Introduction
Preeclampsia (PE) is a major hypertensive and
clinical multisystem disorder during pregnancy, occurring in 3–5%
of pregnancies worldwide between 1980 and 2010 (1,2). PE
has become one of the leading causes of maternal and neonatal
morbidity (2). Dysregulated
differentiation of human trophoblast cells leads to abnormal cell
invasion, causing PE (3–5). Major efforts have been made to
identify the molecular mechanism underlying PE, and the development
of PE is thought to be a multifactorial process, including
placental and endothelial dysfunction, abnormal spiral artery
remodeling, impaired trophoblast cells invasion and increased
trophoblasts apoptosis (6).
Dysfunction of human trophoblast cells has been suggested to be an
important factor in PE pathogenesis, and understanding the
potential mechanisms underlying trophoblast cell behavior may
facilitate the identification of new therapeutic biomarker for
PE.
MicroRNAs (miRNAs) are a class of endogenous and
highly conserved small non-coding RNAs, which serve regulatory
roles by inhibiting the expression of target genes by binding to
specific regions of the 3′-untranslated region (3′-UTR) of the
target mRNAs, thus promoting transcript degradation and/or
translational suppression (7,8).
Accumulating evidence suggested that miRNAs are associated with
many important cellular events, including differentiation, growth,
migration and apoptosis (9).
Dysregulated miRNAs are found in tumor tissues and cell lines, and
miRNAs have shown to act as tumor suppressor genes or oncogenes in
various types of human cancer (10). Accumulating evidence has indicated
that miRNAs serve an important role in the pathogenesis of PE
(11). Xiao et al (12) demonstrated that miR-144 regulates
the proliferation, invasion and migration of human trophoblast
cells by targeting PTEN in PE. Wu et al (13) showed that miR-181a-5p suppresses
the migration and invasion of human trophoblast cells in PE by
directly targeting insulin like growth factor 2 mRNA binding
protein 2. Dai et al (14)
reported that miR-155 inhibits the proliferation and migration of
human trophoblast cells by downregulating CCND1. In addition, a
previous study found that miR-203 contributes to PE development in
the human placenta by targeting vascular endothelial growth factor
A, and miR-203 may be used as a potential therapeutic biomarker in
treating PE (15). Although a
large number of miRNAs have been previously described, the roles
and molecular regulatory mechanisms of specific miRNAs in PE
require further investigation.
A previous study demonstrated that the level of
miR-320a is decreased in different tumor samples, and can function
as a tumor suppressor by downregulating the expression level of
multiple target genes (16).
miR-320a acts as an important regulator in the development of
gastric cancer by directly targeting Ras-related protein Rab-14
(17). miR-320a is downregulated
in human non-small cell lung cancer and suppresses tumor cell
proliferation and invasion by targeting insulin-like growth factor
1 receptor (18). miR-320a
regulates high mobility group box 1 in hepatocellular carcinoma,
limiting cell invasion and metastasis (19). In addition, miR-320a represses the
progression of colorectal cancer by targeting Rac1 (20). However, to the best of our
knowledge, no previous study investigated the biological functions
and mechanisms of miR-320a in PE development.
The aim of the present study was to determine the
potential roles and molecular mechanisms of miR-320a in the
proliferation and invasion of the trophoblast cell line
HTR-8/SVeno. The decreased levels of miR-320a were determined in
placental samples collected from patients with PE by reverse
transcription-quantitative PCR (RT-qPCR). MTT and invasion assays
were performed to detected cell proliferation and invasion in
HTR-8/SVneo cells after transfection with miR-320a mimic or mimic
control. In addition, it was tested whether interleukin (IL)-4 was
a direct target of miR-320a by dual luciferase report assay and
western blots analysis. In addition, following rescue experiments,
miR-320a upregulation was found to suppress proliferation and
invasion of HTR-8/SVneo cells in PE at least partly by directly
regulating IL-4.
Materials and methods
Patients
Placenta samples (gestational age, 32–40 weeks) were
obtained from pregnant patients with PE (n=57, age 19–34 years) and
healthy patients (n=57, age 22–31 years) between January 2012 and
December 2016 at the Department of Obstetrics, The Affiliated
Hospital of Jining Medical University (Jining, China). The criteria
for diagnosis of PE was as follows: Systolic blood pressure ≥140
mmHg or diastolic blood pressure ≥90 mmHg at 20 weeks
post-gestation in a pregnant woman whose blood pressure was
previously normal, 24 h urine protein ≥0.3 g. Exclusion criteria
were as follows: Hypertensive emergencies, blood system diseases
and abnormal pregnancy with other complications of obstetrics. The
clinical characteristics are presented in Table I. The placentas were immediately
snap-frozen in liquid nitrogen after surgery and stored at −80°C
until further experimentation. The diagnosis of PE was based on The
Royal College of Obstetricians and Gynaecologists guidelines
(1). The study was approved by the
Ethics Committee of The Affiliated Hospital of Jining Medical
University (2017TJ05749). Written consent was obtained from all
patients.
 | Table I.Clinical characteristics of patients
with PE and healthy patients. |
Table I.
Clinical characteristics of patients
with PE and healthy patients.
|
Characteristics | PE pregnancies
(n=57) | Normal pregnancies
(n=57) | P-value |
|---|
| Maternal age,
years | 27.12±4.11 | 26.37±3.29 | 0.57 |
| BMI, kg/m2 | 24.91±2.78 | 25.53±3.14 | 0.61 |
| SBP, mmHg | 154.06±7.65 | 109.55±5.93 | <0.001 |
| DBP, mmHg | 102.25±6.48 | 70.69±5.07 | <0.001 |
| 24-h urine protein,
g | 3.67±0.73 | Not detected | <0.001 |
| Gestational age,
weeks | 35.33±3.28 | 39.25±2.16 | 0.04 |
| Infant birth
weight, kg | 2.27±0.50 | 3.06±0.75 | 0.02 |
Biochemical testing
Peripheral blood (5 ml) was collected in EDTA
anticoagulant tubes (Qiagen GmbH) from all patients. Serum was
acquired by centrifugation at 3,000 × g for 10 min at 37°C. Body
mass index was shown as weight divided by height squared
(kg/m2). The 24 h urine protein level was quantified
using a Fully Automated Chemistry Analyzer (AU400; Olympus
Corporation).
Cell culture
The human extravillous trophoblast cells HTR-8/SVneo
were purchased from the Cell Bank of The Chinese Academy of
Sciences. HTR-8/SVneo cells were cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc.), and supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100
U/ml penicillin in a humidified atmosphere with 5% CO2
at 37°C.
RT-qPCR
Total RNA was isolated from placenta samples and
HTR-8/SVneo cells using TRIzol Reagent (Thermo Fisher Scientific,
Inc.). Subsequently, RNA concentration was determined using a
NanoDrop 2000 (Thermo Fisher Scientific, Inc.). RNA was reverse
transcribed into cDNA using a PrimeScript RT Master Mix kit (Takara
Bio, Inc.), according to the manufacturer's protocol. The
thermocycling conditions were as follows: 95°C for 10 min, 42°C for
2 min, 37°C for 15 min, 85°C for 5 sec and 4°C for 30 min. qPCR was
performed using a SYBR PrimeScript RT-qPCR kit (Takara Bio, Inc.)
on a Roche Light-cycler 480 real-time qPCR System (Roche Applied
Science), according to the manufacturer's protocol. The
thermocycling conditions of the qPCR were as follows: Initial
denaturation at 95°C for 30 sec, followed by 40 cycles of 95°C for
10 sec and 60°C for 30 sec. Primers for miR-320a, U6, IL-4 and
GAPDH are presented in Table II.
The primers were designed using Primer Premier software (version
5.0; Premier Biosoft International, Inc.), and purchased from
Takara Biotechnology Co., Ltd. Expression level of miR-320a was
normalized to the U6 expression level. IL-4 expression was
normalized to GAPDH expression. Relative expression was calculated
using the 2−∆∆Cq method (21).
 | Table II.The sequences of primers for reverse
transcription-quantitative PCR assay. |
Table II.
The sequences of primers for reverse
transcription-quantitative PCR assay.
| Gene | Sequence
(5′-3′) |
|---|
| miR-320a | F:
GTTGGATCCGGCGTTTCCTTCCGACATG |
|
| R:
GCTGAATTCGTCCACTGCGGCTGTTCC |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
ACGCTTCACGAATTTGCGT |
| IL-4 | F:
GCAGTTCCACAGGCACAA |
|
| R:
TGGTTGGCTTCCTTCACA |
| GAPDH | F:
CAAGGTCATCCATGACAACT |
|
| R:
GTCCACCACCCTGTTGCTG |
Oligonucleotide and plasmid
transfection
miR-320a mimic, mimic control, pcDNA3.1 empty vector
and IL-4 overexpressing plasmid (pcDNA3.1-IL-4) were purchased from
GenePharma Co., Ltd. The mimic control and pcDNA3.1 empty vector
were used as negative controls for miR-320a mimic and
pcDNA3.1-IL-4, respectively. The sequences of miR-320a mimic and
mimic control were as follows: miR-320a mimic,
5′-AAAAGCUGGGUUGAGAGGGCGA-3′; mimic control,
5′-UUCUCCGAACGUGUCACGUTT-3′. For transfection, HTR-8/SVneo cells
were inoculated in six-well plates at a density of 105
cells/well, and were cultured in a humidified atmosphere with 5%
CO2 at 37°C overnight. Transient transfections of 100
pmol miR-320a mimic, 100 pmol mimic control, 2.5 µg pcDNA3.1 empty
vector and 2.5 µg pcDNA3.1-IL-4 vector were performed using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at
70–80% confluency according to the manufacturer's protocol. Cells
were collected at 48 h after transfection for RT-qPCR analysis.
Target gene prediction
The potential target genes of miR-320a were
identified using three publicly available algorithms, including
TargetScanHuman version 7.2 (22),
PicTar version 2007 (23) and
miRanda version 2010 (24).
Dual luciferase reporter assay
Human IL-4 mRNA 3′-UTR, including the complementary
binding sequence of miR-320a, (5′…UUAUGAGUUUUUGAUAGCUUUAU…-3′), was
obtained from GenePharma Co., Ltd. and inserted into the pmirGLO
vector (Promega Corporation) to construct the wild-type IL-4
reporter plasmid. The mutant IL-4 mRNA 3′-UTR
(5′…UUAUGAGUUUUUGAUGAUCCCAU…-3′) was also cloned into pmirGLO
vector to construct the mutant IL-4 reporter plasmid.
Co-transfection of the wild-type or the mutant IL-4 reporter
plasmid and miR-320a mimic or mimic control was performed in
HTR-8/SVneo cells using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Renilla and Firefly luciferase values were detected at 24 h
post-transfection using a dual luciferase reporter system kit
(Promega Corporation) in a Varioskan Flash Microplate Reader
(Thermo Fisher Scientific, Inc.), following the manufacturer's
protocol. Renilla luciferase activity was used to normalize
Firefly luciferase activity.
MTT assay
The proliferative ability of human trophoblast cells
was assessed using an MTT assay. The transfected HTR-8/SVneo cells
(~8,000 cells/well) were inoculated into each well of 96-well
plates. After 0, 24, 48 and 72 h of transfection, 10 µl MTT (5
mg/ml; Sigma-Aldrich; Merck KGaA) was added into each well. Then,
the cells were incubated in a humidified atmosphere with 5%
CO2 at 37°C for 4 h, and the supernatant was discarded.
Subsequently, 150 µl DMSO (Sigma-Aldrich; Merck KGaA) was added to
dissolve the violet crystals for 10 min at 37°C. A microplate
reader (Bio-Rad Laboratories, Inc.) was used to detect absorbance
of each well at 450 nm wavelength.
Transwell invasion assay
Transwell inserts (Costar; Corning, Inc.) precoated
with Matrigel (BD Biosciences) were used to examine cell invasion.
In total, ~105 transfected HTR-8/SVneo cells were
inoculated in the upper well in 250 µl RPMI 1640 medium, whereas
the lower well contained 500 µl RPMI 1640 medium supplemented with
10% FBS (Sigma-Aldrich; Merck KGaA). After a 24-h incubation at
37°C with 5% CO2, non-invasive cells attached to the
upper side of membranes were removed using cotton swabs. The
invasive cells were fixed with 100% methanol (Beyotime Institute of
Biotechnology) for 20 min at 37°C and stained with 0.1% crystal
violet (Beyotime Institute of Biotechnology) for 10 min at 37°C.
Stained cells were photographed and counted in five
randomly-selected fields of view using a light microscope (DMI6000
B microscope; Leica Microsystems GmbH; magnification, ×200).
Western blot analysis
Total protein was extracted from HTR-8/SVneo cells
using RIPA buffer (Beyotime Institute of Biotechnology), and
concentration of samples was detected using a bicinchoninic acid
assay kit (Beyotime Institute of Biotechnology), according to the
manufacturer's protocol. Subsequently, 10% SDS-PAGE was used to
separate proteins (30 µg) and proteins were transferred onto PVDF
membranes (EMD Millipore). Then, membranes were treated with 5%
non-fat milk for blocking for 60 min at 37°C, and incubated
overnight at 4°C with the following primary antibodies: Anti-IL-4
(cat. no. ab62351; 1:1,000; Abcam) and anti-GAPDH (cat. no.
ab181602; 1:2,000; Abcam). Subsequently, membranes were incubated
with horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (cat. no. ab7090; 1:2,000; Abcam) at 37°C for 60 min.
Specific immunoreactive bands were visualized using a Pierce ECL
western blotting kit (Thermo Fisher Scientific, Inc.). The positive
bands were analyzed using ImageJ software (version 1.49; National
Institutes of Health).
Statistical analysis
Statistical analysis was performed using SPSS 19.0
(IBM Corp.). Data are presented as the mean ± SD. Each assay was
conducted in triplicate. Student's t-test was performed to compare
two groups. One-way ANOVA followed by Bonferroni's post hoc test
was performed to compare multiple groups. P<0.05 was considered
to indicate a statistically significant difference.
Results
Clinical characteristics
Clinical characteristics were obtained from 57
pregnant patients with PE and 57 healthy patients. The clinical
data are presented in Table I. The
analysis of the clinical data suggested that diastolic blood
pressure, systolic blood pressure and the level of 24 h urine
protein in the PE group were higher than those in the healthy
patients (P<0.05). The infant birth weight and gestational age
were lower in the PE group than those in the healthy group
(P<0.05). The maternal age and body mass index did not exhibit
significant differences between the two groups. The high blood
pressure and 24 h urine protein confirmed the diagnosis of PE.
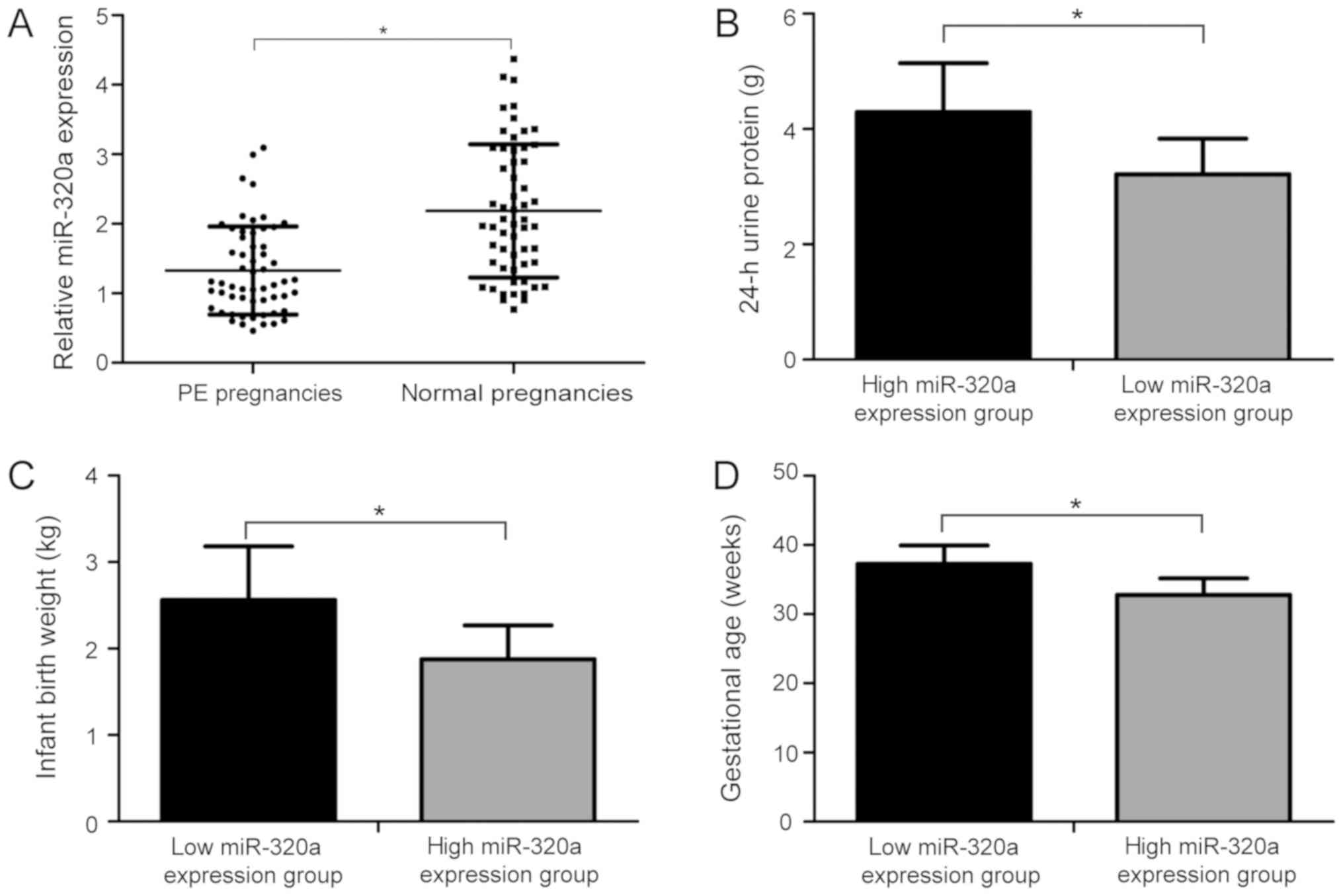
miR-320a expression level is decreased
in placentas of patients with PE
To investigate miR-320a expression level in
placentas from patients with PE and healthy patients, RT-qPCR assay
was performed. The relative levels of miR-320a were markedly
downregulated in placentas from patients with PE compared with
healthy patients (Fig. 1A;
P<0.05). Additionally, patients with PE were divided into low
and high miR-320a expression groups based on the mean
2−∆∆Cq value of miR-320a (1.32). Among the 57 patients
with PE, 33 patients were in the low miR-320a expression group and
24 in the high miR-320a expression group. Interestingly, the
analysis of the two groups suggested that the level of 24-h urine
protein in the high miR-320a expression group was significantly
higher than that in the low miR-320a expression group (Fig. 1B; P<0.05). The birth weight
(Fig. 1C; P<0.05) and
gestational age (Fig. 1D;
P<0.05) were lower in the high miR-320a expression group
compared with the low expression group. The present data suggested
that the expression level of miR-320a was decreased in placentas of
patients with PE and was found to be associated with the
pathogenesis of PE.
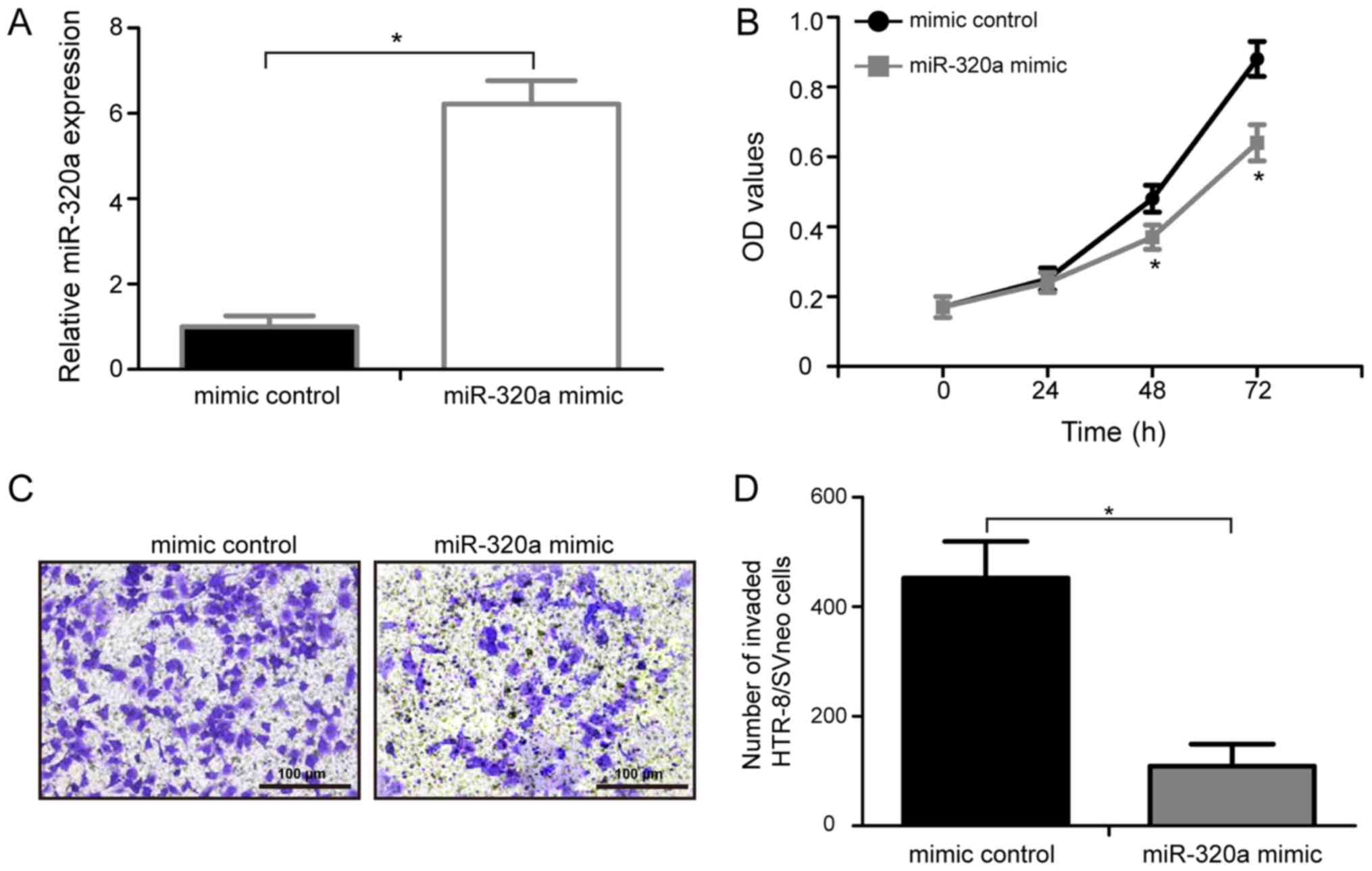
miR-320a overexpression represses the
proliferation of HTR-8/SVneo cells
To identify the biological roles of miR-320a in PE,
miR-320a mimic or mimic control were transfected into HTR-8/SVneo
cells, and a MTT assay was performed to examine the effects of
miR-320a on the proliferation of HTR-8/SVneo cells in vitro.
RT-qPCR analysis showed that miR-320a expression levels in
HTR-8/SVneo treated with miR-320a mimic were significantly
upregulated compared with cells transfected with control mimic
(Fig. 2A; P<0.05). Compared
with cells transfected with control mimic, cells transfected with
miR-320a exhibited a significantly decrease in cell proliferation
at 48 and 72 h (Fig. 2B;
P<0.05). The present data suggested that the upregulation of
miR-320a is involved in the development of PE by inhibiting the
proliferation of HTR-8/SVneo cells.
miR-320a overexpression reduces the
invasion of HTR-8/SVneo cells
To identify the possible function of miR-320a on
human trophoblasts invasion, HTR-8/SVneo cells were transfected
with either miR-320a mimic or mimic control and a Transwell
invasion assay was performed (Fig.
2C). The present data suggested that over-expression of
miR-320a markedly inhibited the invasive ability of HTR-8/SVneo
cells compared with the mimic control (Fig. 2D; P<0.05). The present results
demonstrated that the upregulation of miR-320a is involved in the
development of PE by inhibiting the invasion of HTR-8/SVneo
cells.
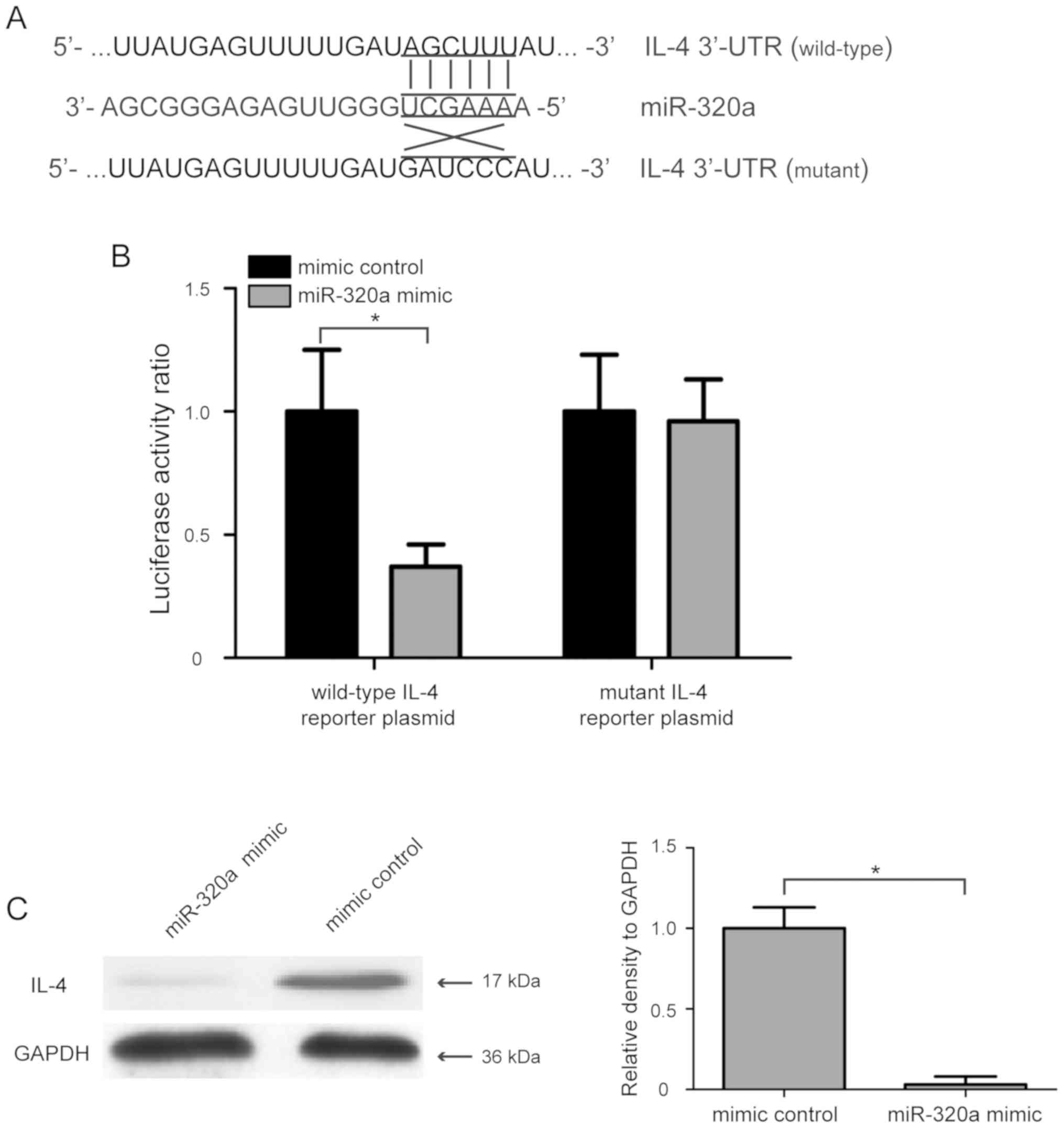
IL-4 is a direct target gene of
miR-320a in HTR-8/SVneo cells
To investigate the molecular mechanism involved in
the miR-320a-mediated regulation of proliferation and invasion of
human trophoblasts, three computational algorithms, including
TargetScanHuman, miRanda and PicTar, were used to analyze the
possible targets of miR-320a. A previous study showed that IL-4
polymorphisms are associated with PE (25), and IL-4 was found to have a high
score in the three computational algorithms. Therefore, IL-4 was
selected as a candidate target gene of miR-320a for further
analysis. To evaluate whether IL-4 was directly regulated by
miR-320a, its 3′-UTR containing the complementary binding sequence
of miR-320a, and a mutant 3′-UTR containing mutations in the
miR-320a binding sequence, were introduced into the pmirGLO vector
(Fig. 3A). After co-transfection
with the wild-type IL-4 reporter plasmid and miR-320a mimic into
HTR-8/SVneo cells, the luciferase activity ratio was significantly
decreased, whereas co-transfection with mutant IL-4 reporter
plasmid and miR-320a mimic did not affect the luciferase activity
of HTR-8/SVneo cells (Fig. 3B).
Additionally, the effect of miR-320a on IL-4 expression was also
determined using western blot analysis, and the present results
suggested that miR-320a mimic transfection significantly suppressed
the protein expression level of IL-4 in HTR-8/SVneo cells (Fig. 3C; P<0.05). The present results
indicated that IL-4 was a direct target gene of miR-320a in
HTR-8/SVneo cells.
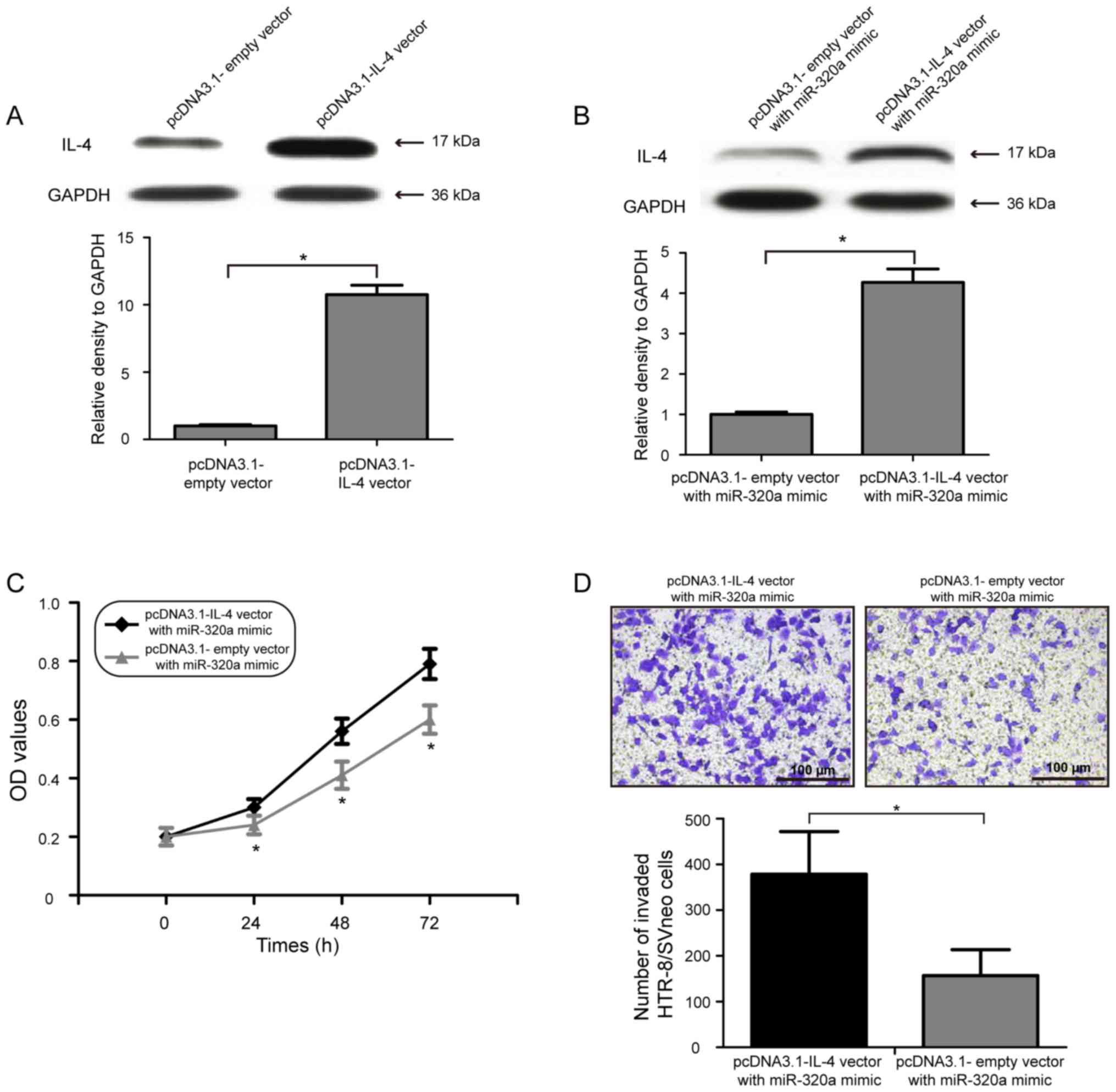
miR-320a inhibits the proliferation
and invasion of HTR-8/SVneo cells by directly targeting IL-4
To examine whether the inhibitory effects of
miR-320a on cell proliferation and invasion of human trophoblasts
were mediated by IL-4 downregulation, IL-4 was overexpressed by
co-transfecting pcDNA3.1-IL-4 vector with miR-320a mimic in
HTR-8/SVneo cells. Western blot analysis suggested that
transfection of pcDNA3.1-IL-4 vector significantly increased the
expression level of IL-4 in HTR-8/SVneo cells (Fig. 4A; P<0.05). In addition, the
protein expression level of IL-4 in HTR-8/SVneo cells was also
increased following co-transfection of the pcDNA3.1-IL-4 vector
with miR-320a mimic (Fig. 4B;
P<0.05). Notably, restoring the expression level of IL-4
partially decreased the inhibitory effect of miR-320a
overexpression on HTR-8/SVneo cell proliferation (Fig. 4C; P<0.05). Consistent with the
cell proliferation results, overexpression of IL-4 in HTR-8/SVneo
cells reduced the inhibitory effect of miR-320a overexpression on
cell invasion (Fig. 4D;
P<0.05). Collectively, the present data suggested that IL-4 was
a functional target gene of miR-320a, and miR-320a upregulation
inhibited the proliferation and invasion of HTR-8/SVneo cells by
directly targeting IL-4.
Discussion
The molecular mechanisms of PE pathogenesis are
complex and are thought to be multifactorial processes (3–5).
Abnormal proliferation and invasion of human trophoblasts serve
crucial roles in the development of PE (26). Therefore, it is important to study
the mechanism of trophoblast proliferation and invasion in order to
identify novel reliable and effective molecular targets for
treating PE. Accumulating evidence has indicated that miRNAs are
dysregulated in placentas of patients with PE (27,28).
The identification of dysregulated miRNAs and their target genes in
the placenta of patients with PE has provided a basis for
investigating the mechanisms underlying PE development (29,30).
To the best of our knowledge, the present study was the first to
suggest that miR-320a may be downregulated in patients with PE, and
IL-4 was identified as a functional target gene of miR-320a. The
present study suggested that miR-320a upregulation was involved in
the development of PE by inhibiting the proliferation and invasion
of trophoblast cells by targeting IL-4.
miR-320a has been found to be dysregulated in
cerebral ischemia, myasthenia gravis and human tumors (31). Recently, several reports have
demonstrated that miR-320a acts as a tumor suppressor gene by
targeting Neuropilin 1 in colorectal cancer (32), Myc in hepatocellular carcinoma
(33), and staphylococcal nuclease
and tudor domain containing 1 in gliomas (34). Since miR-320a regulates multiple
target genes, it may serve an important role in the regulatory
network underlying the development of PE. To the best of our
knowledge, the functions and potential mechanisms of miR-320a in PE
have not been previously identified. The present results were in
line with previous studies (32–34)
suggesting that miR-320a is a downregulated miRNA in patients with
PE. The present statistical analysis showed that patients with PE
with high miR-320a expression had a higher level of 24-h urine
protein, their children presented a lower birth weight, and the
gestational age was decreased compared with patients with PE in the
low miR-320a expression group, indicating that miR-320a may be
associated with the pathogenesis of PE.
Furthermore, the function of miR-320a on cell
proliferation and invasion were analyzed by using MTT and Transwell
invasion assays, respectively, in HTR-8/SVneo cells after
transfection with miR-320a mimic and mimic control. The present
results suggested that transient overexpression of miR-320a induced
a significant decrease in HTR-8/SVneo cell proliferation and
invasion, suggesting that the upregulation of miR-320a is involved
in the development of PE by inhibiting the proliferation and
invasion of HTR-8/SVneo cells. The present results provided novel
insights into the roles of miR-320a in PE development. miRNAs act
by silencing their target genes; therefore, the identification of
the potential targets of miR-320a was required in order to
elucidate the mechanisms underlying miR-320a-mediated PE
development. Using three bioinformatic tools, the potential target
genes of miR-320a have been analyzed. IL-4 was selected for further
experiments as it ranked the highest amongst the potential target
genes. In addition, the association between IL-4 polymorphisms and
PE was previously investigated (25). T helper 2 (Th2) cells produce
IL-13, IL-6, IL-5 and IL-4, and these cytokines are associated with
the production of antibody and inhibition of cell-mediated immunity
(35). A previous study suggested
that PE is associated with an overproduction of Th2-specific
cytokines (36). An increased
expression level of IL-4 is involved in PE during the second half
of the pregnancy, probably by decreasing nitric oxide secretion and
promoting human leukocyte antigen expression (37,38).
To investigate this possibility, wild-type or mutant IL-4 reporter
plasmid and miR-320a mimic or mimic control were transfected into
HTR-8/SVneo cells, and Renilla and Firefly luciferase values
were detected using a dual luciferase reporter assay. The present
results suggested that miR-320a could repress IL-4 expression by
binding to its 3′-UTR. Moreover, western blot assay suggested that
IL-4 protein expression was significantly reduced in HTR-8/SVneo
cells following transfection with miR-320a mimic. The present
results suggested that IL-4 was a direct target gene of miR-320a in
HTR-8/SVneo cells.
In addition, rescue experiments were performed to
investigate whether miR-320a exerted its function via IL-4
downregulation in HTR-8/SVneo cells. The present results suggested
that overexpression of IL-4 partly reversed the inhibitory effects
of miR-320a on the proliferation and invasion of human
trophoblasts. Collectively, the present data suggested that
miR-320a was involved in the development of preeclampsia by
targeting IL-4, suggesting that the miR-320a/IL-4 pathway may be a
novel therapeutic target for PE treatment. Notably, the target
genes of miR-320a are multiple and are not limited to IL-4;
therefore, other potential target genes need to be investigated to
further examine the regulatory network of miR-320a in the
development of PE. The main limitation of the present study is that
the molecular mechanisms underlying IL-4-regulated cell
proliferation and invasion in human trophoblasts were not
investigated and require further investigation.
Collectively, the present study suggested that
miR-320a was lowly expressed in the placentas of patients with PE
and miR-320a may be involved in the development of PE by inhibiting
the proliferation and invasion of trophoblast cells by targeting
IL-4. Therefore, therapeutic approaches targeting the miR-320a/IL-4
pathway may improve the treatment of PE.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NX and LL designed and carried out the experiments.
NX and ZJ analyzed the data. LL and ZJ wrote and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Affiliated
Hospital of Jining Medical University (Jining, China). Written
consent was obtained from all patients.
Patient consent for publication
Written informed consent was obtained from all
patients prior to the start of the study.
Competing interests
The authors declare that they have no competing
interest.
References
|
1
|
Redman CW and Sargent IL: Latest advances
in understanding preeclampsia. Science. 308:1592–1594. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mol BWJ, Roberts CT, Thangaratinam S,
Magee LA, de Groot CJM and Hofmeyr GJ: Pre-eclampsia. Lancet.
387:999–1011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Amaral LM, Wallace K, Owens M and LaMarca
B: Pathophysiology and current clinical management of preeclampsia.
Curr Hypertens Rep. 19:612017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ji L, Brkic J, Liu M, Fu G, Peng C and
Wang YL: Placental trophoblast cell differentiation: Physiological
regulation and pathological relevance to preeclampsia. Mol Aspects
Med. 34:981–1023. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Staun-Ram E and Shalev E: Human
trophoblast function during the implantation process. Reprod Biol
Endocrinol. 3:562005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Noris M, Perico N and Remuzzi G:
Mechanisms of disease: Pre-eclampsia. Nat Clin Pract Nephrol.
1:98–114; quiz 120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: Key players in the immune system, differentiation,
tumorigenesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen DB and Wang W: Human placental
microRNAs and preeclampsia. Biol Reprod. 88:1302013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao J, Tao T, Yin Y, Zhao L, Yang L and
Hu L: miR-144 may regulate the proliferation, migration and
invasion of trophoblastic cells through targeting PTEN in
preeclampsia. Biomed Pharmacother. 94:341–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu L, Song WY, Xie Y, Hu LL, Hou XM, Wang
R, Gao Y, Zhang JN, Zhang L, Li WW, et al: miR-181a-5p suppresses
invasion and migration of HTR-8/SVneo cells by directly targeting
IGF2BP2. Cell Death Dis. 9:162018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai Y, Qiu Z, Diao Z, Shen L, Xue P, Sun H
and Hu Y: MicroRNA-155 inhibits proliferation and migration of
human extravillous trophoblast derived HTR-8/SVneo cells via
down-regulating cyclin D1. Placenta. 33:824–829. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu F, Wu K, Wu W, Chen Y, Wu H, Wang H
and Zhang W: miR203 contributes to preeclampsia via inhibition of
VEGFA expression. Mol Med Rep. 17:5627–5634. 2018.PubMed/NCBI
|
|
16
|
Sun L, Liu B, Lin Z, Yao Y, Chen Y, Li Y,
Chen J, Yu D, Tang Z, Wang B, et al: miR-320a acts as a prognostic
factor and inhibits metastasis of salivary adenoid cystic carcinoma
by targeting ITGB3. Mol Cancer. 14:962015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Liu H, Shao J and Xing G: miR-320a
serves as a negative regulator in the progression of gastric cancer
by targeting RAB14. Mol Med Rep. 16:2652–2658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Shi C, Cao L, Zhong L and Wang D:
MicroRNA-320a is downregulated in non-small cell lung cancer and
suppresses tumor cell growth and invasion by directly targeting
insulin-like growth factor 1 receptor. Oncol Lett. 13:3247–3252.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lv G, Wu M, Wang M, Jiang X, Du J, Zhang
K, Li D, Ma N, Peng Y, Wang L, et al: miR-320a regulates high
mobility group box 1 expression and inhibits invasion and
metastasis in hepatocellular carcinoma. Liver Int. 37:1354–1364.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao H, Dong T, Zhou H, Wang L, Huang A,
Feng B, Quan Y, Jin R, Zhang W, Sun J, et al: miR-320a suppresses
colorectal cancer progression by targeting Rac1. Carcinogenesis.
35:886–895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
23
|
Krek A, Grun D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen J, Zhong M and Yu YH: Association
between interleukin-4 polymorphisms and risk of pre-eclampsia in a
population of Chinese pregnant women. Genet Mol Res. 16:2017.
View Article : Google Scholar
|
|
26
|
Zhang Z, Wang X, Zhang L, Shi Y, Wang J
and Yan H: Wnt/β-catenin signaling pathway in trophoblasts and
abnormal activation in preeclampsia (Review). Mol Med Rep.
16:1007–1013. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pineles BL, Romero R, Montenegro D, Tarca
AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P,
et al: Distinct subsets of microRNAs are expressed differentially
in the human placentas of patients with preeclampsia. Am J Obstet
Gynecol. 196:261, e1–e6. 2007. View Article : Google Scholar
|
|
28
|
Zhu XM, Han T, Sargent IL, Yin GW and Yao
YQ: Differential expression profile of microRNAs in human placentas
from preeclamptic pregnancies vs. normal pregnancies. Am J Obstet
Gynecol. 200:661, e1–e7. 2009. View Article : Google Scholar
|
|
29
|
Jairajpuri DS and Almawi WY: MicroRNA
expression pattern in pre-eclampsia (Review). Mol Med Rep.
13:2351–2358. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Noguer-Dance M, Abu-Amero S, Al-Khtib M,
Lefèvre A, Coullin P, Moore GE and Cavaillé J: The primate-specific
microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol
Genet. 19:3566–3582. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu C, Liao Z, Cai M and Zhang G:
MicroRNA-320a downregulation mediates human liver cancer cell
proliferation through the Wnt/beta-catenin signaling pathway. Oncol
Lett. 13:573–578. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P,
Zhang Q, Dong L, Liu Y and Dong J: microRNA-320a inhibits tumor
invasion by targeting neuropilin 1 and is associated with liver
metastasis in colorectal cancer. Oncol Rep. 27:685–694.
2012.PubMed/NCBI
|
|
33
|
Xie F, Yuan Y, Xie L, Ran P, Xiang X,
Huang Q, Qi G, Guo X, Xiao C and Zheng S: miRNA-320a inhibits tumor
proliferation and invasion by targeting c-Myc in human
hepatocellular carcinoma. Onco Targets Ther. 10:885–894. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li H, Yu L, Liu J, Bian X, Shi C, Sun C,
Zhou X, Wen Y, Hua D, Zhao S, et al: miR-320a functions as a
suppressor for gliomas by targeting SND1 and beta-catenin, and
predicts the prognosis of patients. Oncotarget. 8:19723–19737.
2017.PubMed/NCBI
|
|
35
|
Singh VK and Rai G: Cytokines in posterior
uveitis: An update. Immunol Res. 23:59–74. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kang L, Chen CH, Yu CH, Chang CH and Chang
FM: An association study of interleukin-4 gene and preeclampsia in
Taiwan. Taiwan J Obstet Gynecol. 53:215–219. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hashemi V, Dolati S, Hosseini A, Gharibi
T, Danaii S and Yousefi M: Natural killer T cells in Preeclampsia:
An updated review. Biomed Pharmacother. 95:412–418. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vianna P, Mondadori AG, Bauer ME, Dornfeld
D and Chies JA: HLA-G and CD8+ regulatory T cells in the
inflammatory environment of pre-eclampsia. Reproduction.
152:741–751. 2016. View Article : Google Scholar : PubMed/NCBI
|