Introduction
Acute myeloid leukemia (AML) is a hematological
disorder that results from the abnormal differentiation and
proliferation of hematopoietic cells, and chemotherapy represents
the standard therapeutic treatment for patients with AML (1,2).
Despite the high response rate to chemotherapy, relapse frequently
occurs after remission, lowering the overall response rate in
patients with AML, making post-remission therapy necessary
(3,4). Therefore, it is necessary to find
minimal residual disease (MRD) markers in order to evaluate the
curative effects of therapeutic treatments and predict relapse of
AML. At present, to the best of our knowledge, the main techniques
for detecting MRD are multi-parameter flow cytometry (MFC) and
reverse transcription-quantitative (RT-q)PCR (5,6).
However, due to limitations in detecting the full spectrum of
relapse risks, MFC is not an ideal methodology for MRD monitoring.
Accumulating evidence indicated that RT-qPCR, which is used for
quantitative determination of fusion genes, genetic mutations and
overexpression of oncogenes, is more sensitive than MFC in MRD
monitoring (7,8). Despite the high sensitivity and
accuracy of RT-qPCR, detection of biomarkers is only suitable for
patients with certain fusion genes, such as RUNX family
transcription factor 1 (RUNX1)-RUNX1 translocation partner 1 (ETO)
(9,10). Therefore, it is required to
identify additional molecular markers of MRD suitable for all
patients.
Wilm's tumor 1 (WT1) gene, encoding a zinc-finger
transcription factor, was originally identified as a tumor
suppressor gene regulating hematopoiesis and apoptosis (11). Previous studies have revealed that
WT1 is highly expressed in >80% of patients with AML and may
cause poor clinical outcomes due to an increased resistance to
apoptosis (12–15). Despite these previous studies, to
the best of our knowledge, WT1 has not been used as an established
marker for MRD monitoring in hematological malignancies. Frairia
et al (15) observed that
WT1 is a marker of recurrence after complete remission and prior to
allogeneic hematopoietic stem cell transplantation (allo-HSCT), and
patients with high WT1 expression had a significantly higher 2-year
cumulative incidence of relapse compared to those with low WT1
levels. Therefore, low and high expression of WT1 may be associated
with clinical remission and relapse, respectively, and WT1 may be a
potential prognostic factor and a therapeutic target in patients
with AML. By contrast, other previous studies observed that WT1 may
not be used in MRD detection (16)
or that WT1 upregulation may be associated with favorable outcomes
in patients with AML (17).
In the present study, to clarify these
discrepancies, the expression levels of WT1 and other molecular
markers, such as genetic mutations, were retrospectively analyzed
in 195 patients with AML by RT-qPCR. Additionally, the association
between the expression levels of WT1 and various genetic markers
were investigated, and the prognostic value of WT1 expression in
patients with AML after allo-HSCT was examined.
Materials and methods
Patient samples
A total of 195 patients with AML were enrolled in
the First Affiliated Hospital of Xi'an Jiaotong University. Written
informed consent was obtained from all patients. The present
retrospective study was approved by The Institutional Ethics
Committee of The First Affiliated Hospital of Xi'an Jiaotong
University. Diagnoses were performed according to the
French-American-British diagnostic criteria (FAB) (18) for AML and combined with
immunophenotyping, cytogenetics and molecular biological assays, as
previously described (19). The
195 patients were diagnosed with AML and received therapy between
January 2013 and September 2017. The cohort of patients included
102 men and 93 women; the median age was 45 years, ranging between
7 and 76 years. In total, 169 patients received chemotherapy and 31
received allo-HSCT. Based on FAB criteria, 2, 7, 100, 21, 32, 18
and 3 patients were categorized as M0, M1, M2, M3, M4, M5 and M6,
respectively.
Identification of AML related fusion
genes and mutations
Total RNA was extracted from bone marrow using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. Extracted RNA samples were
treated with DNase I (Applied Biosystems; Thermo Fisher Scientific,
Inc.) to remove any DNA contamination before one-step RT-qPCR.
These RNA samples were then subjected to one-step RT-qPCR to detect
AML-related fusion transcripts using The Leukemia Related Fusion
gene detection kit (Shanghai Yuanqi Bio-Pharmaceutical Co., Ltd.)
according to the manufacturer's protocol. Analysis of positive and
negative controls was performed according to the manufacturer's
protocol. The primers were included in the kit. The mixture of each
reaction contained 15 µl total RNA, 8 µl RT-PCR Buffer, 2 µl
Multiplex Enzyme Mix (Shanghai Yuanqi Bio-Pharmaceutical Co., Ltd.)
in a total volume of 25 µl. The Taqman RT-qPCR reaction was
performed at 42°C for 30 min, at 94°C for 5 min followed by 40
cycles of 94°C for 15 sec and 60°C for 1 min on the 7300 Real Time
PCR System (Thermo Fisher Scientific, Inc.). Sanger sequencing was
conducted to detect AML-related mutations. DNA was extracted from
bone marrow with DNeasy Blood & Tissue Kit (Qiagen, Inc.) for
Sanger sequencing by Leukemia Related Gene Test Kit (Shanghai
Yuanqi Bio-Pharmaceutical Co., Ltd.). The primers were included in
the kit. PCR reactions were carried out in a final volume of 25 µl
containing 3 µl genomic DNA, 9 µl sequencing reaction, and 13 µl
PCR MIX3 (Shanghai Yuanqi Bio-Pharmaceutical Co., Ltd.). Samples
were processed at 42°C for 5 min and at 94°C for 5 min, followed by
40 cycles at 94°C for 30 sec, 58°C for 30 sec, and 72°C for 60 sec,
with a final step for 5 min at 72°C. PCR products were loaded on
agarose gels, purified and sequenced using BigDye Terminators and
ABI 3500 Genetic Analyzer (Thermo Fisher Scientific, Inc.).
Quantitative expression of WT1
In total, 2 ml bone marrow (BM) was extracted from
the patients. Total RNA from BM samples was isolated using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. RNA was treated with Amplification
Grade DNase I (Takara Bio, Inc.) at room temperature for 60 min to
prevent DNA contamination. A total of 5 µl RNA (~500 ng) was used
to detect WT1 expression using a one-step RT-qPCR WT1 kit (Shanghai
Yuanqi Bio-Pharmaceutical Co., Ltd.) according to the
manufacturer's protocol. According to the manufacturer's protocol,
the house-keeping Abelson gene (ABL) was used as the internal
control to evaluate the relative levels of WT1 expression, as
previously described (20,21). The primers were included in the
RT-qPCR WT1 kit (Shanghai Yuanqi Bio-Pharmaceutical Co., Ltd.). The
Taqman PCR mixtures were prepared according to the manufacturer's
protocol and the reactions were performed using an ABI 7500
real-time PCR instrument (Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: Initial incubations at
42°C for 30 min and 94°C for 5 min, followed by 40 cycles of 94°C
for 15 sec and 60°C for 60 sec 40 cycles. All experiments were
repeated three times with appropriate positive and negative
controls. WT1 levels were expressed as the number of WT1 copies per
100 copies of ABL. Relative quantification was performed using
standard reference curves according to the manufacturer's protocol
as previously described (22–24).
The detection threshold of WT1 was 0.02% of ABL gene copies, as
previously described (20,21).
Statistical analysis
All experiments were repeated three times. All data
were analyzed using SPSS software (version 20.0; IBM Corp.). Data
are presented as the mean ± SEM. Differences of WT1 expression
between two groups were analyzed using Student's t-test.
Differences among multiple groups were determined by one-way or
two-way ANOVA followed by Tukey's post hoc test. Repeated measures
ANOVA was performed for the analysis of dependent variables. The
effects of allo-HSCT on overall survival (OS) rates were analyzed
using Kaplan-Meier curves. P<0.05 was considered to indicate a
statistically significant difference.
Results
WT1 expression in AML
The expression level of WT1 in BM samples from
patients with AML was assessed by RT-qPCR analysis. The expression
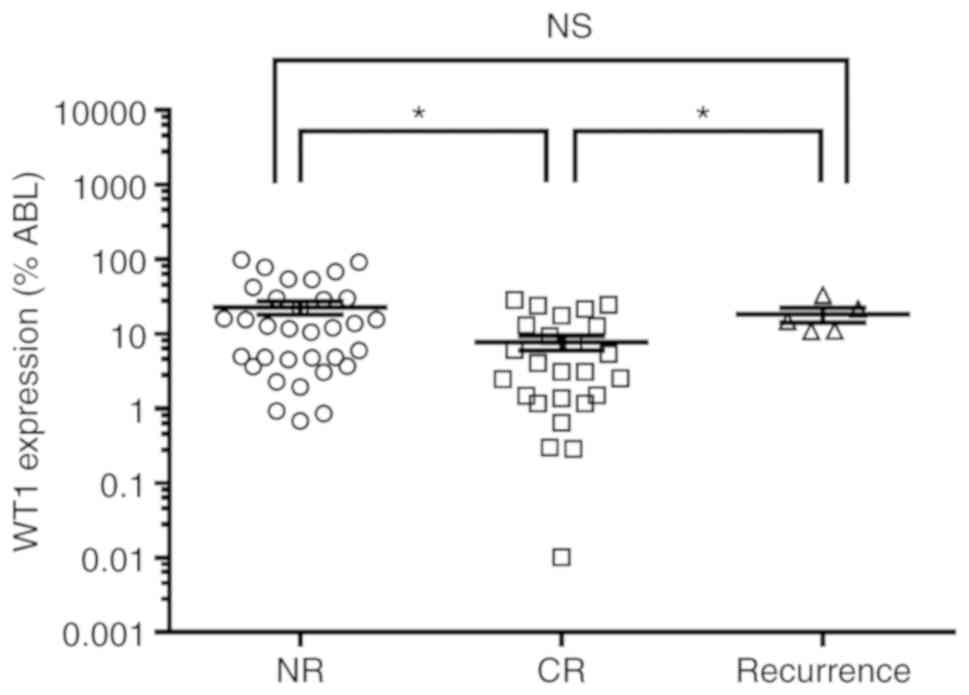
level of WT1 at initial diagnosis was significantly higher in
patients with no response (NR) than in those with complete response
(CR) (Fig. 1). In line with a
previous study (25), CR was
characterized by: i) Bone marrow blasts <5%; ii) the absence of
blasts with Auer rods; iii) the absence of extramedullary disease;
iv) an absolute neutrophil count >1.0×109/l; and v) a
platelet count >100×109/l. Partial remission (PR) was
defined as 5–20% bone marrow blasts and a 50% decrease in bone
marrow blasts during pretreatment. Patients who did not exhibit CR
or PR after chemotherapy were categorized in the NR group.
Moreover, after diagnosis, the expression level of WT1 in patients
with recurrence was significantly higher than in CR patients, but
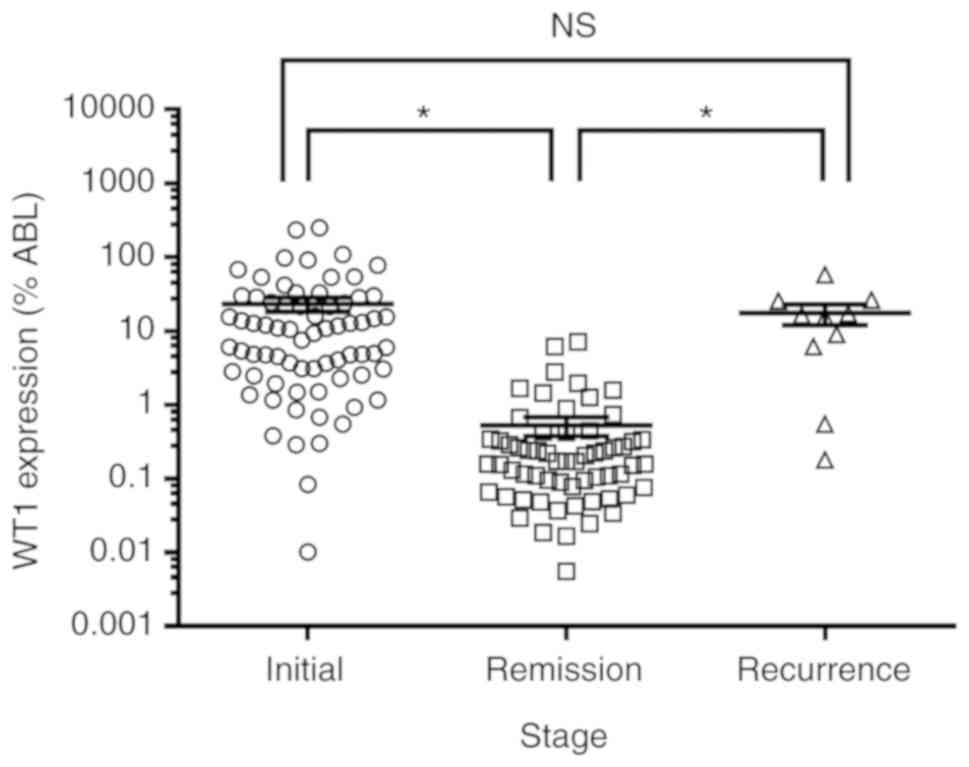
not significantly different than in NR patients (Fig. 1). The expression level of WT1 was
assessed in patients with AML at different stages of the disease:
i) Initial; ii) remission; and iii) recurrence. The present RT-qPCR
results suggested that WT1 expression was significantly lower in
patients with remission than in those with initial and recurrent
AML (Fig. 2). The expression level
of WT1 was not significantly different between patients with
initial and recurrent AML. Furthermore, the expression of WT1 was
analyzed in patients with different AML subtypes according to the
FAB criteria. The expression level of WT1, calculated as a
percentage of ABL expression, was highest in the M3 (48.3%) and M4
(52.4%) subtypes and lowest in the M1 subtype (2.8%). Patients in
the subtypes M6, M2 and M5 exhibited a relative expression level of
WT1 of 34.2, 8.4 and 6.0%, respectively (Table I). There were no significant
differences in ABL expression levels among different subtypes (data
not shown). Importantly, further studies are required to evaluate
the expression level of WT1 in a higher number of patients with
various subtypes of AML.
 | Table I.WT1 expression in AML subtypes at
diagnosis. |
Table I.
WT1 expression in AML subtypes at
diagnosis.
| Subtype | n | Relative WT1
expression, % |
|---|
| M1 | 2 | 2.77±1.272 |
| M2 | 35 | 8.44±1.596 |
| M3 | 10 | 48.30±22.43 |
| M4 | 12 | 52.39±20.74 |
| M5 | 8 | 5.97±2.189 |
| M6 | 3 | 34.22±11.18 |
Association between WT1 expression and
genetic mutations
The present study investigated the association
between the expression level of WT1 and the presence of fusion
genes, including RUNX1-ETO, breakpoint cluster region-ABL,
core-binding factor subunit β-myosin heavy chain 11 and
promyelocytic leukemia (PML)-retinoic acid receptor α (RARA). In
addition, the present study investigated the association between
the expression level of WT1 and prognosis-associated mutations
affecting genes such as CCAAT enhancer binding protein α (CEBPA),
mast/stem cell growth factor receptor, FMS-like tyrosine kinase-3
internal tandem duplication (FLT3-ITD), nucleophosmin 1 and DNA
methyltransferase 3 α. All patients positive for WT1 expression
presented fusion genes or genetic mutations; however, 97.4% of
patients with RUNX1-ETO mutations were positive for WT1 expression,
and 94.7% of patients with FLT3-ITD exhibited a positive expression
of WT1. In addition, the association between the expression level
of WT1 and the aforementioned genetic anomalies was investigated.
Compared with patients without genetic anomalies, WT1 expression
was significantly lower in patients presenting the gene fusion
RUNX1-ETO, but was not significantly altered in patients with other
mutations or fusion genes. However, WT1 expression was higher in
patients with PML-RARA than in those with RUNX1-ETO or CEBPA
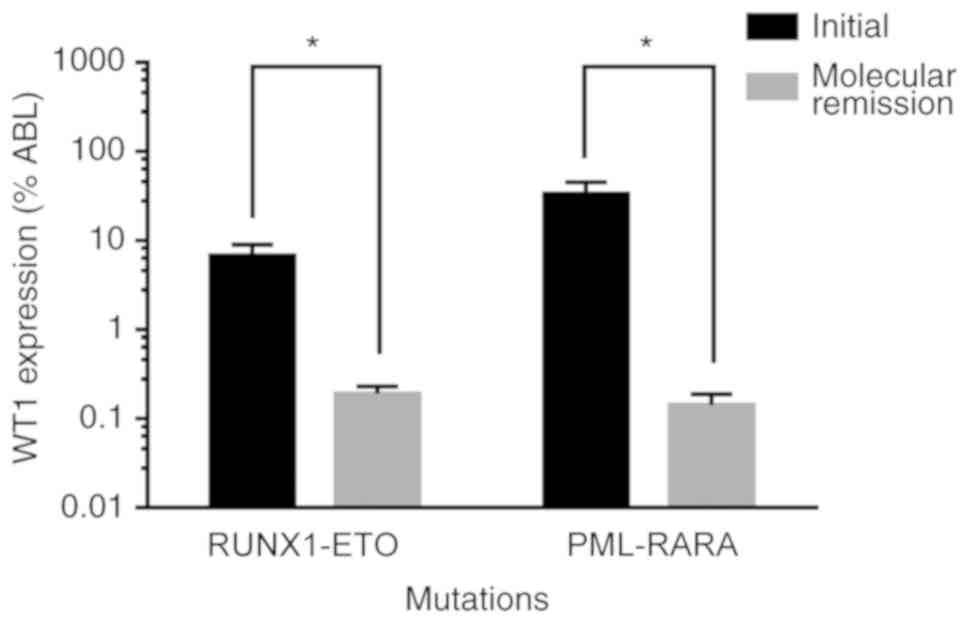
(Table II). In addition, the
level of WT1 expression was significantly reduced after molecular
remission (26) in patients
exhibiting RUNX1-ETO and PML-RARA mutations at initial diagnosis
(Fig. 3).
 | Table II.Comparison between WT1 expression and
various genetic mutations. |
Table II.
Comparison between WT1 expression and
various genetic mutations.
| Genetic mutation | n | Relative WT1
expression, % | P-value |
|---|
| No mutations | 12 | 17.27±4.63 | – |
| RUNX1-ETO | 14 | 6.81±2.23 |
0.0435a |
| PML-RARA | 9 | 52.22±24.69 | 0.1266 |
| CEBPA | 12 | 13.73±4.47 | 0.5877 |
| FLT3-ITD | 12 | 31.85±18.95 | 0.4629 |
WT1 upregulation is associated with
higher mortality rates in patients with AML after allo-HSCT
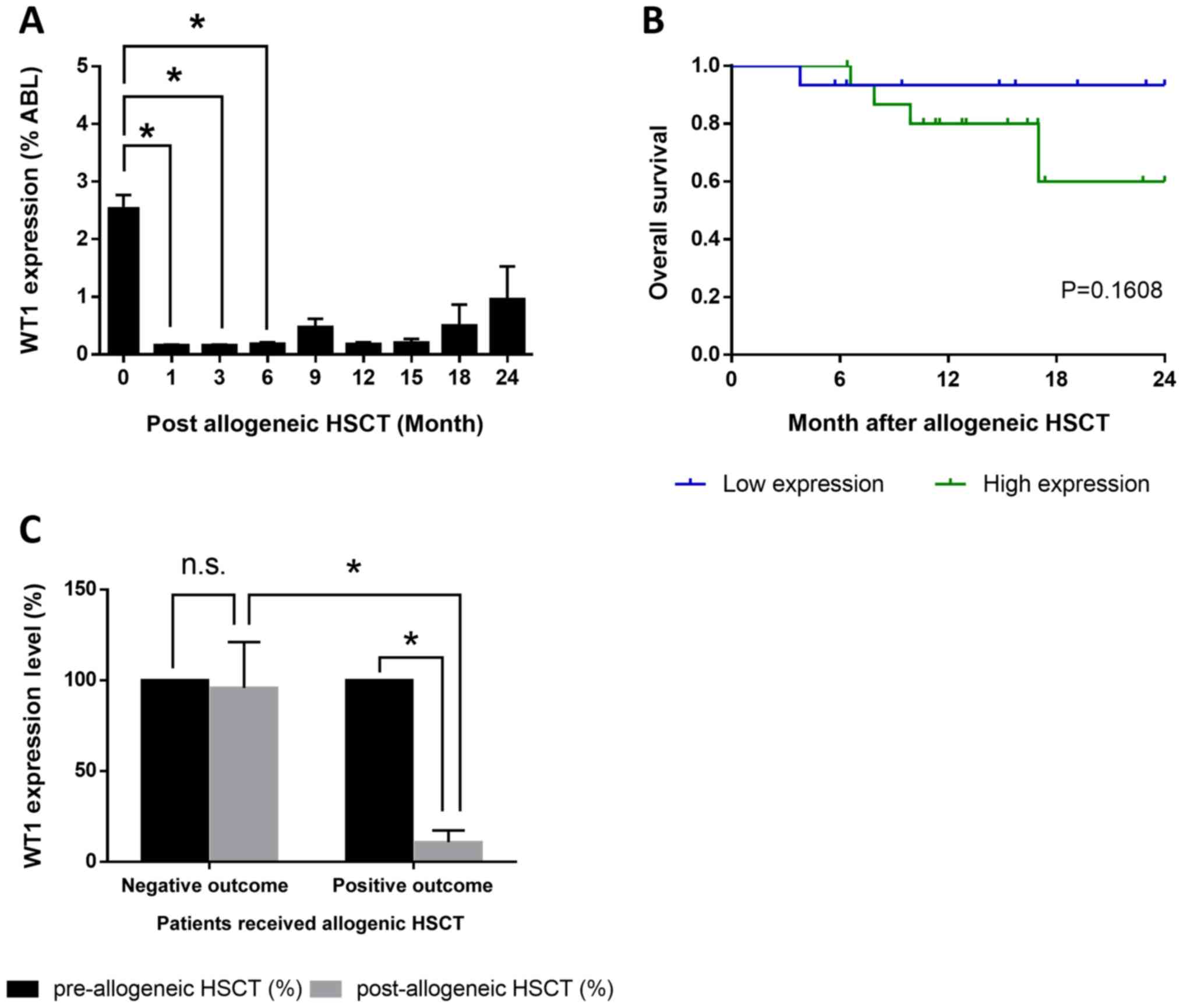
Among the 195 patients with AML, 31 patients
received allo-HSCT. Following allo-HSCT treatment, WT1 expression
decreased significantly after 1, 3 and 6 months compared with
before allo-HSCT treatment (Fig.
4A). The patients who underwent allo-HSCT were divided two
groups: i) High expression level of WT1; and ii) low expression
level of WT1. The mean relative expression level (3.42% compared to
ABL) before allo-HSCT was selected as threshold to divide the two
groups. The 2-year OS after allo-HSCT in the WT1 low expression
group was 93.3% (14/15), whereas in the WT1 high expression group
was ~60% (10/16; Fig. 4B).
Compared with the expression level of WT1 before allo-HSCT, WT1
expression after allo-HSCT was not significantly altered in
patients deceased within 2 years after allo-HSCT. By contrast, the
expression level of WT1 decreased to <10% in surviving patients
(Fig. 4C). The present results
suggested that the dynamic changes in the expression levels of WT1
could be used to predict the treatment outcome and disease state
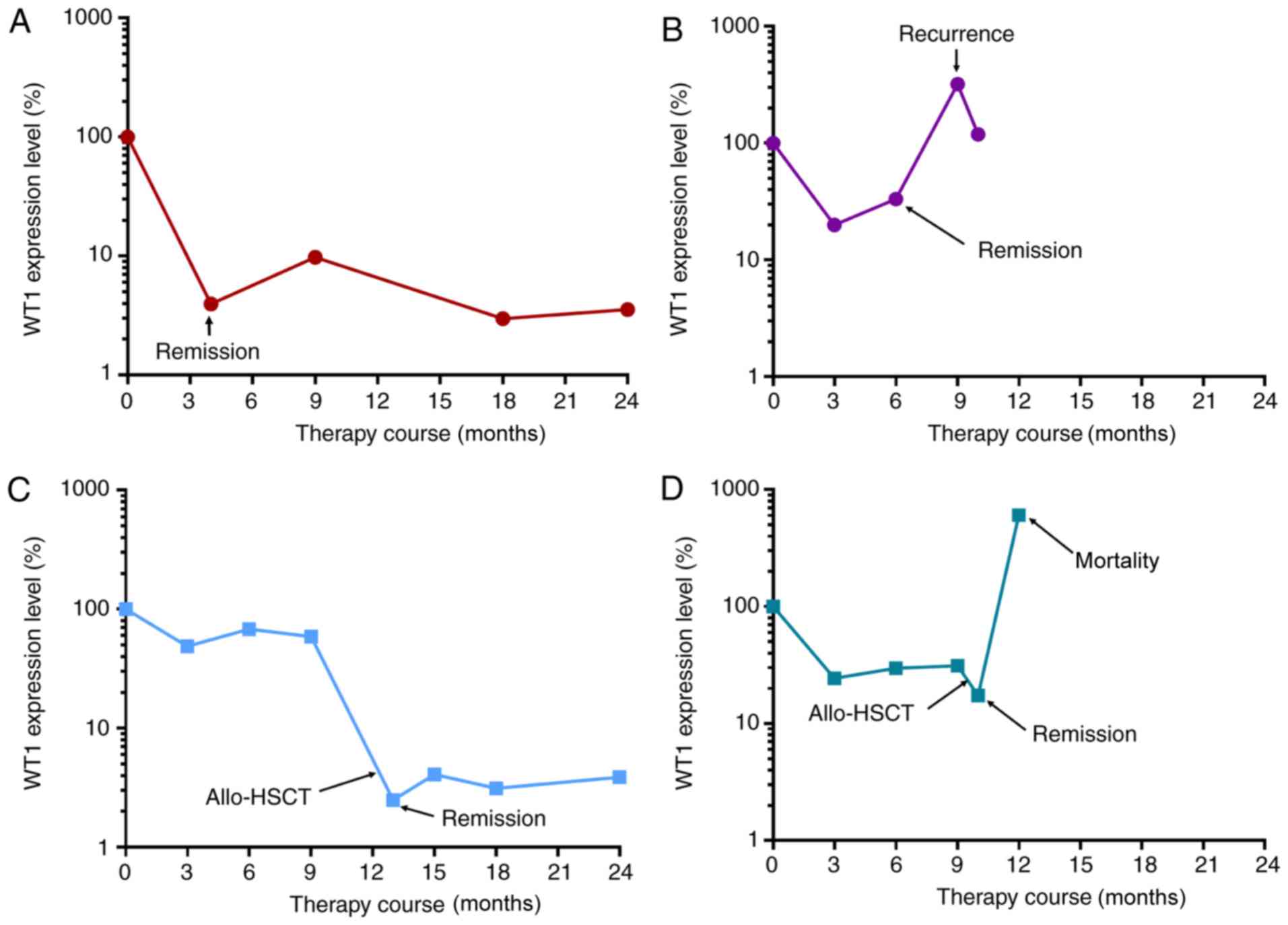
(Fig. 5). WT1 expression decreased
significantly and was maintained at low levels during the remission
stage (Fig. 5A), but increased at
recurrence stage (Fig. 5B).
Following allo-HSCT, patients with AML at remission stage exhibited
low levels of WT1 (Fig. 5C), and
WT1 expression level was significantly increased at recurrence
stage or prior to mortality (Fig.
5D). The present results suggested that the expression level of
WT1 was negatively associated with the therapeutic response in
patients with AML who underwent allo-HSCT.
Discussion
Monitoring MRD has become one of the most effective
approaches to determine prognosis and therapeutic strategies in
patients with AML. WT1 serves an important role in blast cell
survival by enhancing proliferation and inhibiting apoptosis, and
WT1 is upregulated in the majority of patients with initial AML
(27). However, the prognostic
potential of the expression level of WT1 remains unclear. Here, we
retrospectively investigated the prognostic potential of WT1 in a
cohort of patients with AML.
WT1 expression level is higher in patients with AML
at diagnosis compared with healthy patients, decreases at complete
remission, and increases prior to clinical relapse (28,29).
In the present study, the expression levels of WT1 in patients with
AML in initial, remission and recurrence stage are in line with a
previous study indicating that low and high expression of WT1 in
AML are associated with clinical remission and relapse,
respectively (20). Therefore, the
present results suggested that recurrence can be predicted based on
the expression level of WT1 in patients with AML, in line with the
previous study by Mashima et al (30). In addition, WT1 expression is high
in >80% of patients with AML and expression of WT1 decreases
when patients entered remission (31). Therefore, WT1 expression could
potentially be used for MRD detection in patients with AML, in
particular in patients with no cytogenetic or molecular
abnormalities.
Hao et al (32) showed that WT1 expression was
highest in the M3 subtype and lowest in the M1 subtype. In the
present study, lowest WT1 expression was detected in the M1
subtype. However, high levels of WT1 were observed in both the M3
and M4 subtypes. In addition, the expression level of WT1 was
identified to be associated with genetic abnormalities and the
average WT1 expression was higher in patients with CEBPA mutations
compared with patients exhibiting no mutations in the CEBPA gene.
In addition, the expression level of WT1 was increased in patients
with initial AML presenting FLT3-ITD mutation. The present results
are in line with the study by Lyu et al (14), suggesting that patients with
FLT3-ITD mutation presented high levels of WT1 compared with other
patients with wild-type FLT3. Furthermore, compared with patients
without identified mutations, WT1 expression was lower in patients
with RUNX1-ETO and higher in patients with PML-RARA. The present
results suggested that the expression level of WT1 was associated
with the outcome of patients with AML. In addition, the level of
WT1 expression was significantly reduced after molecular remission
in patients exhibiting RUNX1-ETO and PML-RARA mutations at initial
diagnosis, suggesting that WT1 could be used as an MRD marker in
the majority of patients with AML without specific fusion
genes.
Duléry et al (33) reported that the 3-year event-free
survival rate is reduced in WT1-based MRD positive patients
compared with WT1-based MRD negative patients, and patients who
underwent allo-HSCT positive for WT1 expression after 3 months
exhibited an unfavorable prognosis, suggesting a detrimental role
for WT1 in relapse. In the present study, although no patients
exhibited recurrence following allo-HSCT, the mortality rate 2
years after allo-HSCT was higher for patients in the high WT1
expression group compared with those with low expression of WT1.
The present results suggested that WT1 expression was negatively
associated with therapeutic effect of allo-HSCT in patients with
AML. The present results are in line with a previous study by
Candoni et al (34) that
showed a better outcome for WT1-negative patients compared with
WT1-positive patients. Collectively, the present study identified
that WT1 expression may be associated with the prognosis of
patients with AML, including those that received allo-HSCT.
Therefore, WT1 could be considered as an MRD biomarker for AML. Due
to the limitations of the present study, including the inconsistent
number of patients with different AML subtypes, further studies
analyzing a higher number of patients with AML are required in
order to validate the present results.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
Shaanxi Province Science and Technology Research Projects (grant
no. 2016SF-122) and the Clinic Research Award of the First
Affiliated Hospital of Xi'an Jiaotong University (grant no.
XJTU1AHCR2014-032).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL collected the data, wrote and revised the present
manuscript. XW and MZ assisted in writing the manuscript. HZ and JW
performed the RT-qPCR experiments. XW, TM, JS and MZ performed
statistical and data analyses. YC, YK, JX and MW managed the
patient information and contributed to data acquisition. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Institutional
Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong
University. Informed consent was obtained from all patients.
Patient consent for publication
Informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Döhner H, Weisdorf DJ and Bloomfield CD:
Acute Myeloid Leukemia. N Engl J Med. 373:1136–1152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lagunas-Rangel FA, Chávez-Valencia V,
Gómez-Guijosa MA and Cortes-Penagos C: Acute Myeloid
Leukemia-Genetic Alterations and Their Clinical Prognosis. Int J
Hematol Oncol Stem Cell Res. 11:328–339. 2017.PubMed/NCBI
|
|
3
|
Sun Y, Chen BR and Deshpande A: Epigenetic
Regulators in the Development, Maintenance, and Therapeutic
Targeting of Acute Myeloid Leukemia. Front Oncol. 8:412018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Freireich EJ, Wiernik PH and Steensma DP:
The leukemias: A half-century of discovery. J Clin Oncol.
32:3463–3469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ommen HB: Monitoring minimal residual
disease in acute myeloid leukaemia: A review of the current
evolving strategies. Ther Adv Hematol. 7:3–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hourigan CS, Gale RP, Gormley NJ,
Ossenkoppele GJ and Walter RB: Measurable residual disease testing
in acute myeloid leukaemia. Leukemia. 31:1482–1490. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou Y and Wood BL: Methods of Detection
of Measurable Residual Disease in AML. Curr Hematol Malig Rep.
12:557–567. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Elmaagacli AH: Molecular methods used for
detection of minimal residual disease following hematopoietic stem
cell transplantation in myeloid disorders. Methods Mol Biol.
1109:187–207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomlinson B and Lazarus HM: Enhancing
acute myeloid leukemia therapy - monitoring response using residual
disease testing as a guide to therapeutic decision-making. Expert
Rev Hematol. 10:563–574. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ross DM, Watkins DB, Hughes TP and
Branford S: Reverse transcription with random pentadecamer primers
improves the detection limit of a quantitative PCR assay for
BCR-ABL transcripts in chronic myeloid leukemia: Implications for
defining sensitivity in minimal residual disease. Clin Chem.
54:1568–1571. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang L, Han Y, Suarez Saiz F and Minden
MD: A tumor suppressor and oncogene: The WT1 story. Leukemia.
21:868–876. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toska E and Roberts SG: Mechanisms of
transcriptional regulation by WT1 (Wilms' tumour 1). Biochem J.
461:15–32. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morrison AA, Viney RL and Ladomery MR: The
post-transcriptional roles of WT1, a multifunctional zinc-finger
protein. Biochim Biophys Acta. 1785:55–62. 2008.PubMed/NCBI
|
|
14
|
Lyu X, Xin Y, Mi R, Ding J, Wang X, Hu J,
Fan R, Wei X, Song Y and Zhao RY: Overexpression of Wilms tumor 1
gene as a negative prognostic indicator in acute myeloid leukemia.
PLoS One. 9:e924702014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Frairia C, Aydin S, Audisio E, Riera L,
Aliberti S, Allione B, Busca A, D'Ardia S, Dellacasa CM, Demurtas
A, et al: Post-remissional and pre-transplant role of minimal
residual disease detected by WT1 in acute myeloid leukemia: A
retrospective cohort study. Leuk Res. 61:10–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bergmann L, Miething C, Maurer U, Brieger
J, Karakas T, Weidmann E and Hoelzer D: High levels of Wilms' tumor
gene (wt1) mRNA in acute myeloid leukemias are associated with a
worse long-term outcome. Blood. 90:1217–1225. 1997.PubMed/NCBI
|
|
17
|
Miglino M, Colombo N, Pica G, Grasso R,
Clavio M, Bergamaschi M, Ballerini F, Ghiso A, Ghiggi C,
Mitscheunig L, et al: WT1 overexpression at diagnosis may predict
favorable outcome in patients with de novo non-M3 acute myeloid
leukemia. Leuk Lymphoma. 52:1961–1969. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposed revised
criteria for the classification of acute myeloid leukemia. A report
of the French-American-British Cooperative Group. Ann Intern Med.
103:620–625. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A, et al: The 2008 revision of the
World Health Organization (WHO) classification of myeloid neoplasms
and acute leukemia: Rationale and important changes. Blood.
114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ayatollahi H, Sadeghian MH, Naderi M,
Jafarian AH, Shams SF, Motamedirad N, Sheikhi M, Bahrami A and
Shakeri S: Quantitative assessment of Wilms tumor 1 expression by
real-time quantitative polymerase chain reaction in patients with
acute myeloblastic leukemia. J Res Med Sci. 22:542017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Malagola M, Skert C, Ruggeri G, Turra A,
Ribolla R, Cancelli V, Cattina F, Alghisi E, Bernardi S, Perucca S,
et al: Peripheral blood WT1 expression predicts relapse in AML
patients undergoing allogeneic stem cell transplantation. BioMed
Res Int. 2014:1230792014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Inoue K, Sugiyama H, Ogawa H, Nakagawa M,
Yamagami T, Miwa H, Kita K, Hiraoka A, Masaoka T, Nasu K, et al:
WT1 as a new prognostic factor and a new marker for the detection
of minimal residual disease in acute leukemia. Blood. 84:3071–3079.
1994.PubMed/NCBI
|
|
23
|
EN 13641:2002, . Elimination or reduction
of risk of infection related to in vitro diagnostic reagents. In
vitro diagnostic medical devices Directive. EU Declaration of
Conformity. 2017.
|
|
24
|
EP9-A2, . Method Comparison and Bias
Estimation Using Patient Samples: Approved Guideline. 22. 2nd.
NCCLS; Wayne, PA: 2002
|
|
25
|
Zhu HH, Jiang H, Jiang B, Lu J, Jiang Q,
Bao L, Zhang XH, Qin YZ and Huang XJ: Cytarabine, aclarubicin and
granulocyte colony-stimulating factor regimen represents an
effective and safe salvage regimen for patients with acute myeloid
leukemia refractory to first course of induction chemotherapy. Leuk
Lymphoma. 54:2452–2457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gallego Hernanz MP, Torregrosa Diaz JM,
Sorel N, Bobin A, Dindinaud E, Bouyer S, Desmier D, Brizard F,
Leleu X, Maillard N, et al: Long-term molecular remission in a
patient with acute myeloid leukemia harboring a new NUP98-LEDGF
rearrangement. Cancer Med. 8:1765–1770. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Xing C, Zhou B, Ye H, Feng J, Wu J
and Gao S: A regulatory circuitry between miR-193a/miR-600 and WT1
enhances leukemogenesis in acute myeloid leukemia. Exp Hematol.
61:59–68.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Válková V, Polák J, Marková M, Vítek A,
Hájková H, Sálek C, Procházka B, Cetkovský P and Trněný M: Minimal
residual disease detectable by quantitative assessment of WT1 gene
before allogeneic stem cell transplantation in patients in first
remission of acute myeloid leukemia has an impact on their future
prognosis. Clin Transplant. 27:E21–E29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gray JX, McMillen L, Mollee P, Paul S,
Lane S, Bird R, Gill D, Saal R and Marlton P: WT1 expression as a
marker of minimal residual disease predicts outcome in acute
myeloid leukemia when measured post-consolidation. Leuk Res.
36:453–458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mashima K, Oh I, Ikeda T, Toda Y, Ito S,
Umino K, Minakata D, Nakano H, Morita K, Yamasaki R, et al: Role of
Sequential Monitoring of WT1 Gene Expression in Patients With Acute
Myeloid Leukemia for the Early Detection of Leukemia Relapse. Clin
Lymphoma Myeloma Leuk. 18:e521–e527. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Polák J, Hájková H, Maalaufová-Soukupová
J, Marková J, Sálek C, Schwarz J and Haškovec C: Estimation of
molecular upper remission limit for monitoring minimal residual
disease in peripheral blood of acute myeloid leukemia patients by
WT1 expression. Exp Ther Med. 3:129–133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hao Y, Cheng Y, Wu Q, Zhang A, Jiang X and
Xu X: Combined usage of Wilms' tumor gene quantitative analysis and
multiparameter flow cytometry for minimal residual disease
monitoring of acute myeloid leukemia patients after allogeneic
hematopoietic stem cells transplantation. Exp Ther Med.
15:1403–1409. 2018.PubMed/NCBI
|
|
33
|
Duléry R, Nibourel O, Gauthier J,
Elsermans V, Behal H, Coiteux V, Magro L, Renneville A, Marceau A,
Boyer T, et al: Impact of Wilms' tumor 1 expression on outcome of
patients undergoing allogeneic stem cell transplantation for AML.
Bone Marrow Transplant. 52:539–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Candoni A, De Marchi F, Zannier ME,
Lazzarotto D, Filì C, Dubbini MV, Rabassi N, Toffoletti E, Lau BW
and Fanin R: High prognostic value of pre-allogeneic stem cell
transplantation minimal residual disease detection by WT1 gene
expression in AML transplanted in cytologic complete remission.
Leuk Res. 63:22–27. 2017. View Article : Google Scholar : PubMed/NCBI
|