BJ:
Average years of potential life lost in those who have died from
brain and CNS tumors in the USA. Neuroepidemiology. 27:22–27. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pajtler KW, Witt H, Sill M, Jones DT,
Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C,
Johann P, et al: Molecular classification of ependymal tumors
across all CNS compartments, histopathological grades, and age
groups. Cancer Cell. 27:728–743. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pfeffer C and Olsen BR: Editorial: Journal
of negative results in biomedicine. J Negative Results Biomed.
1:22002. View Article : Google Scholar
|
|
6
|
Witt H, Gramatzki D, Hentschel B, Pajtler
KW, Felsberg J, Schackert G, Löffler M, Capper D, Sahm F, Sill M,
et al: DNA methylation-based classification of ependymomas in
adulthood: Implications for diagnosis and treatment. Neuro
Oncology. 20:1616–1624. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma DL, Lin S, Leung KH, Zhong HJ, Liu LJ,
Chan DS, Bourdoncle A, Mergny JL, Wang HM and Leung CH: An
oligonucleotide-based label-free luminescent switch-on probe for
RNA detection utilizing a G-quadruplex-selective iridium(III)
complex. Nanoscale. 6:8489–8494. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He HZ, Chan DS, Leung CH and Ma DL: A
highly selective G-quadruplex-based luminescent switch-on probe for
the detection of gene deletion. Chem Commun (Camb). 48:9462–9464.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leung KH, He HZ, Chan DS, Fu WC, Leungb CH
and Maa DL: An oligonucleotide-based switch-on luminescent probe
for the detection of kanamycin in aqueous solution. Sensors
Actuators B: Chemical. 177:487–492. 2013. View Article : Google Scholar
|
|
10
|
Wu C, Wu KJ, Kang TS, Wang HD, Leung CH,
Liu JB and Ma DL: Iridium-based probe for luminescent nitric oxide
monitoring in live cells. Sci Rep. 8:124672018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kiltschewskij D and Cairns MJ:
Temporospatial guidance of activity-dependent gene expression by
microRNA: Mechanisms and functional implications for neural
plasticity. Nucleic Acids Res. 47:533–545. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao Y, Yu F, Pang L, Zhao H, Liu L, Zhang
G, Liu T, Zhang H, Fan H, Zhang Y, et al: MeSiC: A model-based
method for estimating 5 mC levels at Single-CpG resolution from
MeDIP-seq. Sci Rep. 5:146992015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klutstein M, Nejman D, Greenfield R and
Cedar H: DNA methylation in cancer and aging. Cancer Res.
76:3446–3450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu F, Quan F, Xu J, Zhang Y, Xie Y, Zhang
J, Lan Y, Yuan H, Zhang H, Cheng S, et al: Breast cancer prognosis
signature: Linking risk stratification to disease subtypes. Brief
Bioinform. Sep 3–2018.doi: 10.1093/bib/bby073 (Epub ahead of
print). View Article : Google Scholar
|
|
16
|
Koch A, Joosten SC, Feng Z, de Ruijter TC,
Draht MX, Melotte V, Smits KM, Veeck J, Herman JG, Van Neste L, et
al: Analysis of DNA methylation in cancer: Location revisited. Nat
Rev Clin Oncol. 15:459–466. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng
Y, Zhou T, Zhang MQ, Wu CI and Wang SM: Poly A-transcripts
expressed in HeLa cells. PLoS One. 3:e28032008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu F, Zhang G, Shi A, Hu J, Li F, Zhang X,
Zhang Y, Huang J, Xiao Y, Li X and Cheng S: LnChrom: A resource of
experimentally validated lncRNA-chromatin interactions in human and
mouse. Database (Oxford). 20182018.doi:
10.1093/database/bay039.
|
|
20
|
Zhang Y, Li X, Zhou D, Zhi H, Wang P, Gao
Y, Guo M, Yue M, Wang Y, Shen W, et al: Inferences of individual
drug responses across diverse cancer types using a novel competing
endogenous RNA network. Mol Oncol. 12:1429–1446. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bolha L, Ravnik-Glavac M and Glavac D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:72439682017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L, Li S; Cancer Genome Atlas Research Network, ; Xie W and
Yang D: lncRNA epigenetic landscape analysis identifies EPIC1 as an
oncogenic lncRNA that interacts with MYC and promotes cell-cycle
progression in cancer. Cancer Cell. 33:706–720.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heilmann K, Toth R, Bossmann C, Klimo K,
Plass C and Gerhauser C: Genome-wide screen for differentially
methylated long noncoding RNAs identifies Esrp2 and lncRNA Esrp2-as
regulated by enhancer DNA methylation with prognostic relevance for
human breast cancer. Oncogene. 36:6446–6461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Clough E and Barrett T: The gene
expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou W, Laird PW and Shen H: Comprehensive
characterization, annotation and innovative use of Infinium DNA
methylation BeadChip probes. Nucleic Acids Res.
45:e222017.PubMed/NCBI
|
|
28
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aine M, Sjödahl G, Eriksson P, Veerla S,
Lindgren D, Ringnér M and Höglund M: Integrative epigenomic
analysis of differential DNA methylation in urothelial carcinoma.
Genome Med. 7:232015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li
Y, Liu L, Hou P, Cui T, Tan P, et al: RAID v2.0: An updated
resource of RNA-associated interactions across organisms. Nucleic
Acids Res. 45:D115–D118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X and Ishwaran H: Random forests for
genomic data analysis. Genomics. 99:323–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nguyen TT, Huang JZ and Nguyen TT:
Unbiased feature selection in learning random forests for
high-dimensional data. TheScientificWorldJournal. 2015:4713712015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McLean CY, Bristor D, Hiller M, Clarke SL,
Schaar BT, Lowe CB, Wenger AM and Bejerano G: GREAT improves
functional interpretation of cis-regulatory regions. Nat
Biotechnol. 28:495–501. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Prayson RA and Suh JH: Subependymomas:
Clinicopathologic study of 14 tumors, including comparative MIB-1
immunohistochemical analysis with other ependymal neoplasms. Arch
Pathol Lab Med. 123:306–309. 1999.PubMed/NCBI
|
|
37
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yao J, Zhang L, Hu L, Guo B, Hu X,
Borjigin U, Wei Z, Chen Y, Lv M, Lau JT, et al: Tumorigenic
potential is restored during differentiation in fusion-reprogrammed
cancer cells. Cell Death Dis. 7:e23142016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y, Liu D, Wang L, Wang S, Yu X, Dai
E, Liu X, Luo S and Jiang W: Integrated systems approach identifies
risk regulatory pathways and key regulators in coronary artery
disease. J Mol Med (Berl). 93:1381–1390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang W, Zhang Y, Meng F, Lian B, Chen X,
Yu X, Dai E, Wang S, Liu X, Li X, et al: Identification of active
transcription factor and miRNA regulatory pathways in Alzheimer's
disease. Bioinformatics. 29:2596–2602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ferre F, Colantoni A and Helmer-Citterich
M: Revealing protein-lncRNA interaction. Brief Bioinform.
17:106–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xiong D, Sheng Y, Ding S, Chen J, Tan X,
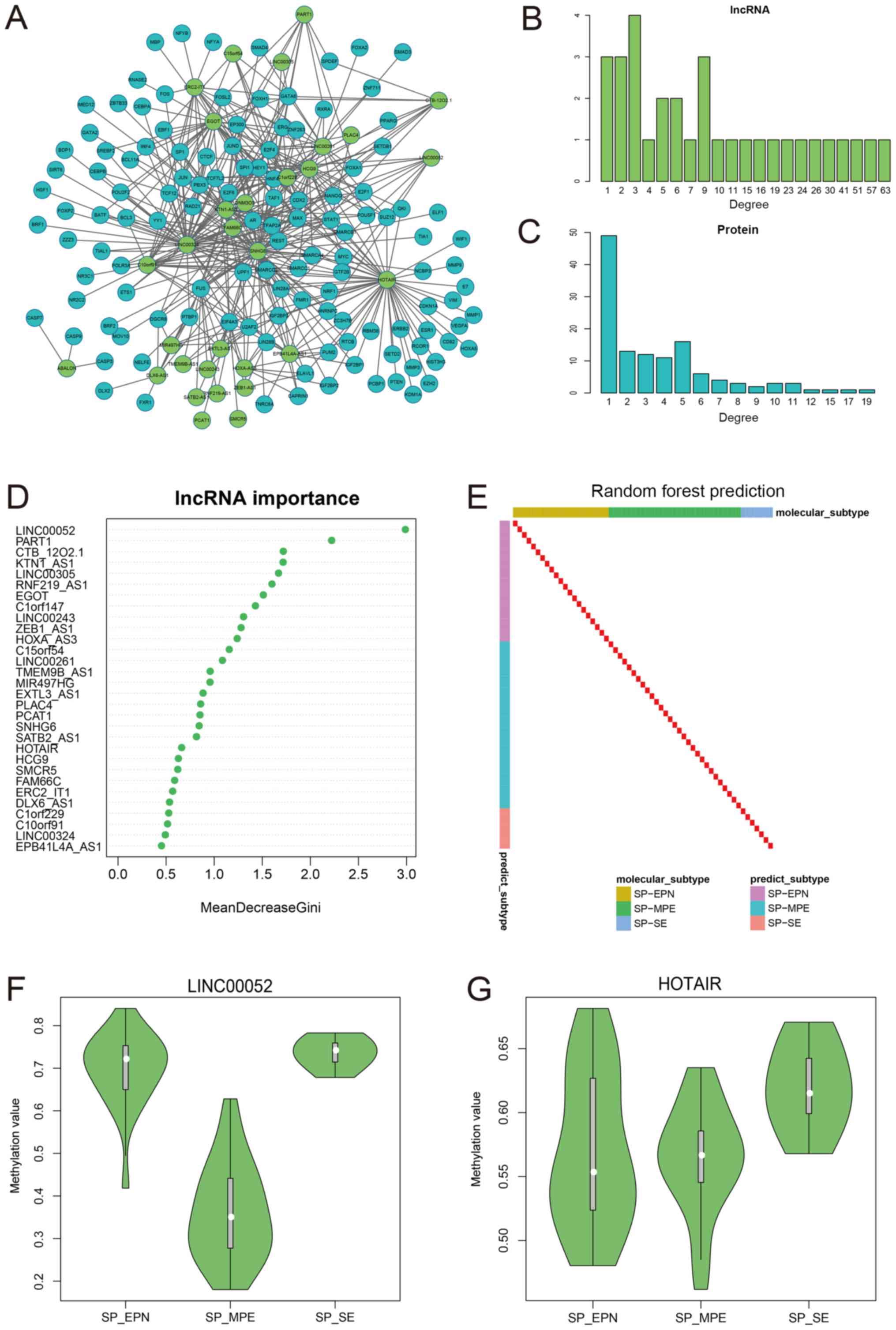
Zeng T, Qin D, Zhu L, Huang A and Tang H: LINC00052 regulates the
expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC
cells invasion and migration. Oncotarget. 7:47593–47608. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Salameh A, Fan X, Choi BK, Zhang S, Zhang
N and An Z: HER3 and LINC00052 interplay promotes tumor growth in
breast cancer. Oncotarget. 8:6526–6539. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hajjari M and Salavaty A: HOTAIR: An
oncogenic long non-coding RNA in different cancers. Cancer Biol
Med. 12:1–9. 2015.PubMed/NCBI
|
|
47
|
Teschendorff AE, Lee SH, Jones A, Fiegl H,
Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, et al:
HOTAIR and its surrogate DNA methylation signature indicate
carboplatin resistance in ovarian cancer. Genome Med. 7:1082015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang JX, Han L, Bao ZS, Wang YY, Chen LY,
Yan W, Yu SZ, Pu PY, Liu N, You YP, et al: HOTAIR, a cell
cycle-associated long noncoding RNA and a strong predictor of
survival, is preferentially expressed in classical and mesenchymal
glioma. Neuro Oncol. 15:1595–1603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Heindryckx F and Li JP: Role of
proteoglycans in neuro-inflammation and central nervous system
fibrosis. Matrix Biol. 68-69:589–601. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Silver DJ and Silver J: Contributions of
chondroitin sulfate proteoglycans to neurodevelopment, injury, and
cancer. Curr Opin Neurobiol. 27:171–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rolls A, Shechter R, London A, Segev Y,
Jacob-Hirsch J, Amariglio N, Rechavi G and Schwartz M: Two faces of
chondroitin sulfate proteoglycan in spinal cord repair: A role in
microglia/macrophage activation. PLoS Med. 5:e1712008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Villano JL, Parker CK and Dolecek TA:
Descriptive epidemiology of ependymal tumours in the United States.
Br J Cancer. 108:2367–2371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Armstrong TS, Vera-Bolanos E, Bekele BN,
Aldape K and Gilbert MR: Adult ependymal tumors: Prognosis and the
M. D. Anderson Cancer Center experience. Neuro Oncol. 12:862–870.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee CH, Chung CK and Kim CH: Genetic
differences on intracranial versus spinal cord ependymal tumors: A
meta-analysis of genetic researches. Eur Spine J. 25:3942–3951.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gittleman H, Boscia A, Ostrom QT, Truitt
G, Fritz Y, Kruchko C and Barnholtz-Sloan JS: Survivorship in
adults with malignant brain and other central nervous system tumor
from 2000–2014. Neuro Oncol. 20 (Suppl 7):vii6–vii16. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Olmos G and Lladó J: Tumor necrosis factor
alpha: A link between neuroinflammation and excitotoxicity.
Mediators Inflamm. 2014:8612312014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mak TW and Yeh WC: Signaling for survival
and apoptosis in the immune system. Arthritis Res. 4 (Suppl
3):S243–S252. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
58
|
Parameswaran N and Patial S: Tumor
necrosis factor-alpha signaling in macrophages. Crit Rev Eukaryot
Gene Expr. 20:87–103. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
van Horssen R, Ten Hagen TL and Eggermont
AM: TNF-alpha in cancer treatment: Molecular insights, antitumor
effects, and clinical utility. Oncologist. 11:397–408. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Probert L: TNF and its receptors in the
CNS: The essential, the desirable and the deleterious effects.
Neuroscience. 302:2–22. 2015. View Article : Google Scholar : PubMed/NCBI
|