Introduction
Metabolic diseases affect millions of people in both
developed and transition countries (1). In addition to conventional genetic
inheritance of risk alleles, emerging evidence has shown that these
diseases are also linked to lifestyle and inherited epigenetic
pattern interactions, which affects gene expression and the
activity of proteins involved in the onset and pathogenesis of
diverse metabolic diseases (2).
The strong link between epigenetics and metabolism
may offer attractive clinical applications to counteract the
escalating prevalence of metabolic diseases, such as obesity, type
2 diabetes mellitus (T2DM), non-alcoholic fatty liver disease
(NAFLD), among others (2,3). Regarding the epigenetic factors,
MicroRNAs (miRNAs) are a class of small non-coding RNAs (ncRNAs)
that regulate the expression of ~60% of protein coding genes. They
control many cellular functions and metabolic pathways, and
subsequently influence the development and progression of a number
of diseases (4–7).
Among several activities, miRNAs are recognized as
regulators of lipid and glucose metabolism and are involved in the
physiopathology of metabolic diseases (6,8). The
liver-enriched miR-122-5p was the first miRNA to be functionally
associated with a metabolic phenotype, regulating cholesterol and
lipid metabolism (9).
Additionally, a miR-122 inhibitor (Miravirsen) was found as a novel
therapeutic strategy against chronic hepatitis C virus (HCV)
infection (10). miR-33a-5p and
miR-33b-5p were also demonstrated as playing crucial roles in
cholesterol and lipid turnover; while miR-34a-5p may be a key
regulator of hepatic lipid homeostasis (11). miR-103a-3p and miR-107 have been
reported as regulators of hepatic insulin sensitivity (6) and as contributors of adipose growth
by accelerating adipocyte differentiation (12). On the other hand, miR-375 is highly
expressed in pancreatic islets and is important for insulin
secretion and β cell development and maintenance (13,14).
Thus, the altered expression of miRNAs and their target genes could
interfere or manage the predisposition to metabolic diseases
(15,16).
Furthermore, some studies have shown the
interactions between miRNAs and other epigenetics mechanisms,
including long ncRNAs (lncRNAs) as described elsewhere (17). Members of the lncRNA family
contribute to intracellular processes by acting as host transcripts
for miRNA (18,19) and lncRNAs can antagonize miRNA
function by competing with miRNAs to bind to target mRNAs (20). Furthermore, lncRNAs may act as
molecular decoys or sponges of miRNAs, affecting the levels and
function of miRNAs (21,22). Otherwise, some lncRNAs are targeted
by miRNAs, repressing lncRNAs expression (23). Besides that, lncRNA-miRNA
interactions can regulate gene expression through a double-negative
feedback loop (24). Moreover,
accumulating evidence associates lncRNAs in the maintenance of
metabolic homeostasis and the dysregulation of certain lncRNAs
promotes the progression of metabolic disorders such as diabetes,
obesity, chronic liver diseases, and cardiovascular diseases
(25,26).
Several small molecules have been suggested as
directly binding to miRNAs, then modifying miRNAs expression, thus
having therapeutic potential (27). In this context, Gumireddy et
al (28) report that the small
molecule diazobenzene modifies the miR-21 expression, suggesting
that miR-21 may become a druggable target.
In this study, we aimed to identify metabolic
disease-related miRNAs and their target genes, and then construct
miRNA-target gene and miRNA-lncRNA networks to find out putative
important biological processes and determine those miRNAs that have
major roles in metabolic diseases. Additionally, we aimed also to
identify small molecules that interact with the miRNAs. These
analyses may provide a theoretical basis for further studies and
contribute to understand important complex mechanisms underlying
metabolic diseases.
Materials and methods
Search for metabolic disease-related
microRNAs
Metabolic disease-related miRNAs were obtained from
two experiment-supported databases: Human MicroRNA Disease Database
(HMDD v3.0) (29) and miR2Disease
(access December 2018) (30). The
miRNAs previously associated with obesity, NAFLD, or T2DM were
incorporated into our analyses. After that, the results obtained
from these two databases were compared to those found in the Matrix
Decomposition and Heterogeneous Graph Inference (MDHGI; access
December 2018) (31), a
miRNA-disease predictor database. For this, we included the top 10
miRNAs predicted as associated with metabolic diseases. Thus, the
inclusion of validated and predicted data increases the power of an
association.
Additionally, we also investigated if the target
genes of these miRNAs were previously associated with metabolic
diseases (T2DM, NAFLD, and obesity) using the DisGeNET v5.0
database (32). The DisGeNET
database is a discovery platform containing one of the largest
publicly available collections of genes and variants associated to
human diseases. For this last approach, we included only the top 10
genes associated with each disease according to the prediction
score. This strategy was used to increase the association evidence
power and to focus on those molecules with potential higher
interest and value.
Evaluation of microRNA target
genes
The list of miRNAs identified as associated with
metabolic diseases was then submitted to bioinformatics analyses to
search for their putative target genes. For this approach, the
information from experimentally validated miRNA-target gene
interactions was combined with the results from target prediction
algorithms in order to retrieve a comprehensive set of target genes
while controlling for false positive rates. CyTargetLinker v3.0.1
web tool (33) was used to search
for validated and predicted miRNA-target gene interactions (MTI)
and visualize them in a graphical way. For this study, we obtained
Homo sapiens MTIs from one experimentally validated database
(miRTarBase v.4.4) and from two predicted miRNA databases
(MicroCosm v.5.0 and TargetScan v.7.0). Moreover, the target genes
were also searched in the miRWalk v.3.0 (34) database and incorporated into the
analysis. The miRNA-mRNA networks were visualized and analyzed
using the Cytoscape software v3.7.0 (35).
Pathways analysis
Functional enrichment analysis of miRNA-target genes
were performed to retrieve Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathways annotations for
the miRNAs target genes, using the plug-ins Biological Networks
Gene Ontology (BiNGO; v3.0.3) (36) and ClueGO/Cluepedia (v.2.3.5)
(37) on Cytoscape environment
(35). The ClueGO/Cluepedia
plug-in permits the visualization of the non-redundant biological
terms for large clusters of gene sets in a functionally grouped
network and the most representative GO term or KEGG pathways was
used to name the module, considering a κ score of 0.3 and q-values
>0.05.
Interactions between microRNAs and
lncRNAs and associations between miRNAs and small molecules
The interactions between miRNAs and lncRNAs were
analyzed using the starBase v2.0 (38) database, and the connections between
miRNAs and small molecules were performed using the SM2miR
(39) and PharmacomiR (40) databases. The miRNet web-tool was
used to perform the search and analysis (41).
Additionally, we also investigated the subcellular
location of miRNAs and lncRNAs associated with metabolic disorders
using RNALocate database (42) as
well as iLoc-lncRNA (43) and
lncLocator (44) web tools.
Statistical analysis and
visualization
Cytoscape v3.7.0 software (35) was used to illustrate the
disease-related networks and analyze the network properties. The
Venn diagrams were constructed using the InteractiveVenn instrument
(45). The names of miRNAs, mRNAs,
and lncRNAs are unified based on miRBase 22 release (46), HUGO gene nomenclature committee
(HGNC), and LNCipedia v5.2 (47),
respectively.
The statistical tests used for the enrichment
analysis were based on the right-sided hypergeometric test and
adjusted for multiple hypotheses using the Benjamini & Hochberg
False Discovery Rate (FDR) test. Interactions with a q-value
<0.05 were considered strongly enriched.
Results
Identification of metabolic
disease-related miRNAs
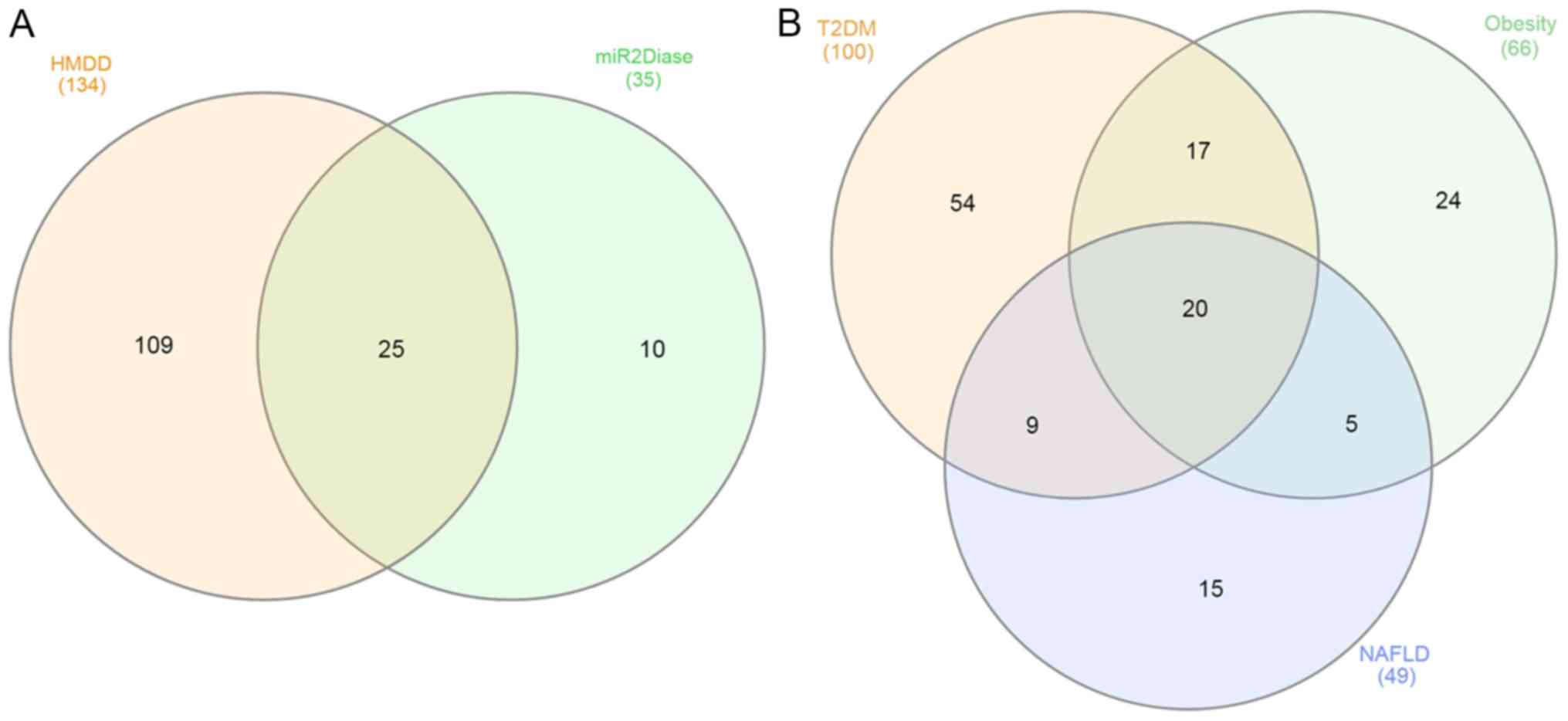
A total of 144 unique miRNAs related to metabolic
disease were found mapping the two databases of human diseases
(Fig. 1A and Table SI). In the HMDD database, 134
unique miRNAs were found; while in the miR2Disease, 35 unique
miRNAs were found. As shown in Fig.
1A, the two databases shared 25 miRNAs. Moreover, the 10 miRNAs
found only in the miR2Disease database were associated with NAFLD.
The miR2Disease database had more data regarding NAFLD than HMDD;
however, there is less information regarding T2DM and obesity
(Table SI). Regarding the 25
miRNAs shared by the 2 databases, these miRNAs were previously
associated with metabolic alterations and other human diseases,
including some types of cancer (16,48,49).
Data from the two resources were systematically
combined according to the metabolic disease criteria. As shown in
Fig. 1B, 100 miRNAs were found to
be associated with T2DM, 66 miRNAs with obesity, and 49 miRNAs with
NAFLD. Moreover, 20 miRNAs were related to the three pathologies
(let-7d-5p, miR-17-5p, miR-21-5p, miR-26a-5p, miR-27a-3p,
miR-27b-3p, miR-29c-3p, miR-30a-5p, miR-33a-5p, miR-34a-5p,
miR-103a-3p, miR-107, miR-122-5p, miR-126-3p, miR-132-3p,
miR-150-5p, miR-155-5p, miR-200a-3p, miR-200b-3p, and miR-375-5p).
Additionally, the results from the two databases were compared with
those from the miRNA-disease predictor, MDHGI (31). Out of the 20 miRNAs, 10 miRNAs
(miR-17-5p, miR-21-5p, miR-29c-3p, miR-34a-5p, miR-103a-3p,
miR-107, miR-122-5p, miR-126-3p, miR-132-3p, and miR-150-5p) were
also found in MDHGI database, increasing the evidence of
association of these miRNAs with metabolic diseases (Table I).
 | Table I.miRNAs associated with metabolic
diseases from distinct databases. |
Table I.
miRNAs associated with metabolic
diseases from distinct databases.
| miRNAs | miR2-Disease | HMDD | MDHGI | DisGe-NET |
|---|
| let-7d-5p | X | X |
| X |
| miR-17-5p | X | X | X | X |
| miR-21-5p | X | X | X |
|
| miR-26a-5p | X | X |
| X |
| miR-27b-3p | X | X |
| X |
| miR-29c-3p | X | X | X | X |
| miR-30a-5p | X | X |
| X |
| miR-33a-5p | X | X |
|
|
| miR-34a-5p | X | X | X | X |
| miR-103a-3p | X | X | X | X |
| miR-107 | X | X | X | X |
| miR-122-5p | X | X | X |
|
| miR-126-3p | X | X | X |
|
| miR-132-3p | X | X | X | X |
| miR-27a-3p |
| X |
| X |
| miR-150 |
| X | X | X |
| miR-200b-3p |
| X |
| X |
| miR-155-5p |
| X |
|
|
| miR-375 |
| X |
|
|
| miR-200a-3p |
| X |
|
|
Putative target genes of the selected
miRNAs associated with metabolic diseases
The 20 miRNAs selected using the strategy described
in the Methods Section regulate together the expression of 10.942
unique target genes (predicted or validated). miR-17-5p had the
largest number of target genes (1181), followed by miR-155-5p (904)
and miR-34a-5p (736). The miR-200a-3p had the lowest number of
target genes (151) (Table SII). A
group of 484 putative target genes was found when we analyzed only
the miRNA-gene interactions reported in three online databases
(Fig. S1). The largest number of
interconnections was found for miR-17-5p and miR-30a-5p. Moreover,
as expected, the miRNAs miR-107 and miR-103a-3p and the miRNAs
miR-27a-3p and miR-27b-3p shared a great number of target genes.
miR-150 did not have common target genes with other miRNAs; and we
could not find target genes for miR-126-3p and miR-375 when
considering only the targets that were reported as validated and
predicted by at least three databases (Fig. S1).
Moreover, of these 20 miRNAs, 13 miRNAs target at
least one of the top 10 candidate genes associated with each
metabolic disorder: T2DM, obesity, or NAFLD, according to DisGeNET
database (Tables I and II). Furthermore, some of these genes are
very well described targets of the selected miRNAs. Regarding the
validated and predicted target genes, PPARG is targeted by
miR-27a-3p and miR-27b-3p, LDLR is targeted by miR-30a-3p,
SIRT1 by miR-132-3p, and NEUROD1 by miR-30a-3p.
However, there are genes in this list that are not regulated by the
selected 20 miRNAs, suggesting that there are more miRNAs involved
in the pathogenesis of metabolic diseases.
 | Table II.Top 10 genes associated with each
analyzed metabolic disease according to the DisGeNET database and
the interactions with the selected miRNAs. |
Table II.
Top 10 genes associated with each
analyzed metabolic disease according to the DisGeNET database and
the interactions with the selected miRNAs.
| A, T2DM |
|---|
|
|---|
| Gene | Gene name | Score | miRNAs |
|---|
| GCK | Glucokinase | 0.899 | – |
| HNF1A | HNF1 homeobox
A | 0.812 | miR-107,
miR-27b-3p |
| HNF4A | Hepatocyte nuclear
factor 4α | 0.729 | miR-27b-3p |
| HNF1B | HNF1 homeobox
B | 0.684 | – |
| AKT2 | AKT
serine/threonine kinase 2 | 0.681 | miR-29c-3p,
miR-103a-3p |
| ABCC8 | ATP binding
cassette subfamily C member 8 | 0.677 | – |
| IRS1 | Insulin receptor
substrate 1 | 0.67 | miR-150-5p,
let-7d-5p, miR-29c-3p |
| NEUROD1 | Neuronal
differentiation 1 | 0.645 | miR-17-5p,
miR-30a-5p |
| PDX1 | Pancreatic and
duodenal homeobox 1 | 0.634 | – |
| PAX4 | Paired box 4 | 0.618 | – |
|
| B,
Obesity |
|
| Gene | Gene
name | Score | miRNAs |
|
| MC4R | Melanocortin 4
receptor | 0.913 | – |
| PPARG | Peroxisome
proliferator activated receptor γ | 0.727 | miR-34a-5p,
miR-27a-3p, miR-27b-3p |
| LEP | Leptin | 0.72 | miR-17-5p,
miR-200b-3p, miR-132-3p, miR-150-5p |
| LEPR | Leptin
receptor | 0.688 | miR-103a-3p,
miR-17-5p, miR-26a-5p |
| POMC |
Proopiomelanocortin | 0.528 | – |
| PCSK1 | Proprotein
convertase subtilisin/kexin type 1 | 0.507 | miR-200b-3p |
| SIM1 | Single-minded
family bHLH transcription factor 1 | 0.492 | miR-27b-3p,
let-7d-5p |
| APOE | Apolipoprotein
E | 0.479 | – |
| UCP3 | Uncoupling protein
3 | 0.475 | miR-17-5p,
miR-200b-3p |
| SH2B1 | SH2B adaptor
protein 1 | 0.439 | – |
|
| C,
NAFLD |
|
| Gene | Gene
name | Score | miRNAs |
|
| ADIPOQ | Adiponectin, C1Q
and collagen domain containing | 0.283 | miR-103a-3p,
miR-107 |
| SIRT1 | Sirtuin 1 | 0.282 | miR-17-5p,
let-7d-5p, miR-132-3p |
| NFE2L2 | Nuclear factor,
erythroid 2 like 2 | 0.281 | – |
| PNPLA3 | Patatin like
phospholipase domain containing 3 | 0.231 | miR-200b-3p,
miR-29c-3p |
| TM6SF2 | Transmembrane 6
superfamily member 2 | 0.205 | – |
| PPARA | Peroxisome
proliferator activated receptor α | 0.205 | miR-17-5p |
| SREBF1 | Sterol regulatory
element binding transcription factor 1 | 0.202 | – |
| LEP | Leptin | 0.202 | miR-17-5p,
miR-200b-3p, miR-132-3p, miR-150-5p |
| FGF21 | Fibroblast growth
factor 21 | 0.202 | – |
| LDLR | Low density
lipoprotein receptor | 0.201 | miR-27b-3p,
miR-150-5p, miR-17-5p, miR-30a-5p |
Additionally, as shown in Table I, of the 20 miRNAs, a subset of 6
miRNAs (miR-17-5p, miR-29c-3p, miR-34a-5p, miR-103a-3p, miR-107,
and miR-132-3p) was found in the four resources (HMDD, miR2Disease,
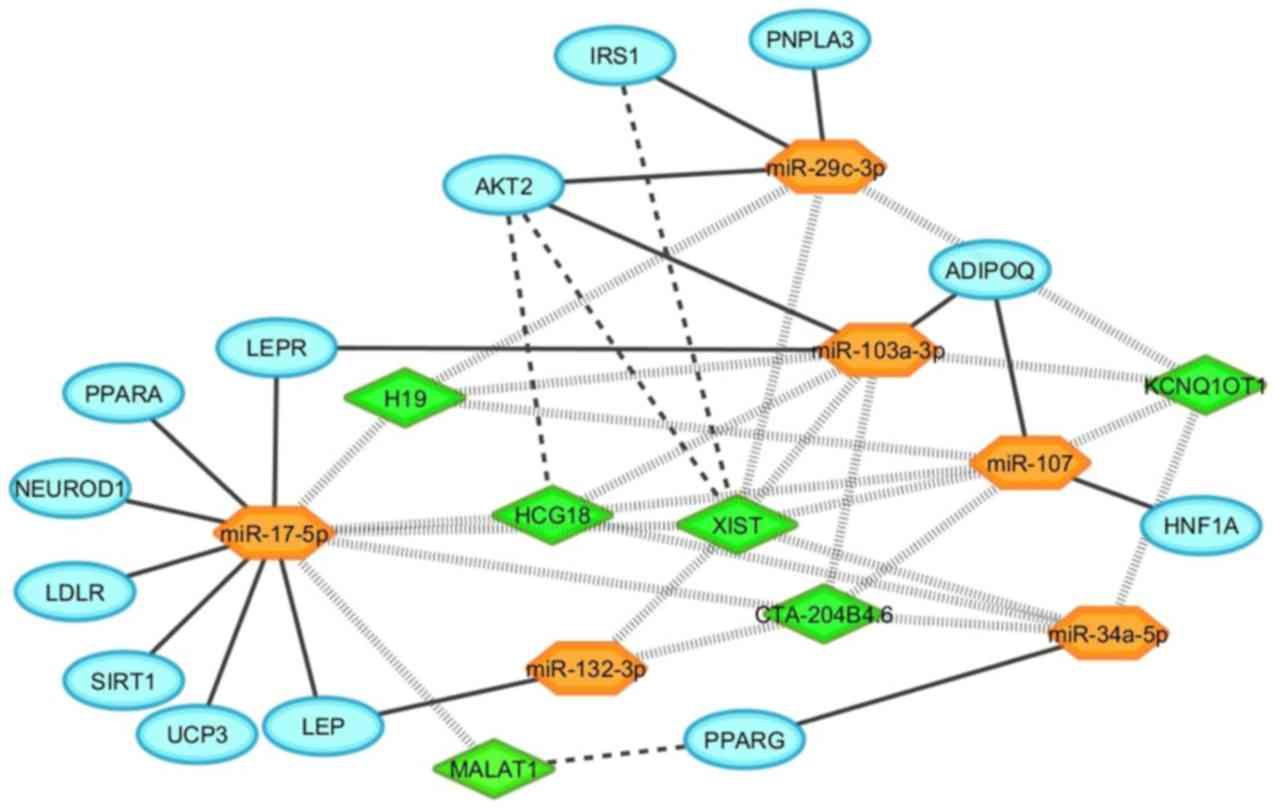
MDHGI, and DisGeNET) used for these analyses (Fig. S2). Fig. 2 summarizes the interaction of the
candidate genes for metabolic diseases and the 6 selected
miRNAs.
Pathway analysis of the selected
miRNAs associated with metabolic diseases
To explore the biological pathways possibly affected
by the target genes of the 20 analyzed miRNAs, functional
enrichment analysis of their target genes using pathway maps from
the GO and KEGG repositories were carried out.
GO terms were investigated for biological, cellular,
and molecular processes associated with the set of predicted and
validated target genes found for the 20 selected miRNAs. As a
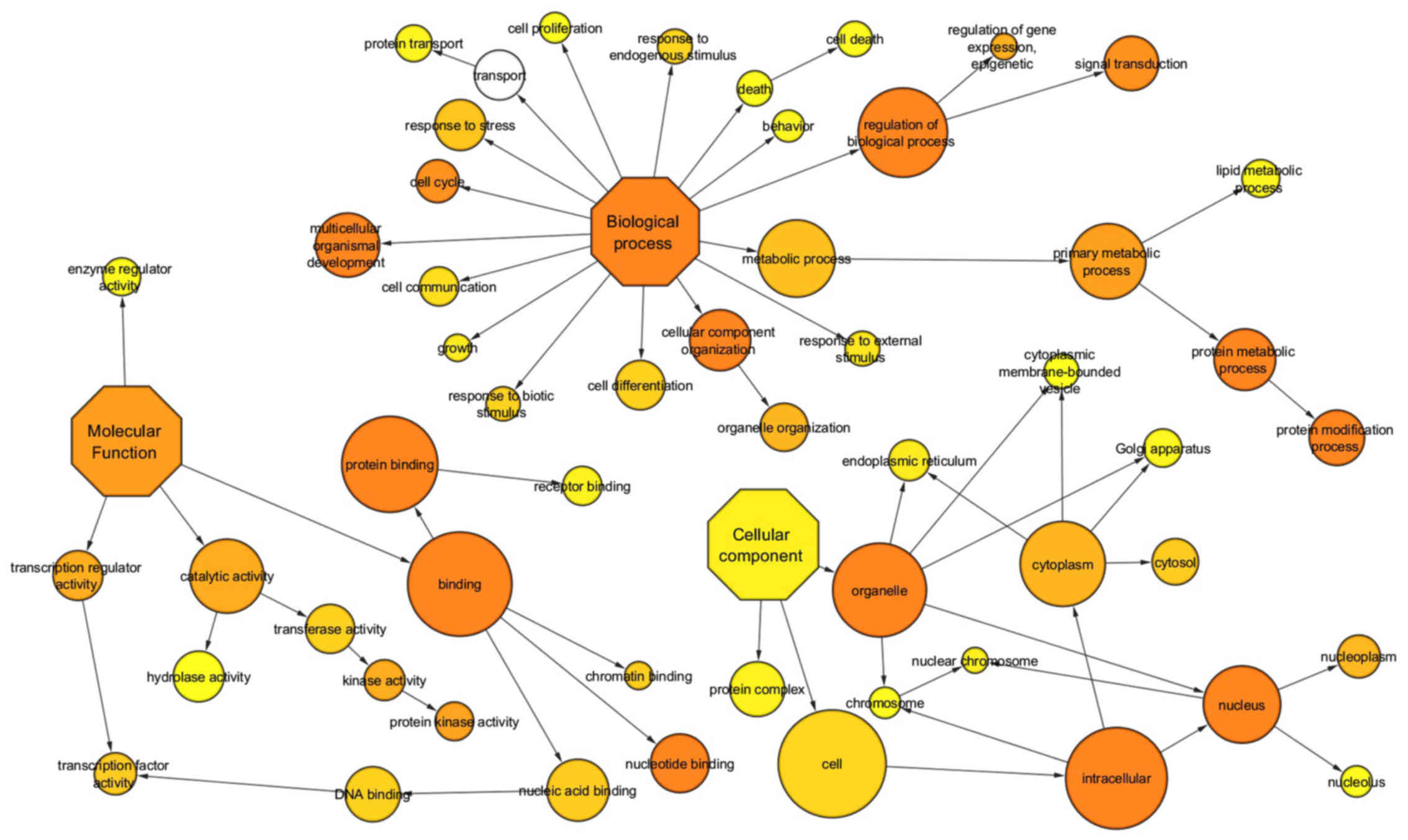
result, a total of 30 unique pathways were enriched for miRNAs.
Many of these pathways are well established to be involved in
metabolic diseases, such as transforming growth factor β receptor,
oxidative stress, apoptosis, VEGF and angiogenesis signaling
pathways (Table SIII and Fig. 3).
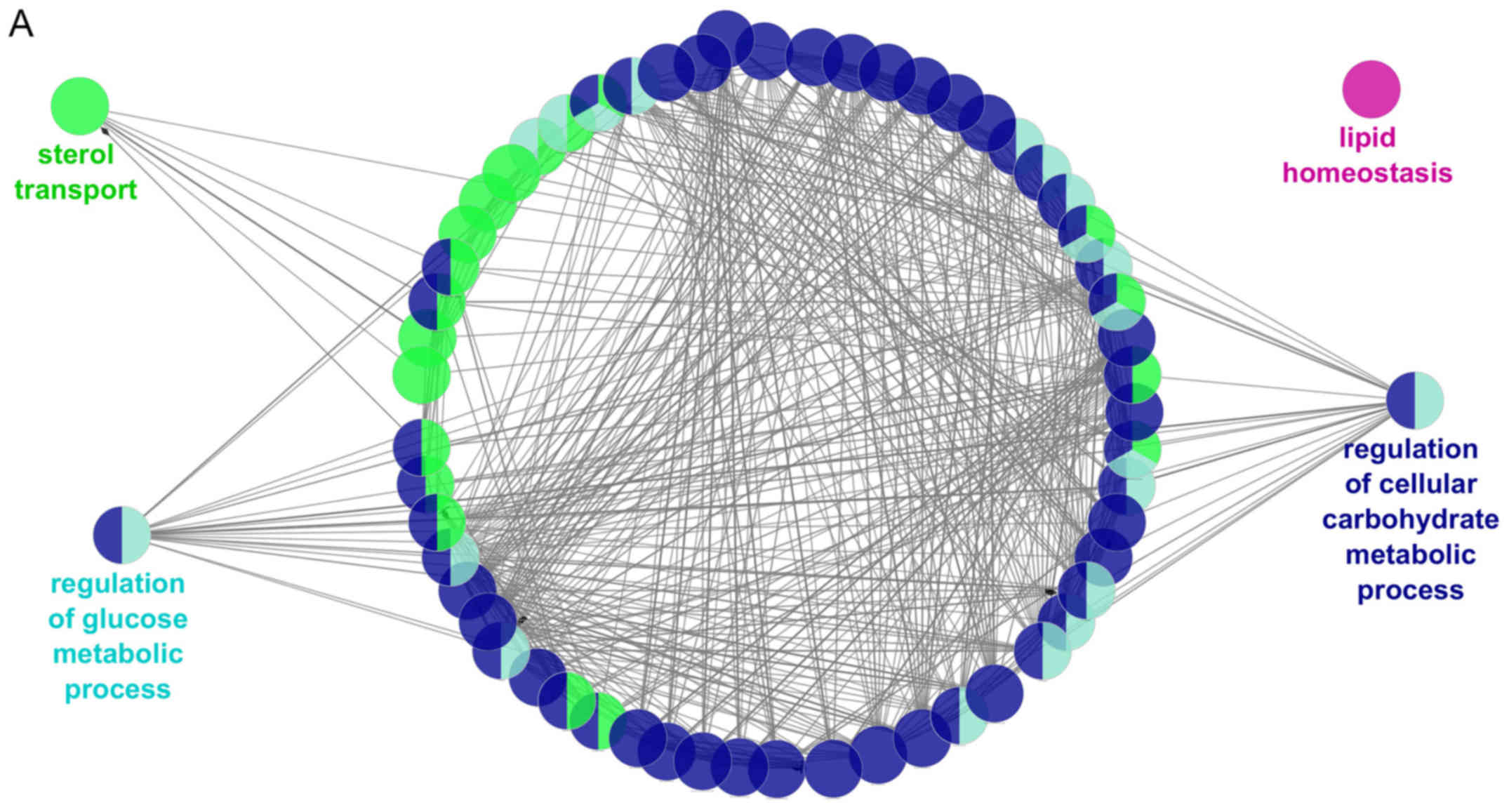
After that, the metabolic pathways in which
participates the subgroup of the 16 genes previously associated
with metabolic diseases (HNF1A, HNF4A, AKT2, IRS1, NEUROD1,
PPARG, LEP, LEPR, PCSK1, SIM1, UCP3, ADIPOQ, SIRT1, PNPLA3,
PPARA and LDLR) were also investigated. These genes are
regulated by the 6 selected miRNAs (as shown in Table II and participate in the
biological processes previously associated with metabolic diseases,
such as regulation of cellular carbohydrate metabolic process,
lipid homeostasis, cholesterol transport, and regulation of glucose
metabolism (Fig. 4A).
Additionally, these target genes were also enriched in some KEGG
pathways such as transcriptional regulation of white adipocyte
differentiation, AMPK signaling pathway, regulation of gene
expression in β cell, and adipocytokine signaling pathway (Fig. 4B).
miRNA-lncRNAs interactions
The 20 miRNAs associated with metabolic diseases
putatively interact with 423 unique lncRNAs (Table SIV). Moreover, the subgroup of 6
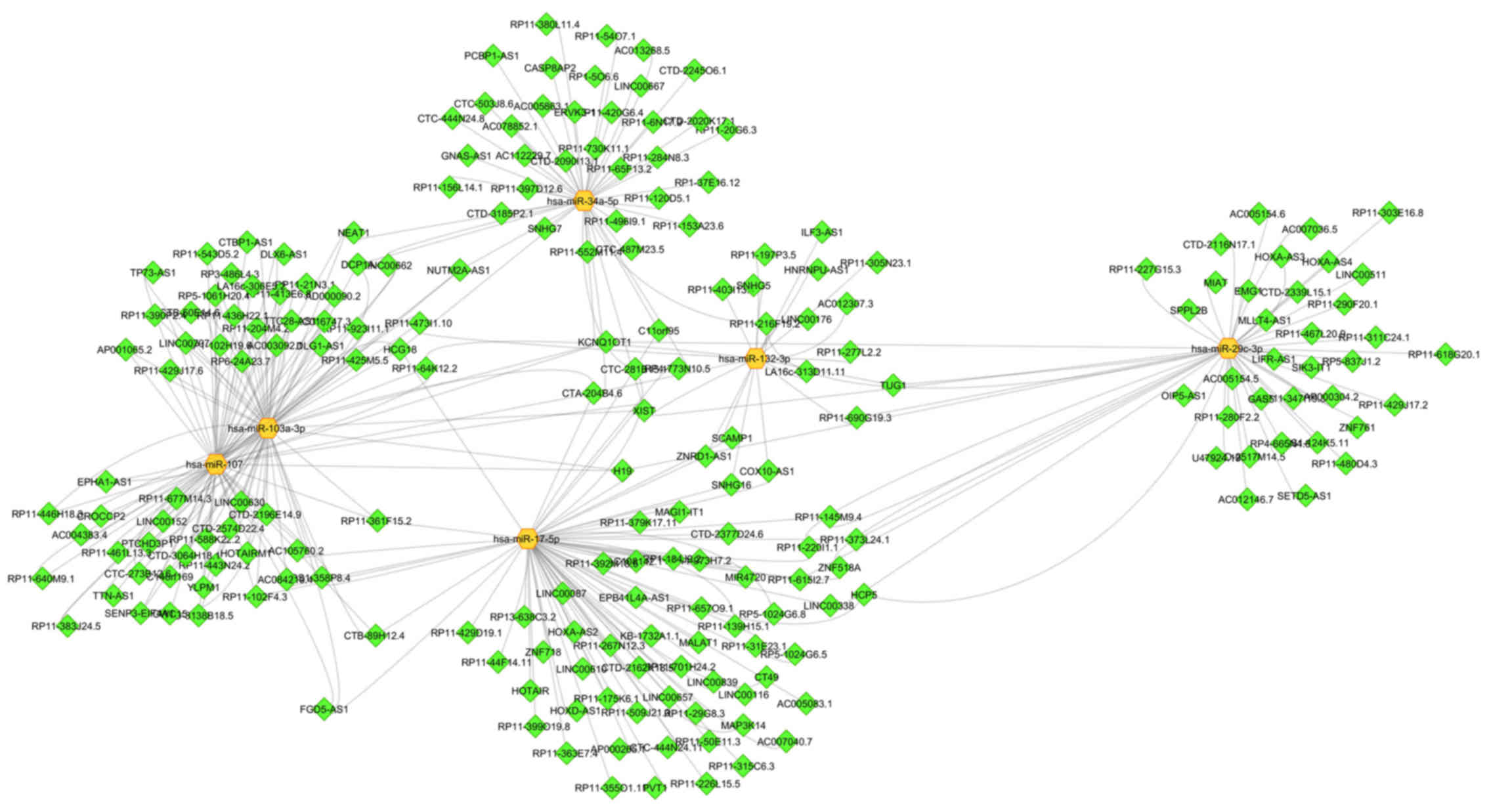
miRNAs putatively interplays with 210 unique lncRNAs (Fig. 5). The miRNA that connects with the
largest number of lncRNAs is miR-17-5p (72 lncRNAs). The
lncRNA-XIST interacts with all sub-selected 6 miRNAs. Moreover,
miR-107 and miR-103a-3p share the largest number of lncRNAs
(Fig. 5). Besides the miRNA-lncRNA
interplay, some relations between lncRNA and genes associated with
metabolic diseases were also presented in Fig. 2. Lnc-RNA-XIST interacts with
AKT2 and IRS1. In the same way, lncRNA-HCG18 also
intercommunicates with AKT2, and lncRNA-MALAT1 interacts
with PPARG (Fig. 2)
Moreover, we also searched subcellular location of
the 6 miRNAs and the 6 lncRNAs associated with metabolic disorders.
The RNAlocate database contains a manually curated RNA-associated
subcellular localization entries with experimental evidence. In
contrast, the iLoc-lncRNA and lncLocator are sequence-based
predictors of subcellular locations. Based on the predicted score
of iLoc-lncRNA, the majority of miRNAs associated with metabolic
disorders is located on exosomes, and the lncRNAs on nucleolus,
nucleus, or nucleoplasm. Similar results were found for lncLocator
for lncRNA location. However, according to RNALocate database, we
noted that the location of ncRNAs depends on the tissue, cell or
condition they are expressed (Table
SV).
Interactions between miRNAs and small
molecules
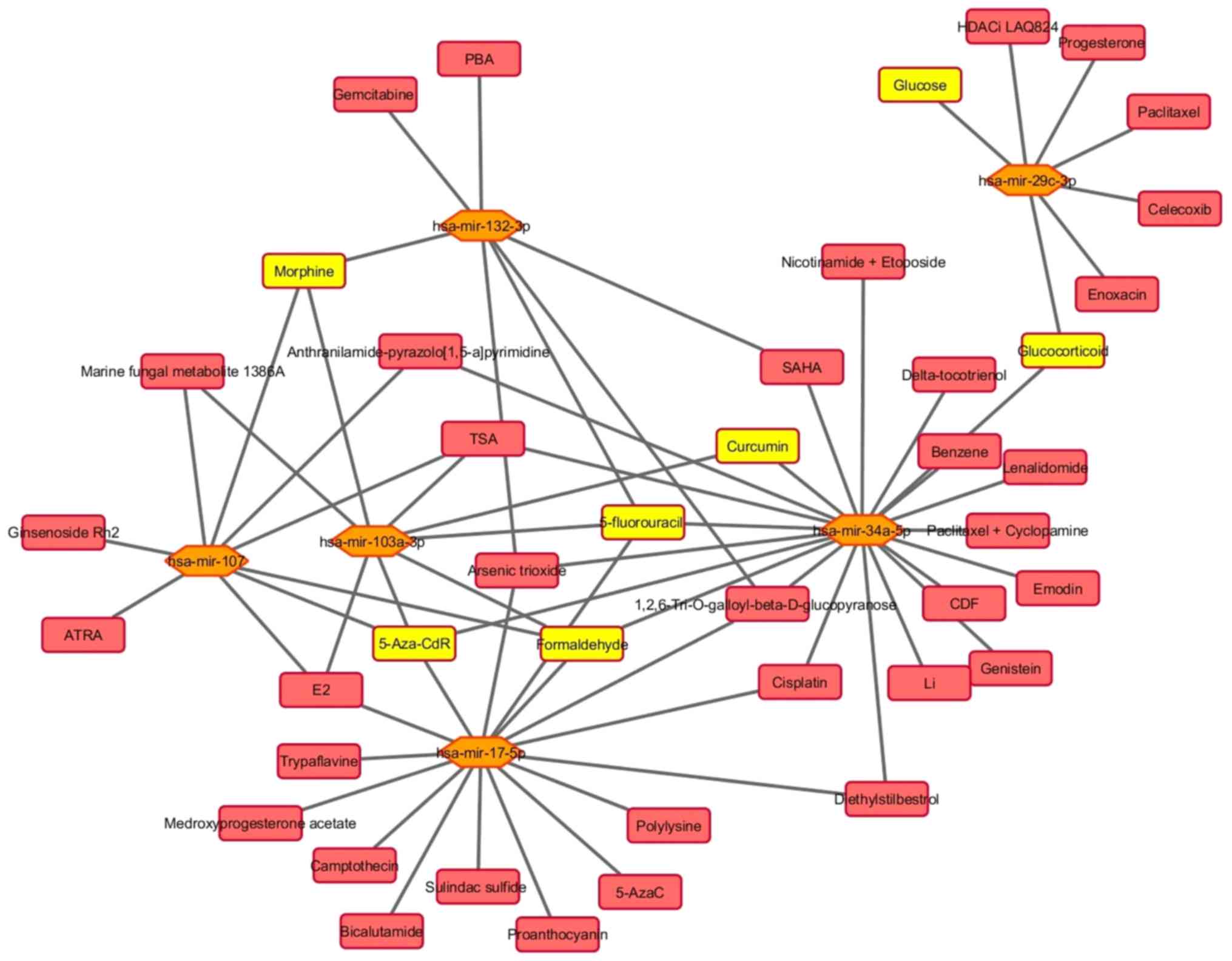
The 20 miRNAs interplay with 102 unique small
molecules (Table SVI). The miRNA
that intercommunicates with the highest number of small molecules
is miR-21-5p (70), and the miRNA
that interacts with the lowest number of molecules is miR-33a-5p
(4). Fig. 6 shows the connections between the 6
selected miRNAs and 42 small molecules. These miRNAs are linked
with different types of molecules, including metabolites, proteins,
chemicals and drugs.
Discussion
In the present study, miRNAs associated with
obesity, T2DM, and NAFLD were identified through a valid text
mining search strategy. For this, several bioinformatics analyses
were conducted to explore the miRNA-mRNA, miRNA-lncRNA, and
miRNA-small molecules interactions involved in the pathogenesis of
metabolic diseases. As main result, we propose an interaction of 6
miRNAs with 13 candidate genes to metabolic diseases, with 6
lncRNAs, and with 7 small molecules. Moreover, the functional
enrichment analysis of miRNAs target genes reflected the complex
biological behavior of metabolic diseases, being associated with
multiple signaling pathways. In general, miRNAs are related to
several human diseases (50). It
is a well-known phenomenon that miRNAs show cooperativity in gene
regulation, i.e. one miRNA binds with many target genes and one
target gene is regulated by many miRNAs [reviewed at (50)].
Among these 6 miRNAs, miR-17-5p demonstrated the
highest degree of connectivity in the present study. Several
reports have linked miR-17-5p expression levels with metabolic
diseases (51–53). Thus, Klöting et al (52) reported a significantly lower
expression of miR-17-5p in the omental adipose tissue of T2DM
patients compared to normal glucose tolerance (NGT) subjects and a
negative correlation with visceral fat area. Also, Heneghan et
al (54) showed a decrease in
miR-17-5p expression in human omental adipose tissue and blood from
obese patients. Additionally, the expression of miR-17-5p was
upregulated in plasma of T2DM with NAFLD compared to those without
NAFLD (51). Contrarily, miR-17-5p
expression was found to be increased in skeletal muscle of T2DM
rats, along with marked downregulation of GLUT4 protein level, and
the miR-17 knockdown ameliorated glucose metabolism, accompanied by
elevation of GLUT4 protein level (53). Moreover, miR-17-5p was reported to
be involved in the adipogenesis process in human adipose-derived
mesenchymal stem cell (55). The
miR-17-5p mimic transfection resulted in enhanced adipogenesis via
repression of bone morphogenetic protein 2 (BMP2) and increased
CCAAT/enhancer-binding protein α and peroxisome
proliferator-activated receptor γ expression (55).
miR-103a-3p and miR-107 belong to the same cluster
of miRNAs that also contains miR-15a/b, miR-16, miR-195, miR-497,
miR-503, miR-424, and miR-646 (56). miR-103a-3p and miR-107 have been
shown to be upregulated in the liver of T2DM patients (16), acting in the insulin signaling
pathway by primarily targeting caveolin-1, which is located in
lipid rafts and affects insulin receptor viability (57). Hence, the silence these two miRNAs
in mice improved glucose homeostasis and insulin sensitivity
(57). miR-107 is a
lipid-modulated miRNA involved in modifications of the circadian
system (58); interestingly, it
has been reported that gut microbiota may be involved in the
regulation of intestinal miR-107 levels (59). Moreover, some of the effects of
miR-103a-3p and miR-107 might be mediated through other miRNAs
since they strongly inhibit the miRNA-processing enzyme Dicer
(60).
The miR-29 family is among the most abundantly
expressed miRNA in the pancreas and liver in mice and humans
(61). Moreover, the miR-29 family
has been reported as a critical regulator of cholesterol turnover,
fatty acid synthesis, and glucose handling (61,62).
The knockdown of miR-29 family members (miR-29a, b and c) in a
murine model led to a significant reduction of cholesterol and
triglyceride plasma levels, reduced fatty acid content in the
liver, and increased gene and protein expression levels of Ahr,
Foxo3, and Sirt1 (62). Moreover,
miR-29c-3p expression was increased in skeletal muscle from T2DM
patients and decreased in healthy young men following exercise
training. In addition to reduced IRS1 protein abundance, miR-29c-3p
also decreased insulin signaling downstream of PI3K at the level of
Akt and GSK3 phosphorylation in human skeletal muscle cells
(63).
The main functions described for miR-34a include
cell cycle arrest, apoptosis, and senescence promotion (64). Furthermore, a meta-analysis of
profiling studies found that miR-34a was upregulated in T2DM
patients (16). Also, the
expression of this miRNA in subcutaneous fat tissue significantly
correlated with BMI (kg/m2) values (52). Similar results were found in ob/ob
mice with NAFLD compared to their corresponding controls (65). Moreover, the exposure to
perfluorononanoic acid (an organic pollutant with toxicological
impact on the liver) induced hepatic miR-34a expression in mice
(66,67).
miR-132 expression was upregulated in both blood and
liver of T2DM patients (16).
miR-132 targets insulin-mediated regulation of CYP2E1 (cytochrome
P450, family 2, subfamily E, polypeptide 1), which is involved in
the metabolism of xenobiotics in the liver (68). In omental fat, the expression
levels of miR-132-3p were decreased in T2DM patients compared to
NGT subjects and the number of macrophages infiltrating the fat
depot was significantly associated with miR-132 expression
(52).
Besides indicating a group of miRNAs associated with
metabolic diseases, the present study also provides new insights
into the complex molecular mechanisms involved in metabolic
diseases by revealing some pathways that may be regulated by the
selected miRNAs. These miRNAs potentially control genes from
several important processes, including cancer, apoptosis,
transcriptional regulation of white adipocyte differentiation,
regulation of gene expression in β cells, AMPK, and adipocytokine
signaling pathways.
Additionally, these six miRNAs have target genes
previously associated with metabolic diseases, indicating that the
differential expression of this set of miRNAs could lead to
metabolic diseases via dysregulation of metabolism-associated
genes, including PPARG, LDLR, SIRT1 and NEUROD1. In
this sense, PPARG gene regulates fatty acid storage and
glucose metabolism and has been implicated in the pathophysiology
of several diseases, including obesity, T2DM, atherosclerosis, and
cancer [reviewed at (69)]. The
LDLR gene mediates the endocytosis of LDL-cholesterol,
contributing to maintain the LDL plasma levels (70). SIRT1 gene is downregulated
in cells that have high insulin resistance and the overexpression
of SIRT1 increases insulin sensitivity (71,72).
In the same way, NEUROD1 gene is related to increased
expression of glucokinase, suggesting that this gene may play
important roles in the regulation of insulin synthesis and
secretion (73). Moreover,
NeuroD1 is required for normal development and maintenance
of pancreatic endocrine cells and the nervous system (74).
An increasing number of publications demonstrate
that miRNAs interact with lncRNAs, thereby triggering decay of
lncRNA or repressing its function (18,19).
Thus, it was reasonable to investigate the pathogenesis and
treatment of metabolic diseases by studying the specific
miRNA-lncRNA co-regulation effect. In the present study,
lncRNA-XIST was found to interact with all 6 miRNAs and some other
genes associated with metabolic diseases, suggesting that this
lncRNA may have a physiopathological role in these diseases. In
this context, lncRNA-XIST was increased in patients with T2DM
compared to controls, and its expression positivity correlated with
HbA1c, HOMA-IR, and fasting insulin levels (25). Based on online biology websites, we
found that miR-17-5p may be targeted by lncRNA-XIST, and a study
carried out on a linage of cancer cells (NSCLC) suggested that
lncRNA-XIST may regulate autophagy via the miR-17/ATG7 signaling
pathway (75).
It is known that the subcellular location of ncRNAs,
especially lncRNAs, correlated with functionality, which could
influence disease susceptibility; however, the location of ncRNAs
is still controversial and little is known regarding metabolic
diseases (76). LncRNA transcripts
can be found in many different sites within the cell, including the
chromatin, nucleus, cytoplasm, and exosomes (19,76).
LncRNA subcellular location is likely dependent on several factors,
such as sequence and structural motifs which can facilitate binding
to proteins involved in location (77).
Small molecules can regulate multiple biological
processes and have been proposed and used for therapeutic purpose
in different human diseases (78).
Recently, several drug-like compound libraries were screened
successfully against different miRNAs in cellular assays
demonstrating the possibility to target miRNAs with small molecules
(79). The present article
evidenced that some molecules can modulate miRNA expression, and
this could be a way to indirectly regulate gene expression.
Some strengths and limitations of our study should
be considered. As strengths, a comprehensive search of multiple
databases was conducted. Additionally, we performed robust
bioinformatic analyses to investigate the pathways in which these
miRNAs are participating, explaining the association with metabolic
diseases. Even though these methods are already powerful, this
evaluation had some limitations. First, the results are based on
the available literature about this topic. Second, we could not
exclude the possibility that other miRNAs should be associated with
the metabolic disorders; moreover, the results found in the online
databases change over time. Third, the lack of standardization of
the official nomenclature of miRNAs, without the description of
which miRNA straight was analyzed (−3p or −5p). Fourth, this is an
association study and because of that we could not describe the
events order. These limitations should be considered when
interpreting the results. Although limitations exist in the current
data, the patterns uncovered here are important for understanding
the association of miRNAs and metabolic diseases, and for
identifying new miRNAs, pathways and target genes putatively
involved in disease onset and progression.
Taken together, the present analyses demonstrate
that the molecular mechanisms of metabolic diseases can be
understood, and that biomarker prediction can be achieved through
data mining and integration analysis. Overall, 20 candidate miRNAs
were screened by bioinformatics analysis, and 6 of them (miR-17-5p,
miR-29c-3p, miR-34a-5p, miR-103a-3p, miR-107 and miR-132-3p)
presented the strongest association with metabolic diseases. The
construction of miRNA-mRNA, miRNA-lncRNA and miRNA-small molecules
networks provides a novel approach to the study of the metabolic
disease pathogenesis and establishes solid knowledge for the
personalized treatment of these disorders in the future. However,
more studies are needed to validate these results.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
TSA is a recipient of scholarships from Coordenação
de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; grant no.
88881.170123/2018-01). The present study was also supported by
CIBERobn (grant no. CB12/03/30002).
Availability of data and materials
The datasets analyzed during the current study are
available in the HMDD v3.0 (www.cuilab.cn/hmdd), miR2Disease (www.miR2Disease.org), DisGeNET v5.0 (www.disgenet.org/), MDHGI (chengroup.cumt.edu.cn/tool/mdhgi/), and miRNet
(www.mirnet.ca/) databases, which were accessed in
December 2018.
Authors' contributions
TSA designed the study, analyzed and interpreted the
data, and drafted the manuscript. FIM interpreted the data and
critically reviewed the manuscript. JAM designed and supervised the
study, interpreted the data and critically revised the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Collaborators GRF: Global, regional, and
national comparative risk assessment of 79 behavioural,
environmental and occupational, and metabolic risks or clusters of
risks, 1990–2015: A systematic analysis for the global burden of
sisease study 2015. Lancet. 388:1659–1724. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martínez JA, Milagro FI, Claycombe KJ and
Schalinske KL: Epigenetics in adipose tissue, obesity, weight loss,
and diabetes. Adv Nutr. 5:71–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nilsson EE, Sadler-Riggleman I and Skinner
MK: Environmentally induced epigenetic transgenerational
inheritance of disease. Environ Epigenet. 4:dvy0162018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vienberg S, Geiger J, Madsen S and
Dalgaard LT: MicroRNAs in metabolism. Acta Physiol (Oxf).
219:346–361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O'Brien KP, Ramphul E, Howard L, Gallagher
WM, Malone C, Kerin MJ and Dwyer RM: Circulating MicroRNAs in
cancer. Methods Mol Biol. 1509:123–139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rotllan N, Price N, Pati P, Goedeke L and
Fernández-Hernando C: microRNAs in lipoprotein metabolism and
cardiometabolic disorders. Atherosclerosis. 246:352–360. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esau C, Davis S, Murray SF, Yu XX, Pandey
SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al: miR-122
regulation of lipid metabolism revealed by in vivo antisense
targeting. Cell Metab. 3:87–98. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Janssen HL, Reesink HW, Lawitz EJ, Zeuzem
S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A,
Zhou Y, et al: Treatment of HCV infection by targeting microRNA. N
Engl J Med. 368:1685–1694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie H, Lim B and Lodish HF: MicroRNAs
induced during adipogenesis that accelerate fat cell development
are downregulated in obesity. Diabetes. 58:1050–1057. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Poy MN, Hausser J, Trajkovski M, Braun M,
Collins S, Rorsman P, Zavolan M and Stoffel M: miR-375 maintains
normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci
USA. 106:5813–5818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
El Ouaamari A, Baroukh N, Martens GA,
Lebrun P, Pipeleers D and van Obberghen E: miR-375 targets
3′-phosphoinositide-dependent protein kinase-1 and regulates
glucose-induced biological responses in pancreatic beta-cells.
Diabetes. 57:2708–2717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zampetaki A, Kiechl S, Drozdov I, Willeit
P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E,
et al: Plasma microRNA profiling reveals loss of endothelial
miR-126 and other microRNAs in type 2 diabetes. Circ Res.
107:810–817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu H and Leung SW: Identification of
microRNA biomarkers in type 2 diabetes: A meta-analysis of
controlled profiling studies. Diabetologia. 58:900–911. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA-LncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamamura S, Imai-Sumida M, Tanaka Y and
Dahiya R: Interaction and cross-talk between non-coding RNAs. Cell
Mol Life Sci. 75:467–484. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moran VA, Perera RJ and Khalil AM:
Emerging functional and mechanistic paradigms of mammalian long
non-coding RNAs. Nucleic Acids Res. 40:6391–6400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Franco-Zorrilla JM, Valli A, Todesco M,
Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA and
Paz-Ares J: Target mimicry provides a new mechanism for regulation
of microRNA activity. Nat Genet. 39:1033–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang K, Sun T, Li N, Wang Y, Wang JX, Zhou
LY, Long B, Liu CY, Liu F and Li PF: MDRL lncRNA regulates the
processing of miR-484 primary transcript by targeting miR-361. PLoS
Genet. 10:e10044672014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Legnini I, Morlando M, Mangiavacchi A,
Fatica A and Bozzoni I: A feedforward regulatory loop between HuR
and the long noncoding RNA linc-MD1 controls early phases of
myogenesis. Mol Cell. 53:506–514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chiyomaru T, Yamamura S, Fukuhara S,
Yoshino H, Kinoshita T, Majid S, Saini S, Chang I, Tanaka Y,
Enokida H, et al: Genistein inhibits prostate cancer cell growth by
targeting miR-34a and oncogenic HOTAIR. PLoS One. 8:e703722013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sathishkumar C, Prabu P, Mohan V and
Balasubramanyam M: Linking a role of lncRNAs (long non-coding RNAs)
with insulin resistance, accelerated senescence, and inflammation
in patients with type 2 diabetes. Hum Genomics. 12:412018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji E, Kim C, Kim W and Lee EK: Role of
long non-coding RNAs in metabolic control. Biochim Biophys Acta
Gene Regul Mech. (pii): S1874-9399(18)30460-7. 2018.
|
|
27
|
Deiters A: Small molecule modifiers of the
microRNA and RNA interference pathway. AAPS J. 12:51–60. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gumireddy K, Young DD, Xiong X, Hogenesch
JB, Huang Q and Deiters A: Small-molecule inhibitors of microrna
miR-21 function. Angew Chem Int Ed Engl. 47:7482–7484. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li
J, Zhou Y and Cui Q: HMDD v3.0: A database for experimentally
supported human microRNA-disease associations. Nucleic Acids Res.
47:D1013–D1017. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, Li M, Wang G and Liu Y: miR2Disease: A manually curated
database for microRNA deregulation in human disease. Nucleic Acids
Res. 37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen X, Yin J, Qu J and Huang L: MDHGI:
Matrix decomposition and heterogeneous graph inference for
miRNA-disease association prediction. PLoS Comput Biol.
14:e10064182018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Piñero J, Bravo À, Queralt-Rosinach N,
Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz
F and Furlong LI: DisGeNET: A comprehensive platform integrating
information on human disease-associated genes and variants. Nucleic
Acids Res. 45:D833–D839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kutmon M, Kelder T, Mandaviya P, Evelo CT
and Coort SL: CyTargetLinker: A cytoscape app to integrate
regulatory interactions in network analysis. PLoS One.
8:e821602013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maere S, Heymans K and Kuiper M: BiNGO: A
cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2004. View Article : Google Scholar
|
|
37
|
Bindea G, Galon J and Mlecnik B: CluePedia
cytoscape plugin: Pathway insights using integrated experimental
and in silico data. Bioinformatics. 29:661–663. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu X, Wang S, Meng F, Wang J, Zhang Y,
Dai E, Yu X, Li X and Jiang W: SM2miR: A database of the
experimentally validated small molecules' effects on microRNA
expression. Bioinformatics. 29:409–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rukov JL, Wilentzik R, Jaffe I, Vinther J
and Shomron N: Pharmaco-miR: Linking microRNAs and drug effects.
Brief Bioinform. 15:648–659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fan Y, Siklenka K, Arora SK, Ribeiro P,
Kimmins S and Xia J: miRNet-dissecting miRNA-target interactions
and functional associations through network-based visual analysis.
Nucleic Acids Res. 44:W135–W141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang T, Tan P, Wang L, Jin N, Li Y, Zhang
L, Yang H, Hu Z, Zhang L, Hu C, et al: RNALocate: A resource for
RNA subcellular localizations. Nucleic Acids Res. 45:D135–D138.
2017.PubMed/NCBI
|
|
43
|
Su ZD, Huang Y, Zhang ZY, Zhao YW, Wang D,
Chen W, Chou KC and Lin H: iLoc-lncRNA: Predict the subcellular
location of lncRNAs by incorporating octamer composition into
general PseKNC. Bioinformatics. 34:4196–4204. 2018.PubMed/NCBI
|
|
44
|
Cao Z, Pan X, Yang Y, Huang Y and Shen HB:
The lncLocator: A subcellular localization predictor for long
non-coding RNAs based on a stacked ensemble classifier.
Bioinformatics. 34:2185–2194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Heberle H, Meirelles GV, da Silva FR,
Telles GP and Minghim R: InteractiVenn: A web-based tool for the
analysis of sets through Venn diagrams. BMC Bioinformatics.
16:1692015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Volders PJ, Anckaert J, Verheggen K,
Nuytens J, Martens L, Mestdagh P and Vandesompele J: LNCipedia 5:
Towards a reference set of human long non-coding RNAs. Nucleic
Acids Res. 47:D135–D139. 2018. View Article : Google Scholar :
|
|
48
|
Peng C, Ye Y, Wang Z, Guan L, Bao S, Li B
and Li W: Meta-analysis of circulating microRNAs for the diagnosis
of hepatocellular carcinoma. Dig Liver Dis. 51:621–631. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shi C, Huang F, Gu X, Zhang M, Wen J, Wang
X, You L, Cui X, Ji C and Guo X: Adipogenic miRNA and
meta-signature miRNAs involved in human adipocyte differentiation
and obesity. Oncotarget. 7:40830–40845. 2016.PubMed/NCBI
|
|
50
|
Paul P, Chakraborty A, Sarkar D, Langthasa
M, Rahman M, Bari M, Singha RS, Malakar AK and Chakraborty S:
Interplay between miRNAs and human diseases. J Cell Physiol.
233:2007–2018. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ye D, Zhang T, Lou G, Xu W, Dong F, Chen G
and Li Y: Plasma miR-17, miR-20a, miR-20b and miR-122 as potential
biomarkers for diagnosis of NAFLD in type 2 diabetes mellitus
patients. Life Sci. 208:201–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Klöting N, Berthold S, Kovacs P, Schön MR,
Fasshauer M, Ruschke K, Stumvoll M and Blüher M: MicroRNA
expression in human omental and subcutaneous adipose tissue. PLoS
One. 4:e46992009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xiao D, Zhou T, Fu Y, Wang R, Zhang H, Li
M, Lin Y, Li Z, Xu C, Yang B, et al: MicroRNA-17 impairs glucose
metabolism in insulin-resistant skeletal muscle via repressing
glucose transporter 4 expression. Eur J Pharmacol. 838:170–176.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Heneghan HM, Miller N, McAnena OJ, O'Brien
T and Kerin MJ: Differential miRNA expression in omental adipose
tissue and in the circulation of obese patients identifies novel
metabolic biomarkers. J Clin Endocrinol Metab. 96:E846–E850. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li H, Li T, Wang S, Wei J, Fan J, Li J,
Han Q, Liao L, Shao C and Zhao RC: miR-17-5p and miR-106a are
involved in the balance between osteogenic and adipogenic
differentiation of adipose-derived mesenchymal stem cells. Stem
Cell Res. 10:313–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Finnerty JR, Wang WX, Hébert SS, Wilfred
BR, Mao G and Nelson PT: The miR-15/107 group of microRNA genes:
Evolutionary biology, cellular functions, and roles in human
diseases. J Mol Biol. 402:491–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Trajkovski M, Hausser J, Soutschek J, Bhat
B, Akin A, Zavolan M, Heim MH and Stoffel M: MicroRNAs 103 and 107
regulate insulin sensitivity. Nature. 474:649–653. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Daimiel-Ruiz L, Klett-Mingo M,
Konstantinidou V, Micó V, Aranda JF, García B, Martínez-Botas J,
Dávalos A, Fernández-Hernando C and Ordovás JM: Dietary lipids
modulate the expression of miR-107, a miRNA that regulates the
circadian system. Mol Nutr Food Res. 59:1865–1878. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xue X, Cao AT, Cao X, Yao S, Carlsen ED,
Soong L, Liu CG, Liu X, Liu Z, Duck LW, et al: Downregulation of
microRNA-107 in intestinal CD11c(+) myeloid cells in response to
microbiota and proinflammatory cytokines increases IL-23p19
expression. Eur J Immunol. 44:673–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Roggli E, Britan A, Gattesco S, Lin-Marq
N, Abderrahmani A, Meda P and Regazzi R: Involvement of microRNAs
in the cytotoxic effects exerted by proinflammatory cytokines on
pancreatic beta-cells. Diabetes. 59:978–986. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Dooley J, Garcia-Perez JE, Sreenivasan J,
Schlenner SM, Vangoitsenhoven R, Papadopoulou AS, Tian L,
Schonefeldt S, Serneels L, Deroose C, et al: The microRNA-29 family
dictates the balance between homeostatic and pathological glucose
handling in diabetes and obesity. Diabetes. 65:53–61. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kurtz CL, Fannin EE, Toth CL, Pearson DS,
Vickers KC and Sethupathy P: Inhibition of miR-29 has a significant
lipid-lowering benefit through suppression of lipogenic programs in
liver. Sci Rep. 5:129112015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Massart J, Sjögren RJO, Lundell LS, Mudry
JM, Franck N, O'Gorman DJ, Egan B, Zierath JR and Krook A: Altered
miR-29 expression in Type 2 diabetes influences glucose and lipid
metabolism in skeletal muscle. Diabetes. 66:1807–1818. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen F and Hu SJ: Effect of microRNA-34a
in cell cycle, differentiation, and apoptosis: A review. J Biochem
Mol Toxicol. 26:79–86. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li S, Chen X, Zhang H, Liang X, Xiang Y,
Yu C, Zen K, Li Y and Zhang CY: Differential expression of
microRNAs in mouse liver under aberrant energy metabolic status. J
Lipid Res. 50:1756–1765. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cui R, Li C, Wang J and Dai J: Induction
of hepatic miR-34a by perfluorooctanoic acid regulates
metabolism-related genes in mice. Environ Pollut. 244:270–278.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang J, Yan S, Zhang W, Zhang H and Dai J:
Integrated proteomic and miRNA transcriptional analysis reveals the
hepatotoxicity mechanism of PFNA exposure in mice. J Proteome Res.
14:330–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Shukla U, Tumma N, Gratsch T, Dombkowski A
and Novak RF: Insights into insulin-mediated regulation of CYP2E1:
miR-132/-212 targeting of CYP2E1 and role of phosphatidylinositol
3-kinase, Akt (protein kinase B), mammalian target of rapamycin
signaling in regulating miR-132/-212 and miR-122/-181a expression
in primary cultured rat hepatocytes. Drug Metab Dispos.
41:1769–1777. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yahaya TO and Salisu TF: A review of Type
2 diabetes mellitus predisposing genes. Curr Diabetes Rev. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Go GW and Mani A: Low-density lipoprotein
receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J
Biol Med. 85:19–28. 2012.PubMed/NCBI
|
|
71
|
Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X
and Zhai Q: SIRT1 improves insulin sensitivity under
insulin-resistant conditions by repressing PTP1B. Cell Metab.
6:307–319. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kauppinen A, Suuronen T, Ojala J,
Kaarniranta K and Salminen A: Antagonistic crosstalk between NF-κB
and SIRT1 in the regulation of inflammation and metabolic
disorders. Cell Signal. 25:1939–1948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Moates JM, Nanda S, Cissell MA, Tsai MJ
and Stein R: BETA2 activates transcription from the upstream
glucokinase gene promoter in islet beta-cells and gut endocrine
cells. Diabetes. 52:403–408. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo
FJ, Leiter AB and Tsai MJ: Diabetes, defective pancreatic
morphogenesis, and abnormal enteroendocrine differentiation in
BETA2/neuroD-deficient mice. Genes Dev. 11:2323–2334. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sun W, Zu Y, Fu X and Deng Y: Knockdown of
lncRNA-XIST enhances the chemosensitivity of NSCLC cells via
suppression of autophagy. Oncol Rep. 38:3347–3354. 2017.PubMed/NCBI
|
|
76
|
van Heesch S, van Iterson M, Jacobi J,
Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E and
Simonis M: Extensive localization of long noncoding RNAs to the
cytosol and mono- and polyribosomal complexes. Genome Biol.
15:R62014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Goff LA and Rinn JL: Linking RNA biology
to lncRNAs. Genome Res. 25:1456–1465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Rizvi NF and Smith GF: RNA as a small
molecule druggable target. Bioorg Med Chem Lett. 27:5083–5088.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Warner KD, Hajdin CE and Weeks KM:
Principles for targeting RNA with drug-like small molecules. Nat
Rev Drug Discov. 17:547–558. 2018. View Article : Google Scholar : PubMed/NCBI
|