Introduction
The Src homology and collagen (Shc) family of
adaptor proteins assists in propagating extracellular signals to
the internal cellular environment by acting as a substrate for
different receptors (1–3). In addition, Shc proteins are known
for their role in establishing crosstalk between different
signalling pathways (4,5). Shc proteins are capable of performing
this transduction function as they possess a unique structural
homologue [collagen homology (CH)2-phosphotyrosine binding domain
(PTB)-CH1-Src homology-2 (SH2)] (4,5). The
family comprises four members: ShcA, ShcB, ShcC and the most
recently identified member ShcD (4,6,7).
Expression analysis has revealed high levels of ShcD in the
vertical growth phase, as well as in metastatic melanoma (6). ShcD has been proposed to mediate cell
motility by activating both Ras-dependent and Ras-independent
migratory pathways in melanoma (6). In the same study, when the Ras/MAPK
signalling pathway was inhibited, ShcD-overexpressing cells
maintained their ability to mediate cell migration (6); the mechanisms underlying ShcD-induced
cell migration have require further investigation.
For cells to acquire a motile phenotype, they
initially transition to a mesenchymal phenotype, and this results
in the ability to invade the extracellular matrix (8). Additionally, cancer cells require new
blood vessel formation to assist in metastasis (9,10).
Epithelial-mesenchymal transition (EMT) is required for normal
cells to migrate to aid in embryonic development and wound healing,
but it is also employed by cancer cells (9). Transforming growth factor (TGF)β
signalling plays a major role in epithelial cell morphology changes
and alterations in gene expression patterns to acquire a motile
phenotype (11–13).
Various studies have reported that TGFβ signalling
is transduced via Smad and non-Smad signalling. TGFβ binds to TGFβ
receptor type II, which then transphosphorylates TGFβ receptor type
I. Two of the proteins recruited by TGFβ receptor type I activation
are Smad2 and Smad3, and the complex they form is responsible for
Smad4 translocation to the nucleus (14,15).
Smad4 promotes the transcription of different genes responsible for
acquiring the EMT phenotype (16).
TGFβ signalling is involved in promoting the expression of stemness
genes, such as snail family transcriptional repressor 2 (SLUG) and
snail family transcriptional repressor 1 (SNAIL), which help
initiate the EMT process (12,17).
It has been reported that TGFβ triggers cell invasion and
metastasis by inducing the expression of vascular endothelial
growth factor (VEGF) and matrix metalloproteinase 2 (MMP2)
(18).
In the present study, we proposed that ShcD may have
a role in inducing EMT and thus mediate cell migration.
Investigating the expression of EMT genes in ShcD-overexpressing
cells could aid in determining the function of ShcD and improve our
knowledge of the mechanisms by which ShcD induces melanoma cell
migration.
Materials and methods
Antibodies and reagents
The cells were lysed using 1% Triton lysis buffer
supplemented with 1mM Na3VO4, 50 mM NaF, 1 mM
PMSF and protease inhibitors cocktail. All the lysis buffer
reagents were obtained from Sigma Aldrich (Merck KGaA). The
extracted proteins were quantified by Pierce BCA protein assay from
Invitrogen (23225; Thermo Fisher Scientific, Inc.). The proteins
were then resolved in a 10% SDS-PAGE gel. Immunoblotting was
performed by probing proteins transferred onto PVDF membranes with
the following antibodies: Anti-Smad2 (ab40855; Abcam),
anti-phosphorylated-Smad2 (ab184557; Abcam), anti-histone H3
(ab1791; Abcam), anti-extracellular signal-regulated kinase (ERK;
cat. no. 9102; Cell Signaling Technology, Inc. (CST)],
anti-phosphorylated-ERK (pERK; cat. no. 9101; CST), Anti-TGFβ
receptor I antibody (ab31013; Abcam), anti-β actin (A5441; Sigma
Aldrich; Merck KGaA) and anti-green fluorescent protein (GFP;
sc-9996; Santa Cruz Biotechnology, Inc.). To enable detection of
the primary antibody, a rabbit or a mouse secondary antibody
coupled to HRP was used according to the primary antibody species
(ab7628 and ab191866, respectively; Abcam). All the primary
antibodies were incubated over night at 4°C, while the secondary
antibodies were incubated for 1 h at RT. The membranes were
developed employing ECL Western Blotting substrate kit from
Promega, USA (W1015). The images were acquired by Bio-Rad ChemiDoc
touch imaging system. Human TGFβ (T2815; Sigma-Aldrich; Merck KGaA)
was used for cell treatment at 5 ng/ml at 37°C.
Cell culture
293 and G5 cells were a kind gift from Dr Prigent
(University of Leicester, Leicester, UK). The G5 cell line was
generated by Samrein Ahmed, 2013, (University of Leicester). The GF
cell line was generated by Samrein Ahmed at the University of
Sharjah (UAE). The method of how stable cell lines were generated
is described by Ahmed et al (19). FM-55p (13012417) and MM138
(10092321) melanoma cell lines were supplied from Sigma Aldrich
(Merck KGaA) from the ECACC collection. The two cell lines were
maintained as indicated by ECACC instructions.
The 293, G5, and GF cell lines were cultured in
Dulbecco's Modified Eagle's medium (D6429; Sigma Aldrich; Merck
KGaA) supplemented with 10% FBS (F9665; Sigma Aldrich; Merck KGaA)
and 1% penicillin/streptomycin. G5 and GF cells were cultured with
200 µg/ml neomycin or hygromycin, respectively for selection. The
cells were incubated at 37°C and 5% CO2. Before and
during the experiments, the cells were maintained without selection
pressure to eliminate any effect of the selection treatment.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using a total RNA
Purification Kit 1700 (Norgen Biotek Corp.). mRNA was then
converted into cDNA using a TruScript Reverse Transcriptase kit
following the manufacturer's protocol (cat. no. 54440; Norgen
Biotek Corp.). qPCR was performed using a SYBR Green PCR kit
(204145; Qiagen GmbH) and the following primers:
Homo sapiens VEGF forward,
5′-CTACCTCCACCATGCCAAGT-3′, and reverse,
5′-GCAGTAGCTGCGCTGATAGA-3′; homo sapiens MMP-2 forward,
5′-TCTCCTGACATTGACCTTGGC-3′, and reverse,
5′-CAAGGTGCTGGCTGAGTAGATC-3′; Homo sapiens SNAIL forward,
5′-ACCACTATGCCGCGCTCTT-3′, and reverse, 5′-GGTCGTAGGGCTGCTGGAA-3′;
homo sapiens SLUG forward, 5′-TGTTGCAGTGAGGGCAAGAA-3′, and reverse,
5′-GACCCTGGTTGCTTCAAGGA-3′; and homo GAPDH forward,
5′-AGGGCTGCTTTTAACTCTGGT-3′, and reverse,
5′-CCCCACTTGATTTTGGAGGGA-3′. The RT-qPCR parameters for each of the
genes are demonstrated in Table I.
Fluorescence signals were detected using a Qiagen Rotor Gene Q PCR
fluorescence analyser (Qiagne GmbH). The obtained quantification
cycle (Cq) values were analysed using the 2−ΔΔCq method
(20).
 | Table I.RT-qPCR parameters for the tested
genes. |
Table I.
RT-qPCR parameters for the tested
genes.
| Gene Name | RT-qPCR
parameters |
|---|
| SLUG | -Denaturation at
95C for 15 min |
|
| −45 Cycles of: |
|
| −94C
for 15 sec |
|
| −64C
for 30 sec |
|
| −72C
for 30 sec |
|
| -Dissociation at
60-95C |
| SNAIL | -Denaturation at
95C for 15 min |
|
| −45 Cycles of: |
|
| −94C
for 15 sec |
|
| −65.7C
for 30 sec |
|
| −72C
for 30 sec |
|
| -Dissociation at
60-95C |
| VEGF | -Denaturation at
95C for 15 min |
|
| −45 Cycles of: |
|
| −94C
for 15 sec |
|
| −60C
for 30 sec |
|
| −72C
for 30 sec |
|
|
−Dissociation at 60-95C |
| MMP2 | -Denaturation at
95C for 15 min |
|
| −45 Cycles of: |
|
| −94C
for 15 sec |
|
| −61C
for 30 sec |
|
| −72C
for 60 sec |
|
| -Dissociation at
60-95C |
| GAPDH | -Denaturation at
95C for 15 min |
|
| −45 Cycles of: |
|
| −94C
for 15 sec |
|
| −58C
for 30 sec |
|
| −72C
for 30 sec |
|
| -Dissociation at
60-95C |
Transwell assay
Briefly, 1.25×105 cells were resuspended
in DMEM containing 0.1% serum and then added to upper Boyden
chambers. Conditioned medium was made by adding fresh DMEM with 10%
FBS to TGFβ-treated or untreated GF, or G5 cell-derived medium at a
ratio of 1:1. The lower chambers contained the conditioned medium,
and the cells were allowed to migrate for 16 h at 37°C. After the
incubation time, the Boyden chamber membranes were stained with
0.2% crystal violet in 10% ethanol for 30 min at room temperature,
and absorbance readings were obtained at 570 nm using Thermo
Scientific Varioskan Flash-Elisa microplate reader (Thermo Fisher
Scientific, Inc.).
Subcellular fractionation
The steps conducted to separate the nuclear fraction
from the cytoplasmic fraction are described by Ahmed and Prigent
(21). Briefly, cells were
pelleted at 4°C at 122 × g for 5 min and the pellets were treated
with hypotonic buffer (10 mM HEPES pH 7.8, 25 mM
β-glycerophosphate, 25 mM MgCl2, 0.1 mM
Na3VO4, 0.5 mM EDTA and 0.1% protease
inhibitors). Next, 10% NP-40 was added and accompanied with
vigorous vortexing for 15 sec at room temperature. This was
followed by 30 sec centrifugation at 13,000 × g at 4°C. Nuclear
protein extraction was performed by adding a high salt buffer (50
mM HEPES pH 7.8, 50 mM KCl, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 10%
glycerol, 0.2 mM NaF, 0.2 mM Na3VO4, and 0.1%
protease inhibitor cocktail). Nuclear fractions were then analyzed
by western blotting.
Statistical analysis
Each experiment was performed at least twice.
Western blotting band analysis was performed using ImageJ-Version
1.50b (National Institutes of Health) (22) and Excel version 365. In the present
study, the error bars represent the standard error of the mean.
One-tailed student tests were used to calculate statistical
differences. For the analysis of SLUG expression, one-way ANOVA was
used as well as Dunnett's multiple comparison test for differences
between multiple groups, using GraphPad Prism 7.04 (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
TGFβ receptor I expression in the
generated stable cell lines
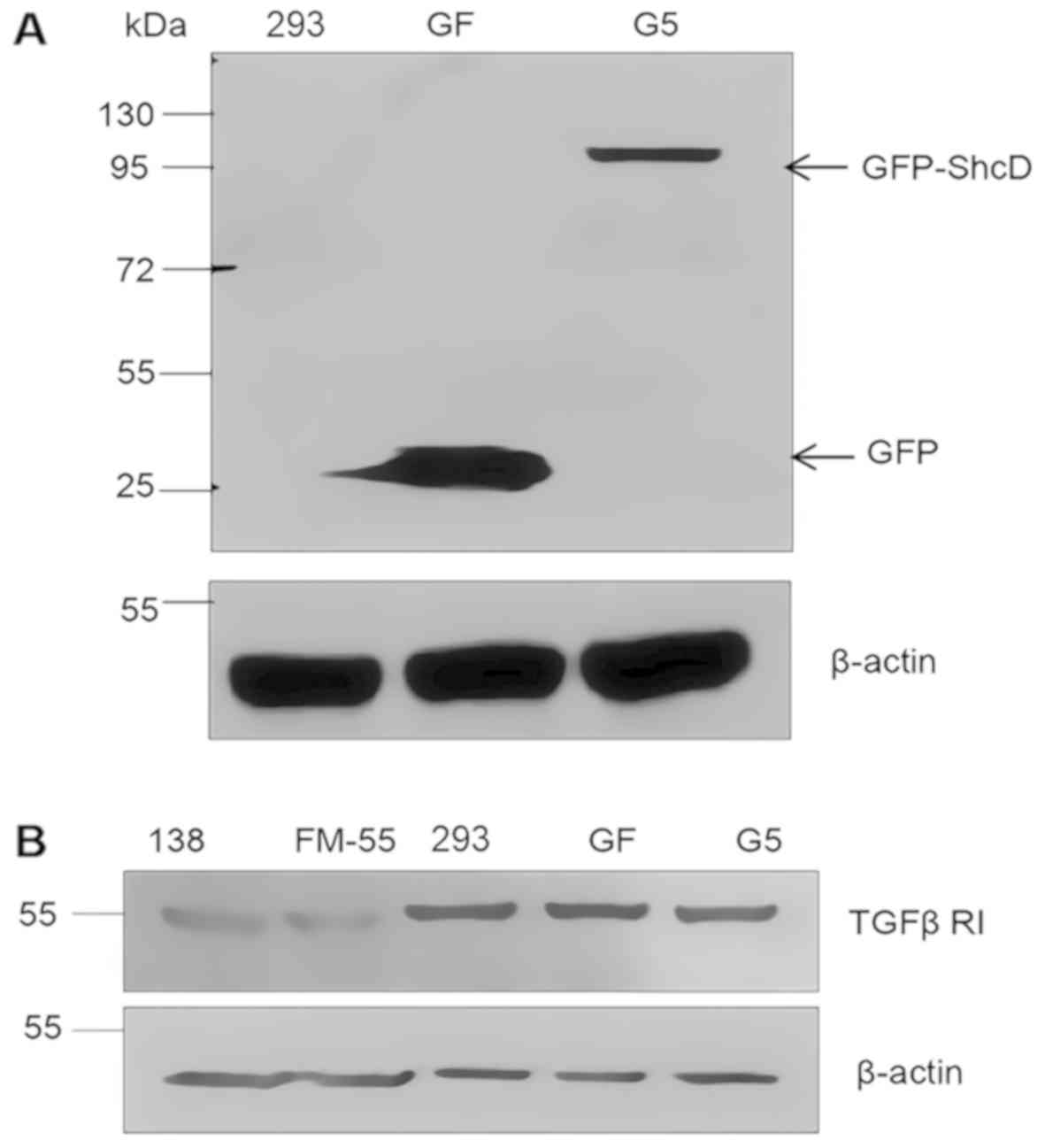
To determine the effects of ShcD on downstream TGFβ
signalling, stable cell lines expressing GFP-ShcD or GFP were
generated (G5 and GF, respectively), and the expression of GFP-ShcD
or GFP was confirmed by immunoblotting with an anti-GFP antibody
(Fig. 1). Next-generation
sequencing showed that the ShcD expression level was 296 times
higher in G5 cells than in GF cells (data not shown).
The 293, GF, G5, and two melanoma cell lines (FM-55p
and MM138) were analysed to determine their TGFβ receptor type I
expression profile to ensure cell responsiveness to TGFβ treatment
(Fig. 1).
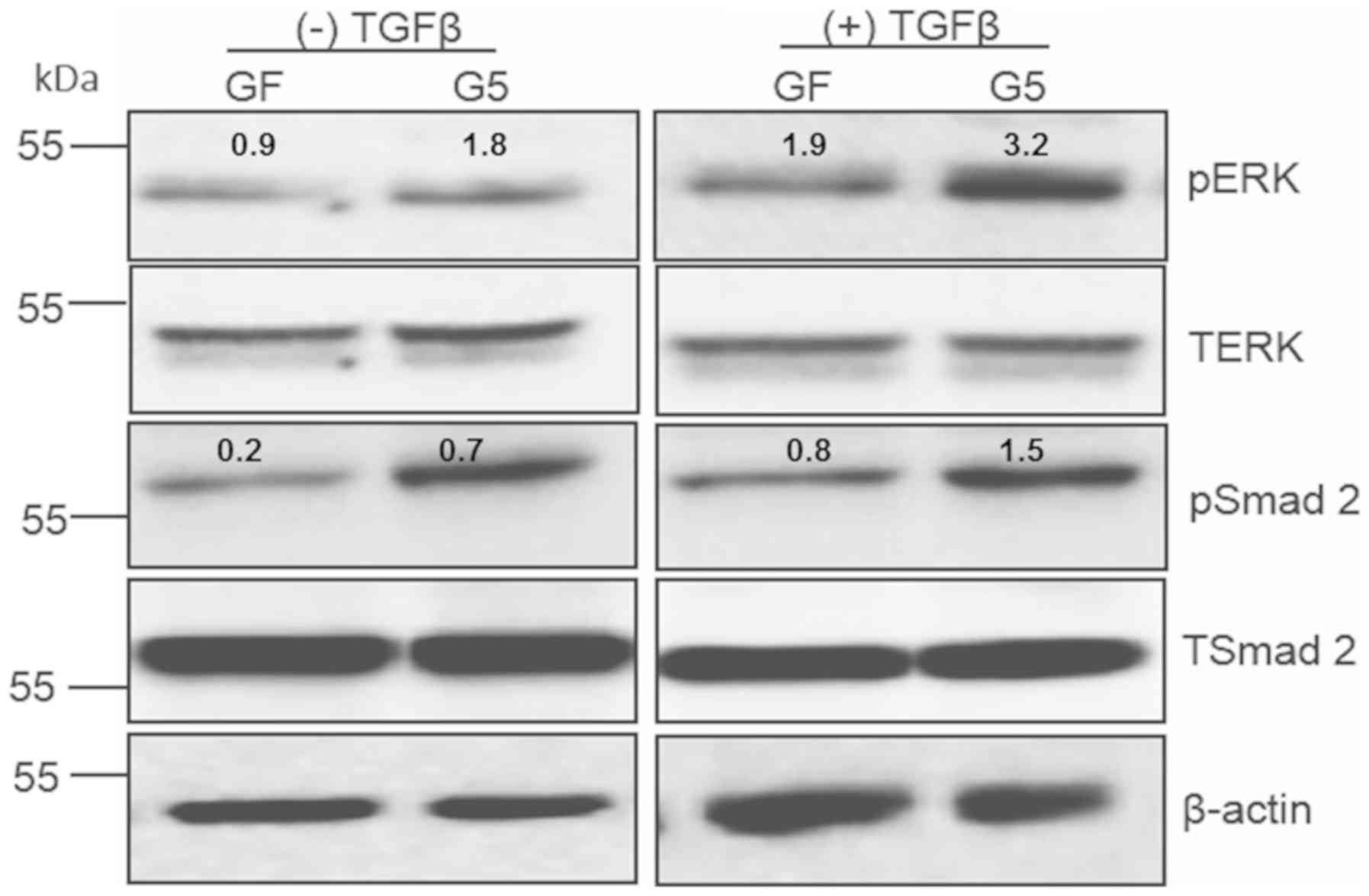
TGFβ treatment promotes ERK
phosphorylation in GFP-ShcD-expressing cells
To examine whether ShcD contributes to TGFβ
signalling transduction, 293, GF and G5 cells were treated with or
without TGFβ. pERK was shown to be notably upregulated in
GFP-ShcD-expressing cells than in GFP-expressing cells. Similar to
pERK, but to a lesser extent (23), the levels of pSmad2 were increased
in GFP-ShcD-expressing cells (Fig.
2). TGFβ treatment caused elevation of pERK by 1.8 fold in
GFP-ShcD-expressing cells, while it resulted in increased pERK by
1-fold in GFP-expressing cells. The TGFβ-mediated Smad2
phosphorylation was less evident in GFP-ShcD-expressing cells than
that of ERK phosphorylation. It was accordingly hypothesized that
TGFβ treatment causes an increase in pERK and, to a lesser extent,
in Smad2 phosphorylation in GFP-ShcD-expressing cells.
Expression of cell motility-related
genes in GF and G5 cells
The role of TGFβ in establishing and maintaining EMT
was reported to be partially achieved by inducing the transcription
of various genes, such as the stemness genes SLUG and SNAIL
(17). Therefore, SNAIL and SLUG
expression was assessed via RT-qPCR. Upon TGFβ treatment,
GFP-ShcD-expressing cells exhibited significantly upregulated
expression of the stemness genes with or without the addition of
10% serum (Fig. 3A and B).
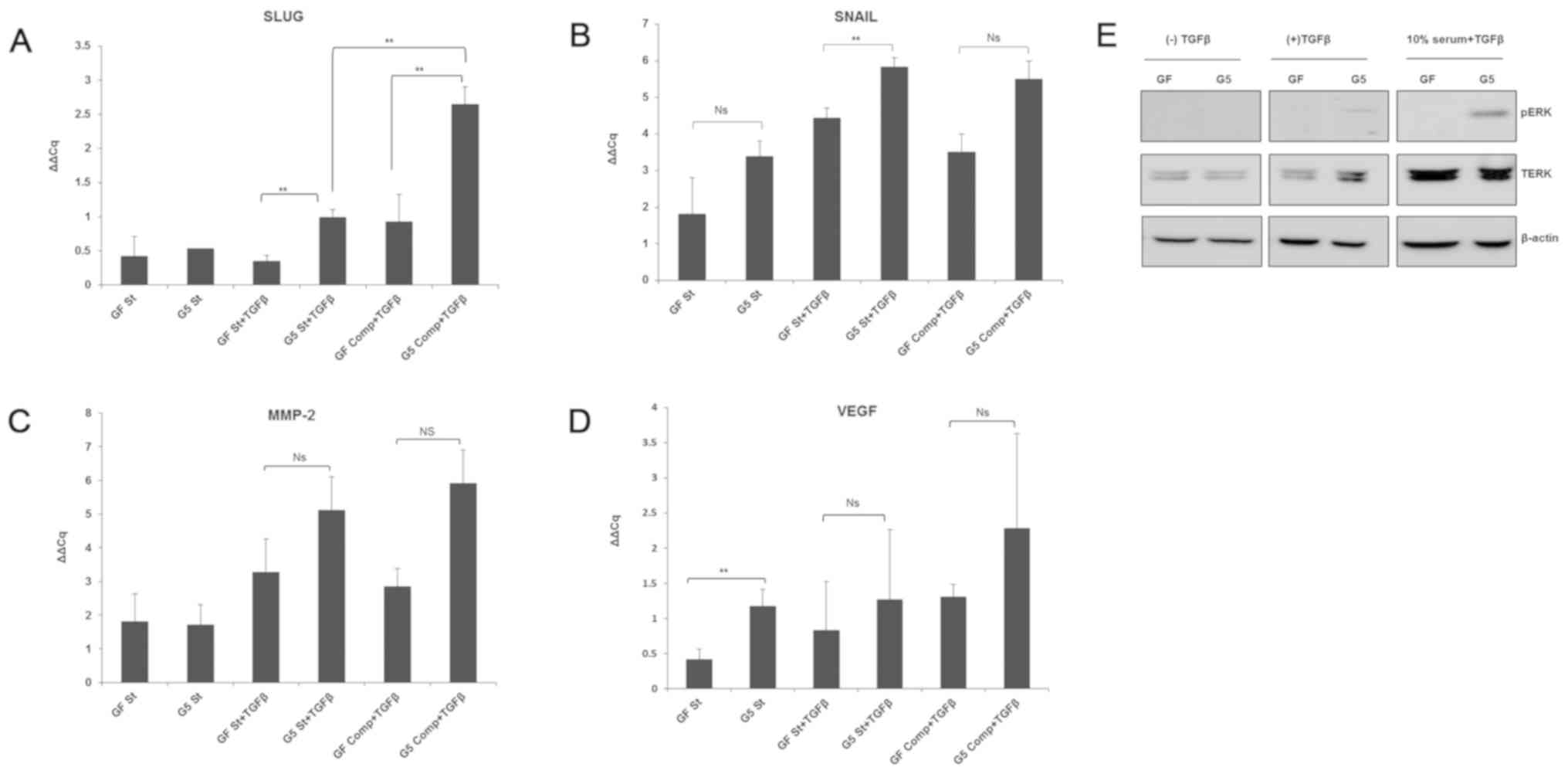 | Figure 3.Expression of cell motility-related
genes in GF and G5 cells. (A-D) GF and G5 cells were incubated with
0.1% serum, 0.1% serum plus 5 ng/ml TGFβ2, or complete medium plus
TGFβ2 for 24 h. Total RNA was extracted, and the mRNA was then
converted to cDNA. Reverse transcription-quantitative-PCR was then
performed for VEGF, MMP-2, SLUG and SNAIL using Qiagen Rotor Gene Q
PCR. Ns, no significance, **P<0.05. (E) Parallel sets of GF and
G5 cells were treated similarly for 1 h as aforementioned. The cell
lysates obtained were resolved on an SDS-PAGE gel, and
immunoblotting was performed using an anti-pERK, anti-TERK or
anti-β-actin antibody. TERK, total extracellular signal-regulated
kinase; p, phosphorylated; SLUG, snail family transcriptional
repressor 2; SNAIL, snail family transcriptional repressor 1; TGFβ,
transforming growth factor β. |
After cancer cells detach from neighbouring cells,
resistance from the extracellular cellular matrix hinders cell
motility; the secretion of extracellular proteinases, such as MMPs
(23), is thus crucial for
overcoming this resistance. MMP2 was found to be secreted and
upregulated by various types of cancer cells to facilitate invasion
(24). Furthermore, MMP2
expression was shown to be significantly increased in
GFP-ShcD-expressing cells compared with corresponding GF cells when
incubated with TGFβ and 10% serum but not with TGFβ alone (Fig. 3C).
Cancer metastasis is facilitated by new blood vessel
formation (25). A previous report
revealed that TGFβ induces angiogenesis via VEGF upregulation
(26). Our findings failed to
demonstrate any significant increase in VEGF gene expression in
TGFβ-treated G5 cells, with or without complete medium addition,
but interestingly, VEGF expression was upregulated in
serum-deprived GFP-ShcD-expressing cells than in corresponding
GFP-expressing cells (Fig.
3D).
In summary, SNAIL and SLUG expression was
upregulated in GFP-ShcD-expressing cells in response to TGFβ
treatment, unlike VEGF expression, which demonstrated higher levels
when the cells were deprived of serum. MMP2 upregulation in
GFP-ShcD-expressing cells was induced by the combination of 10%
serum and TGFβ, but not by TGFβ alone (Table II).
 | Table II.Alterations in gene expression under
different conditions. |
Table II.
Alterations in gene expression under
different conditions.
|
| Cell culture
conditions |
|---|
|
|
|
|---|
| Gene | (−) TGFβ | (+) TGFβ | TGFβ + serum |
|---|
| SNAIL |
−a |
+b | +/−c |
| SLUG | − | + | + |
| MMP2 | − | − | + |
| VEGF | + | − | − |
A parallel set of cells was obtained for western
blotting to assess ERK phosphorylation. pERK levels were notably
upregulated in GFP-ShcD-expressing cells than in control cells upon
TGFβ treatment regardless to complete addition (Fig. 3E).
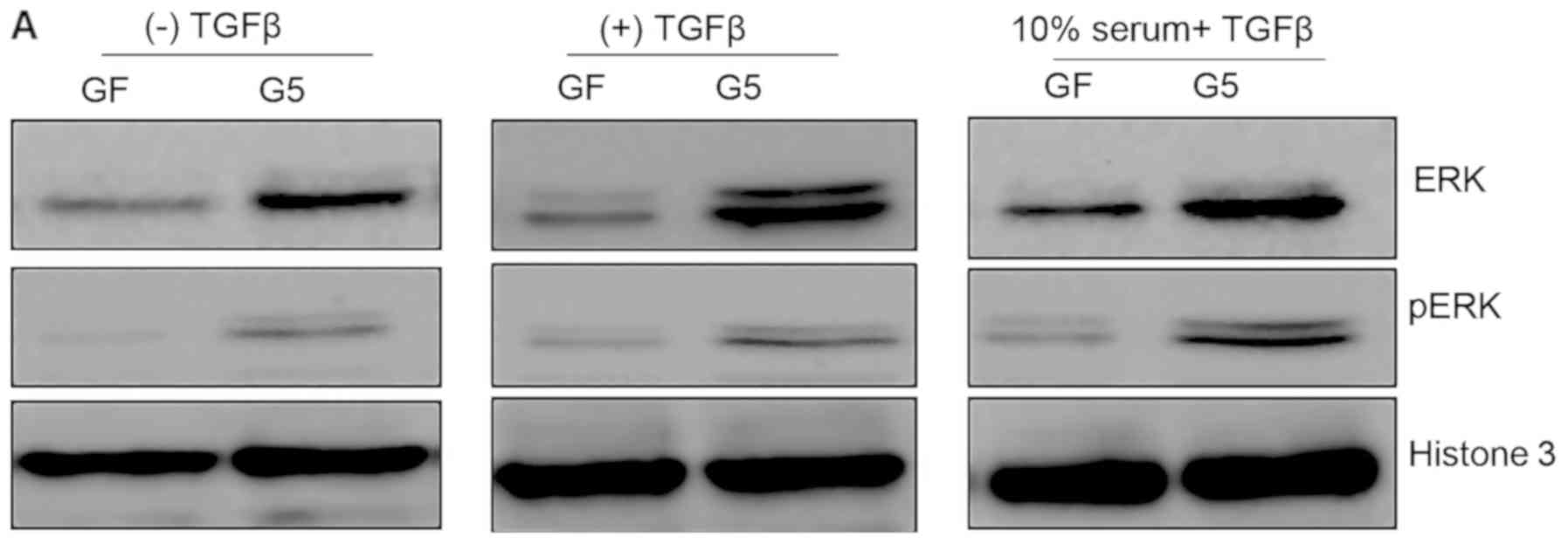
GFP-ShcD-expressing cells have higher
nuclear levels of pERK than GF cells
GFP-ShcD-expressing cells were found to have higher
expression levels of SNAIL, SLUG and MMP2 than control cells; thus,
we investigated the nuclear levels of pERK upon TGFβ treatment. GF
and G5 cells were either starved or treated with TGFβ with or
without 10% serum. The nuclear fraction of GFP-ShcD-expressing
cells exhibited higher nuclear pERK levels than that of
GFP-expressing cells in starvation, TGFβ, or TGFβ treatment with
complete medium conditions (P>0.05; 0.2, 0.18 and 0.09,
respectively; Fig. 4A and B).
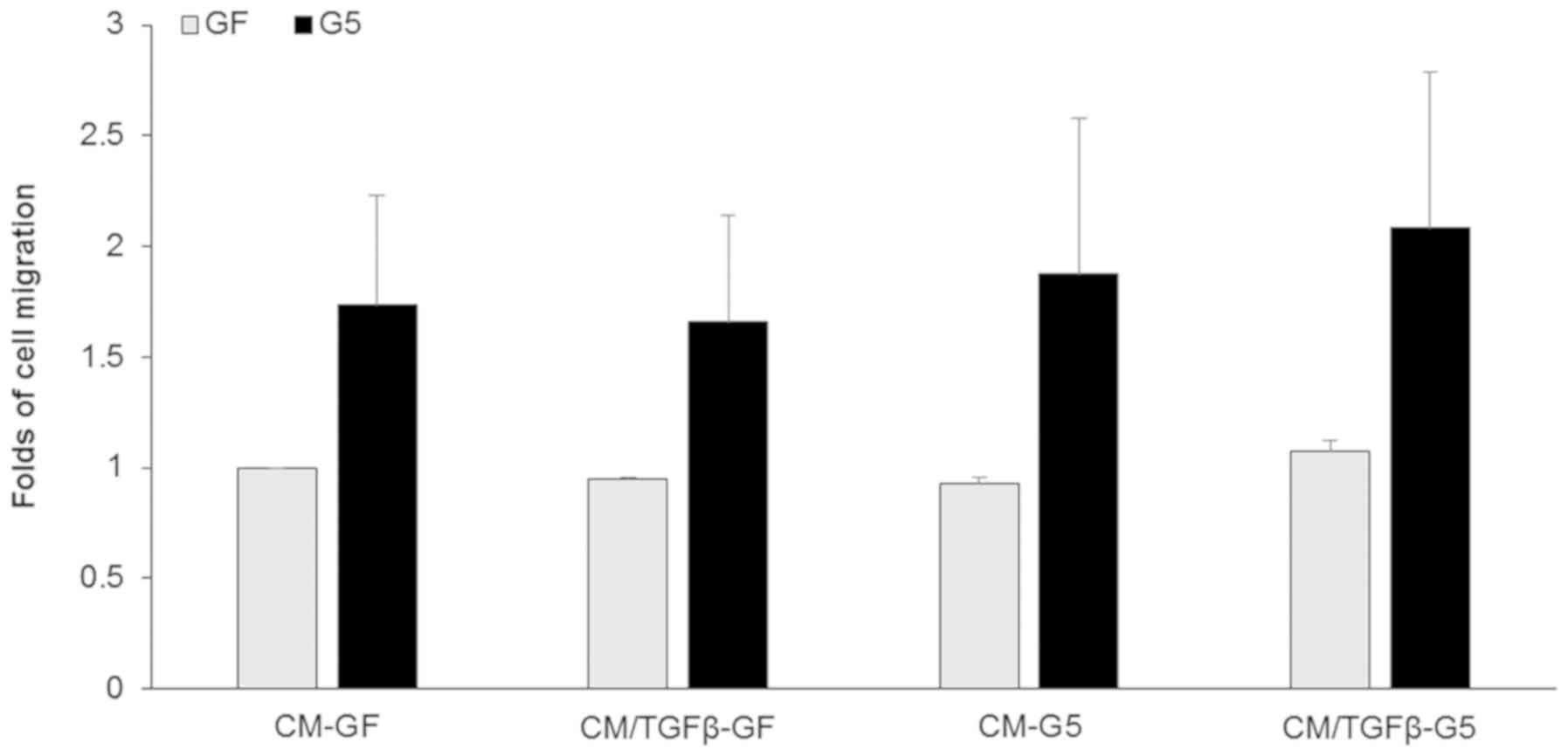
GFP-ShcD-expressing cells exhibit
greater migration than GFP-expressing cells
To determine the migration ability of
ShcD-overexpressing cells, Transwell assays were employed. The
cells were allowed to migrate against a gradient created by adding
conditioned medium derived from TGFβ-treated or untreated cells
expressing GFP-ShcD or GFP. GFP-ShcD-expressing cells exhibited
increased migration than their control counterparts, while a slight
increase in cell migration towards conditioned medium derived from
TGFβ-treated G5 cells was observed (Fig. 5). Despite GFP-ShcD-expressing cells
showed higher migratory abilities, the data was not statistically
significant.
Discussion
The role of ShcD in inducing melanoma cell migration
was determined to be partially related to MAPK pathway activation
(6). At present, few studies have
investigated the role of ShcD in melanoma cell migration.
Therefore, we aimed to determine the mechanisms underlying
ShcD-mediated cell invasion and metastasis.
Stable protein expression is advantageous as the
entire cell population expresses the exogenous protein, which makes
it a more reliable protein expression system than others.
Additionally, by analyzing stable protein expression, we generated
a cellular system of ShcD upregulation, which is the mechanism by
which 50% of melanomas acquire their invasive phenotype (6).
As TGFβ is a key factor in the acquisition of cell
mobility, it was proposed that ShcD may exert its role in migration
downstream of TGFβ signalling. This assumption regarding the Shc
family is not novel as ShcA has been reported to play an essential
role in non-Smad TGFβ signalling by inducing ERK phosphorylation
(27–29). ERK regulates gene transcription
through its downstream transcription network to mediate EMT
(30–32). In the present study, SNAIL and SLUG
expression levels were higher in TGFβ-treated GFP-ShcD-expressing
cells than in their control counterparts. Furthermore, our findings
revealed that upon TGFβ stimulation, ShcD overexpression enhanced
ERK phosphorylation, subsequently inducing SLUG and SNAIL
upregulation.
SLUG and SNAIL belong to the zinc finger-like family
of transcription factors that are responsible for transcribing
genes associated with cell migration (33,34).
Previous studies have shown that the overexpression of SLUG and
SNAIL is related to the invasive and metastatic phenotypes of
various types of cancer, which are associated with the expression
of different genes, particularly MMPs (17,35–37).
MMPs are extracellular proteases that contribute to
tissue remodelling and angiogenesis, and assist in cancer cell
migration (38). MMP2 upregulation
is one of the molecular changes required to acquire invasion
capacity in cells (39). Notably,
ERK activation was reported to mediate MMP2 promoter stimulation
(40). However, ERK
phosphorylation was determined to lead to MMP2 upregulation, and
SLUG and SNAIL were also shown to promote MMP2 upregulation
(41,42). These findings could support our
observation, in which we demonstrated that GFP-ShcD-expressing
cells exhibited upregulated MMP2 expression upon TGFβ stimulation.
In contrast to the role of TGFβ in stimulating VEGF expression
(12,26), our data indicated that VEGF
expression was significantly increased (P=0.04) without TGFβ
treatment. This finding is supported by a previous study, which
demonstrated VEGF upregulation under conditions of serum starvation
in HT29 colon cancer cells via the ERK pathway (43). Notably, in this study, ERK
phosphorylation was detected in the nuclei of starved G5 cells,
which could explain the significant increase in VEGF expression in
the serum-starved ShcD-expressing cells. These cells also
demonstrated enhanced migration ability in the presence or absence
of TGFβ treatment. A slight, yet not statistically significant,
increase was observed with TGFβ treatment. This observation is
likely due to the higher levels of phosphorylated ERK in
ShcD-expressing cells regardless of TGFβ treatment. Conclusively,
ShcD promoted ERK phosphorylation, which positively affected cell
migration-related genes, such as SLUG, SNAIL, MMP2 and VEGF;
however, only the first two genes were significantly affected by
TGFβ treatment. Thus, it was proposed that ShcD acts downstream
TGFβ, leading to the phosphorylation of ERK, and the subsequent
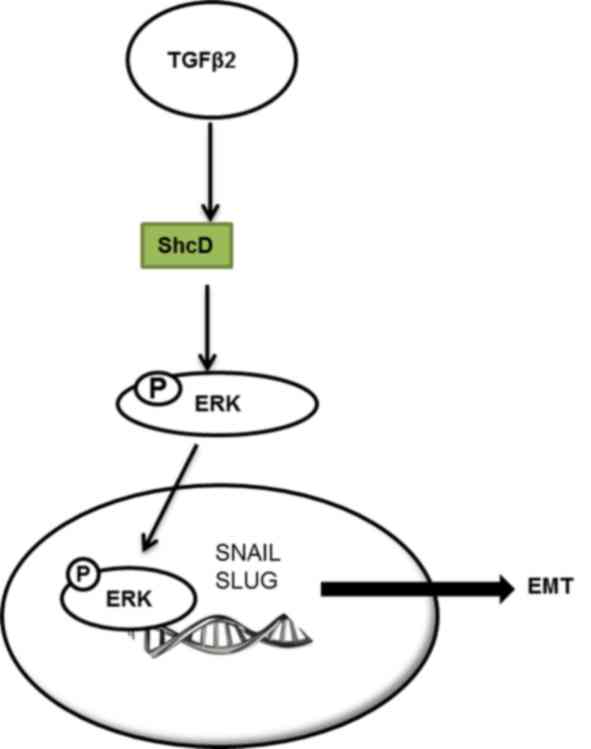
induction of SNAIL and SLUG expression, promoting EMT (Fig. 6).
In the present study, ShcD was found to be
associated with the migration of melanoma cells, which was achieved
via MAPK activation; nevertheless, a MAPK-independent mechanism was
proposed. We demonstrated that ShcD overexpression induces the
expression of certain EMT-related genes by favouring crosstalk
between ERK and TGFβ signalling. Furthermore, ShcD overexpression
was determined to promote MMP2 upregulation. These findings suggest
a novel mechanism underlying the potential of ShcD to promote cell
migration and invasion.
Acknowledgements
The authors would like to thank Dr Sally Prigent
(University of Leicester, Leicester, UK) for providing the 293 and
G5 cell lines.
Funding
The present study was supported by the 5th round of
Bohroengir Ingelhiem grants and by a targeted grant from the
University of Sharjah (grant no. 16010901011-P).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SBMA designed the experiments, performed some of the
experiments, analysed the data and wrote the manuscript. SA, FAS,
ZM, SM, NS and KR performed some of the experiments. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TGFβ
|
transforming growth factor β
|
|
Shc
|
Src homology and collagen
|
|
MMP
|
matrix metalloproteinase
|
|
VEGF
|
vascular endothelial growth factor
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
GFP
|
green fluorescent protein
|
References
|
1
|
Takahashi Y, Tobe K, Kadowaki H, Katsumata
D, Fukushima Y, Yazaki Y, Akanuma Y and Kadowaki T: Roles of
insulin receptor substrate-1 and Shc on insulin-like growth factor
I receptor signaling in early passages of cultured human
fibroblasts. Endocrinology. 138:741–150. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Polk DB: Shc is a substrate of the rat
intestinal epidermal growth factor receptor tyrosine kinase.
Gastroenterology. 109:1845–1851. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stephens RM, Loeb DM, Copeland TD, Pawson
T, Greene LA and Kaplan DR: Trk receptors use redundant signal
transduction pathways involving SHC and PLC-gamma 1 to mediate NGF
responses. Neuron. 12:691–705. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ahmed SBM and Prigent SA: Insights into
the Shc family of adaptor proteins. J Mol Signal. 12:22017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ravichandran KS: Signaling via Shc family
adapter proteins. Oncogene. 20:6322–6330. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fagiani E, Giardina G, Luzi L, Cesaroni M,
Quarto M, Capra M, Germano G, Bono M, Capillo M, Pelicci P and
Lanfrancone L: RaLP, a new member of the Src homology and collagen
family, regulates cell migration and tumor growth of metastatic
melanomas. Cancer Res. 67:3064–3073. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jones N, Hardy WR, Friese MB, Jorgensen C,
Smith MJ, Woody NM, Burden SJ and Pawson T: Analysis of a Shc
family adaptor protein, ShcD/Shc4, that associates with
muscle-specific kinase. Mol Cell Biol. 27:4759–4773. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wells A, Chao YL, Grahovac J, Wu Q and
Lauffenburger DA: Epithelial and mesenchymal phenotypic switchings
modulate cell motility in metastasis. Front Biosci (Landmark Ed).
16:815–837. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Son H and Moon A: Epithelial-mesenchymal
transition and cell invasion. Toxicol Res. 26:245–252. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie L, Law BK, Chytil AM, Brown KA, Aakre
ME and Moses HL: Activation of the Erk pathway is required for
TGF-beta1-induced EMT in vitro. Neoplasia. 6:603–610. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferrari G, Cook BD, Terushkin V, Pintucci
G and Mignatti P: Transforming growth factor-beta 1 (TGF-beta1)
induces angiogenesis through vascular endothelial growth factor
(VEGF)-mediated apoptosis. J Cell Physiol. 219:449–458. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang YE: Non-Smad pathways in TGF-beta
signaling. Cell Res. 19:128–139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hata A and Chen YG: TGF-β signaling from
receptors to Smads. Cold Spring Harb Perspect Biol. 8(pii):
a0220612016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Denis JF, Sader F, Gatien S, Villiard é,
Philip A and Roy S: Activation of Smad2 but not Smad3 is required
to mediate TGF-β signaling during axolotl limb regeneration.
Development. 143:3481–3490. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Naber HP, Drabsch Y, Snaar-Jagalska BE,
ten Dijke P and van Laar T: Snail and Slug, key regulators of
TGF-β-induced EMT, are sufficient for the induction of single-cell
invasion. Biochem Biophys Res Commun. 435:58–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim ES, Sohn YW and Moon A:
TGF-beta-induced transcriptional activation of MMP-2 is mediated by
activating transcription factor (ATF)2 in human breast epithelial
cells. Cancer Lett. 252:147–156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ahmed SBM, Amer S, Emad M, Rahmani M and
Prigent SA: Studying the ShcD and ERK interaction under acute
oxidative stress conditions in melanoma cells. Int J Biochem Cell
Biol. 112:123–133. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ahmed SB and Prigent SA: A nuclear export
signal and oxidative stress regulate ShcD subcellular localisation:
A potential role for ShcD in the nucleus. Cell Signal. 26:32–40.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schindelin J, Arganda-Carreras I, Frise E,
Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S,
Schmid B, et al: Fiji: An open-source platform for biological-image
analysis. Nat Methods. 9:676–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Friedl P and Bröcker EB: The biology of
cell locomotion within three-dimensional extracellular matrix. Cell
Mol Life Sci. 57:41–64. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong W, Li H, Zhang Y, Yang H, Guo M, Li L
and Liu T: Matrix metalloproteinase 2 promotes cell growth and
invasion in colorectal cancer. Acta Biochim Biophys Sin (Shanghai).
43:840–848. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zetter BR: Angiogenesis and tumor
metastasis. Annu Rev Med. 49:407–424. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fantozzi A, Gruber DC, Pisarsky L, Heck C,
Kunita A, Yilmaz M, Meyer-Schaller N, Cornille K, Hopfer U,
Bentires-Alj M and Christofori G: VEGF-mediated angiogenesis links
EMT-induced cancer stemness to tumor initiation. Cancer Res.
74:1566–1575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee MK, Pardoux C, Hall MC, Lee PS,
Warburton D, Qing J, Smith SM and Derynck R: TGF-beta activates Erk
MAP kinase signalling through direct phosphorylation of ShcA. EMBO
J. 26:3957–3967. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Northey JJ, Chmielecki J, Ngan E, Russo C,
Annis MG, Muller WJ and Siegel PM: Signaling through ShcA is
required for transforming growth factor beta- and
Neu/ErbB-2-induced breast cancer cell motility and invasion. Mol
Cell Biol. 28:3162–3176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hudson J, Ha JR, Sabourin V, Ahn R, La
Selva R, Livingstone J, Podmore L, Knight J, Forrest L, Beauchemin
N, et al: p66ShcA promotes breast cancer plasticity by inducing an
epithelial-to-mesenchymal transition. Mol Cell Biol. 34:3689–3701.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Navandar M, Garding A, Sahu SK, Pataskar
A, Schick S and Tiwari VK: ERK signalling modulates epigenome to
drive epithelial to mesenchymal transition. Oncotarget.
8:29269–29281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chiu LY, Hsin IL, Yang TY, Sung WW, Chi
JY, Chang JT, Ko JL and Sheu GT: The ERK-ZEB1 pathway mediates
epithelial-mesenchymal transition in pemetrexed resistant lung
cancer cells with suppression by vinca alkaloids. Oncogene.
36:242–253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng H, Li W, Wang Y, Liu Z, Cai Y, Xie
T, Shi M, Wang Z and Jiang B: Glycogen synthase kinase-3 beta
regulates Snail and β-catenin expression during Fas-induced
epithelial-mesenchymal transition in gastrointestinal cancer. Eur J
Cancer. 49:2734–2746. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Barrallo-Gimeno A and Nieto MA: The Snail
genes as inducers of cell movement and survival: Implications in
development and cancer. Development. 132:3151–3161. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ganesan R, Mallets E and Gomez-Cambronero
J: The transcription factors Slug (SNAI2) and Snail (SNAI1)
regulate phospholipase D (PLD) promoter in opposite ways towards
cancer cell invasion. Mol Oncol. 10:663–676. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Medici D, Hay ED and Olsen BR: Snail and
Slug promote epithelial-mesenchymal transition through
beta-catenin-T-cell factor-4-dependent expression of transforming
growth factor-beta3. Mol Biol Cell. 19:4875–4887. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Uygur B and Wu WS: SLUG promotes prostate
cancer cell migration and invasion via CXCR4/CXCL12 axis. Mol
Cancer. 10:1392011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun Y, Song GD, Sun N, Chen JQ and Yang
SS: Slug overexpression induces stemness and promotes
hepatocellular carcinoma cell invasion and metastasis. Oncol Lett.
7:1936–1940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Page-McCaw A, Ewald AJ and Werb Z: Matrix
metalloproteinases and the regulation of tissue remodelling. Nat
Rev Mol Cell Biol. 8:221–233. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lochter A, Galosy S, Muschler J, Freedman
N, Werb Z and Bissell MJ: Matrix metalloproteinase stromelysin-1
triggers a cascade of molecular alterations that leads to stable
epithelial-to-mesenchymal conversion and a premalignant phenotype
in mammary epithelial cells. J Cell Biol. 139:1861–1872. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang D, Bar-Eli M, Meloche S and Brodt P:
Dual regulation of MMP-2 expression by the type 1 insulin-like
growth factor receptor: the phosphatidylinositol 3-kinase/Akt and
Raf/ERK pathways transmit opposing signals. J Biol Chem.
279:19683–19690. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Merikallio H, T TT, Pääkkö P, Mäkitaro R,
Kaarteenaho R, Lehtonen S, Salo S, Salo T, Harju T and Soini Y:
Slug is associated with poor survival in squamous cell carcinoma of
the lung. Int J Clin Exp Pathol. 7:5846–5854. 2014.PubMed/NCBI
|
|
42
|
Li Y, Klausen C, Zhu H and Leung PC:
Activin a increases human trophoblast invasion by inducing
SNAIL-mediated MMP2 Up-regulation through ALK4. J Clin Endocrinol
Metab. 100:E1415–E1427. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jung YD, Nakano K, Liu W, Gallick GE and
Ellis LM: Extracellular signal-regulated kinase activation is
required for up-regulation of vascular endothelial growth factor by
serum starvation in human colon carcinoma cells. Cancer Res.
59:4804–4807. 1999.PubMed/NCBI
|