Introduction
Osteoporosis is a systemic skeletal disease common
in older women, which is characterized by low bone mass and
micro-architectural deterioration of bone tissue, followed by
increasing bone fragility and susceptibility to fracture (1). It is estimated that >50% of women
aged 50 years and older will sustain an osteoporotic fracture
during their lifetime (2).
However, at present, it is difficult to achieve satisfactory
clinical efficacy for the treatment of osteoporosis due to the side
effects of drugs and patient compliance (1,3).
Promoting osteogenic differentiation is an important strategy to
enhance bone mineral density and slow the development of
osteoporosis (4). Bone marrow
mesenchymal stem cells (BMSCs) are mesodermal cells that can be
obtained from multiple sources, including adipose tissue,
periosteum and bone marrow (5).
Due to their unique capacity to differentiate into osteoblasts,
chondrocytes and adipocytes, BMSCs are widely used in osteoporosis
research worldwide (6,7). Psoralea corylifolia L., a
traditional Chinese herbal medicine, has a long history of clinical
efficacy against conditions such as fractures, bone defects and
osteoporosis (8). Psoralen, as the
main flavonoid active ingredient in Psoralea corylifolia L.,
was demonstrated to promote BMSCs to undergo osteogenic
differentiation through activation of the bone morphogenetic
protein (BMP) signalling pathway (9). However, the mechanisms underlying the
osteogenic differentiation effects of psoralen have not yet been
fully elucidated, to the best of the authors' knowledge, and the
epigenetic regulatory mechanisms required investigation.
In recent decades, it has been demonstrated that
short non-coding RNAs, microRNAs (miRNAs/miRs), are involved in
many biological processes, including cell survival, proliferation
and differentiation, while their aberrant expression can lead to
the development of diseases (10).
Notably, miRNAs are also the central regulators of BMSCs in
regulating osteogenic differentiation. miR-203 and miR-320
negatively regulated BMP-2-induced osteogenic differentiation by
suppressing homeobox protein DLX-5 (11). miR-195 inhibited the abnormal
activation of osteogenic differentiation in MC3T3-E1 cells by
targeting RAF proto-oncogene serine/threonine-protein kinase
(12). miR-495 inhibited new bone
regeneration by targeting high mobility group at-hook 2 (13). Therefore, the regulation of
osteogenic transcription factors by miRNAs was demonstrated to be
an effective strategy in regulating osteogenic differentiation.
Runt-related transcription factor 2 (Runx2) is a transcription
factor that is indispensable for skeletal development, and controls
bone formation by acting as a signalling hub and transcriptional
regulator to coordinate target gene expression (14,15).
However, the role of miRNAs in targeting Runx2 requires
clarification in the osteogenic differentiation of BMSCs.
In the present study, BMSCs treated with psoralen
were used to identify differentially expressed miRNAs and their
target genes via miRNA microarray and bioinformatics analysis.
Using overexpression or inhibition methods in vitro, the
underlying mechanisms of miRNAs in psoralen-induced BMSC osteogenic
differentiation were examined, providing a potential molecular
therapeutic strategy for osteoporosis.
Materials and methods
Isolation and culture of BMSCs
20 male Sprague-Dawley rats, 3 weeks old (weighing
140±20 g), obtained from the Experimental Animal Center of
Guangzhou University of Traditional Chinese Medicine (Guangzhou,
China), were sacrificed by decollation and were sterilised using
75% ethanol. The marrow from the femur and tibia was mixed with
complete medium (low-glucose DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.)) and was gradient centrifuged at 900 × g for 30 min at room
temperature with Percoll at a density of 1.073 g/ml. The cells were
cultivated with complete medium, with fresh media changes every 3
days, and were incubated at 37°C in 5% CO2. Primary rat
BMSCs were passaged 6–7 days later. As the BMSCs were obtained from
rats, there were many other cells in the primary, first-passage and
second-passage cells. The purity (>95%) and cell viability were
high in the third-passage BMSCs, and thus were used for subsequent
experiments. All the rats received humane care in accordance with
the guidelines set by the Care of Experimental Animals Committee of
Guangzhou University of Chinese Medicine. Additionally, the present
study was approved by the Ethics Committee of Guangzhou University
of Chinese Medicine. All rats were housed in a
specific-pathogen-free facility (22±2°C, relative humidity 60±10%),
under a 12/12 h light/dark cycle (lights on from 6:00 am), without
restriction to food and water, and fasted 12 h before
sacrifice.
Cell Counting Kit-8 (CCK-8) assay of
cell viability
Third-passage BMSCs were collected and seeded into
96-well plates at a density of 1×104 cell/ml and were
co-cultured with 20 µg/ml psoralen continuously (37°C), which was
purchased from The National Institute for the Control of
Pharmaceutical and Biological Products. Next, 10 µl CCK-8 solution
(Dojindo Molecular Technologies, Inc.) was added into each well
after cell culture for 12, 24, 36, 48 and 72 h. The cells were
incubated at 37°C for 2 h in the dark. The absorbance values at a
wavelength of 490 nm were detected using a microplate reader
(Bio-Rad Laboratories, Inc.). BMSCs that were not co-cultured with
psoralen were considered the control group. Each experiment was
repeated three times.
Alizarin red staining (AR-S) after
osteogenic differentiation
Third-passage BMSCs were seeded into 24-well plates
at a density of 5×104 cell/ml and then were divided into
three groups: Psoralen group, positive control group and blank
control group. BMSCs co-cultured with 20 µg/ml psoralen were
considered the psoralen group. The positive control group was
induced by α-Minimum Essential Medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 5×10−5 mol/l isobutyl
xanthine, 2×10−4 mol/l indomethacin, 1×10−5
mol/l dexamethasone and 10 mg/l insulin. BMSCs that were not
co-cultured with psoralen or osteogenic-inducing fluid were treated
as the blank control group. Medium containing psoralen or
osteogenic-inducing fluid was changed every 3 days. After 2 weeks,
the BMSCs were fixed with 95% ethanol 10 min at room temperature
and then were stained with 40 mM AR-S solution for 10 min at pH 4.2
(room temperature). Subsequently, the BMSCs were treated with 10%
cetylpyridinium chloride in 10 mM sodium phosphate for 15 min at
room temperature and then were washed with PBS for 15 min. Calcium
mineral deposition was observed using a light microscope at three
different views (magnifications, ×100 and 200) to compare the
degree of osteogenic differentiation between the different
groups.
miRNA microarray analysis
Third-passage BMSCs were seeded at a density of
1×105 cell/ml in a 6-well plate. After reaching 80–85% confluence,
the BMSCs treated with 20 µg/ml psoralen represented the psoralen
group, while the BMSCs that did not receive treatment with psoralen
represented the control group. After 72 h, the total RNA of the two
groups was extracted using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. miRNA microarray analysis was then performed by Guangzhou
RiboBio Co., Ltd. The differentially expressed miRNAs were verified
by reverse transcription-quantitative PCR (RT-qPCR).
RT-qPCR
RT-qPCR was performed using the Prime Script™ RT
reagent kit and SYBR Premix EX Taq II kit (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The thermocycling conditions for qPCR were as follows:
Pre-denatured at 95°C for 10 min, following by 40 cycles of 95°C
denaturation for 10 sec, annealing at 60°C for 20 sec and finally
extension 70°C for 15 sec. U6 was used as the internal reference
for miRNA, and GAPDH was used as the internal reference for
osteogenic-specific factors, and the relative expression levels
were calculated using the 2−ΔΔCq method (16). The primers were synthesized by
Sangon Biotech Co., Ltd. and were as follows: Runx2 (forward:
5′-TCTTAGAACAAATTCTGCCCTTT-3′; reverse:
5′-TGCTTTGGTCTTGAAATCACA-3′); Osterix (forward:
5′-AGAGATCTGAGCTGGGTAGAGG-3′; reverse: 5′-AAGAGAGCCTGGCAAGAGG-3′);
alkaline phosphatase (ALP; forward: 5′-CCAACTCTTTTGTGCCAGAGA-3′;
reverse: 5′-GGCTACATTGGTGTTGAGCTTTT-3′); GAPDH (forward:
5′-ATTTGGTCGTATTGGGCG-3′; reverse: 5′-TGGAAGATGGTGATGGGATT-3′);
miR-122: (forward: 5′-GCGAAAGCATTTGCCAAGAA-3′, reverse:
5′-CATCACAGACCTGTTATTGC-3′); miR-154 (forward:
5′-TAGGTTATCCGTGTTGCCTTCG-3′; reverse:
5′-AAGGCAACACGAUAACCUAUU-3′); miR-488 (forward:
5′-CGGGGCAGCUCAGUACAG-3′; reverse: 5′-CAGTGCGTGTCGTGGAGT-3′);
miR-205 (forward: 5′-CCTCCCTAAATCCTCCATCC-3′; reverse:
5′-TCTAGGAAGGACAGCCTCCA-3′); U6 (forward: 5′-CTCGCTTCGGCAGCACA-3′;
reverse: 5′-AACGCTTCACGAATTTGCGT-3′).
Bioinformatics analysis
TargetScan (http://www.targetscan.org), PicTar (http://pictar.mdc-berlin.de) and miRanda (http://www.microrna.org) were used to analyse
potential miR-488 binding sites on the Runx2 3′-untranslated region
(3′UTR). The consistency of the analyses and predictions using
these three websites suggests that they are reliable, and the
results were further verified by in vitro experiments.
RNA oligoribonucleotide synthesis and
transfection
The RNA oligoribonucleotides [miR-488 mimics
(5′-GGGTCTATTACCGTGAGAGTT), miR-488 inhibitor
(TTGAGAGTGCCATTATCTGGG-3′), mimics control
(5′-TACGTCCAAGGTCGGGCAGGAAGA-3′), inhibitor control
(5′-UCCUCCGAACGUGUCACGUTT-3′), pcDNA 3.1-Runx2 and pcDNA
3.1-control] used in the present study were synthesized by Shanghai
GenePharma Co., Ltd. Prior to transfection, BMSCs were isolated and
seeded (2×106 cells/l) into 6-well plates and grown
until they were 60–80% confluent. The cells were then transfected
with 100 nM miR-488 mimics, miR-488 inhibitor, mimics control,
inhibitor control, pcDNA 3.1-Runx2 or pcDNA 3.1-control for 6 h,
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The cells were then digested with 0.025% trypsin
for 24 h and collected for further analyses, including RTq-PCR,
western blotting, a luciferase reporter assay and
immunocytochemistry.
Luciferase reporter assay
Third-passage BMSCs were collected following 0.25%
trypsin digestion when the BMSCs reached 80% confluence. The cells
were transfected with 0.2 µg pLUC-Runx2 plasmid construct
(Guangzhou RiboBio Co., Ltd.) and miR-488 mimics or inhibitor for 6
h using Lipofectamine® 2000. The activity in each well
was measured using the Dual Luciferase Reporter Assay System
(Promega Corporation) to quantify the luminescent signal. Firefly
luciferase was selected as the internal reference. The relative
luciferase activity was calculated as the value of each group/the
value of miR-control group.
Rescue assays
The open reading frame (ORF) fragments of Runx2
without the 3′UTRs were amplified by PCR as aformentioned using Taq
DNA Polymerase (Beijing Solarbio Science & Technology Co.,
Ltd.), and then Runx2 recombinant vectors were constructed using
the pcDNA 3.1 plasmid (Shanghai GenePharma Co., Ltd.). The
thermocycling conditions were as follows: Pre-denatured at 94°C for
5 min, followed by 35 cycles at 94°C for 10 sec, 60°C for 45 sec
and 72°C for 1 min. Third-passage BMSCs were seeded at a density of
2×104 cell/cm2 into 6-well plates and then
were transfected with pcDNA 3.1-Runx2 or miR-488 mimics or
co-transfected with both constructs. Cells transfected without any
plasmid were treated as the control group. RT-qPCR, western
blotting, AR-S staining and immunocytochemistry were then used to
detect the degree of osteogenic differentiation.
Western blotting
The protein was extracted from BMSCs by
radioimmunoprecipitation assay lysis buffer (50 mM Tris-HCl (pH
7.4), 150 mM NaCl, 1% NP-40, 0.1% SDS). The supernatant was
obtained by centrifugation at 12,000 × g at 4°C for 20 min, and the
concentration of protein was determined by a bicinchoninic acid
assay (Pierce; Thermo Fisher Scientific, Inc.). Then 20 µg protein
was added to 10% SDS-PAGE for electrophoresis at a constant
electric current of 25 mA/gel to the bottom of separation gel. The
proteins were then transferred onto a PVDF membrane through
electrophoresis at normal temperature with constant voltage of 80 V
for 100 min, and the membrane was blocked with 5% non-fat milk in
PBS with Tween-20 solution at 4°C overnight. The blots were probed
with primary antibodies against Runx2 (Abcam, ab114133, 1:2,000),
Osterix (Abcam, ab209484, 1:2,000), ALP (Abcam, ab83259, 1:2,000)
and β-actin (Abcam, ab8227, 1:2,000) at room temperature for 2 h,
followed by incubation with relevant secondary antibodies (Abcam,
ab97051, ab150118, 1:2,000) labelled with horseradish peroxidase at
room temperature for 1 h and washing with TBS with 0.1% Tween-20.
The protein expression was detected using a SuperSignal West Femto
Maximum Sensitivity Substrate kit (Roche Applied Science). The
relative value of the target protein was calculated by comparing
with the corresponding internal reference. Densitometry was
conducted using ImageJ (v1.8.0; National Institutes of Health).
Immunocytochemistry
After transfection, the BMSCs were fixed with 4%
polyoxymethylene for 20 min at room temperature, permeabilized with
0.25% Triton X-100 (Sigma-Aldrich; Merck KGaA), and blocked with
0.1% bovine serum albumin (Roche Diagnostics) at room temperature
for 30 min. Subsequently, the BMSCs were incubated overnight at 4°C
with the following primary antibodies: Runx2 (Abcam, ab114133,
1:500), Osterix (Abcam, ab209484, 1:500), ALP (Abcam, ab83259,
1:500). After washing three times with PBS, the BMSCs were reacted
with the appropriate secondary antibody (Bioss, Beijing, PV-0024,
PV-0023, 1:100) for 30 min at 37°C. The cell nuclei were stained
with haematoxylin (Abcam) at room temperature for 1 min and were
visualized by light microscopy (magnification, ×200, Leica
Microsystems GmbH). The BMSCs were observed at three different
views to compare the degree of osteogenic differentiation between
the different groups.
Statistical analysis
Each experiment was repeated three times. SPSS 20.0
software (IBM Corp.) was used for the statistical analyses. The
data are presented as the mean ± standard deviation. Student's
t-test was used to compare the differences between two groups.
One-way ANOVA was used to compare the differences between three or
more groups, and Tukey's test was used as a post hoc test to
determine which groups were significantly different from each
other. P<0.05 was considered to indicate a statistically
significant difference. The plots were constructed using GraphPad
Prism 6.0 (GraphPad Software, Inc.).
Results
Psoralen promotes the viability and
osteogenic differentiation of BMSCs
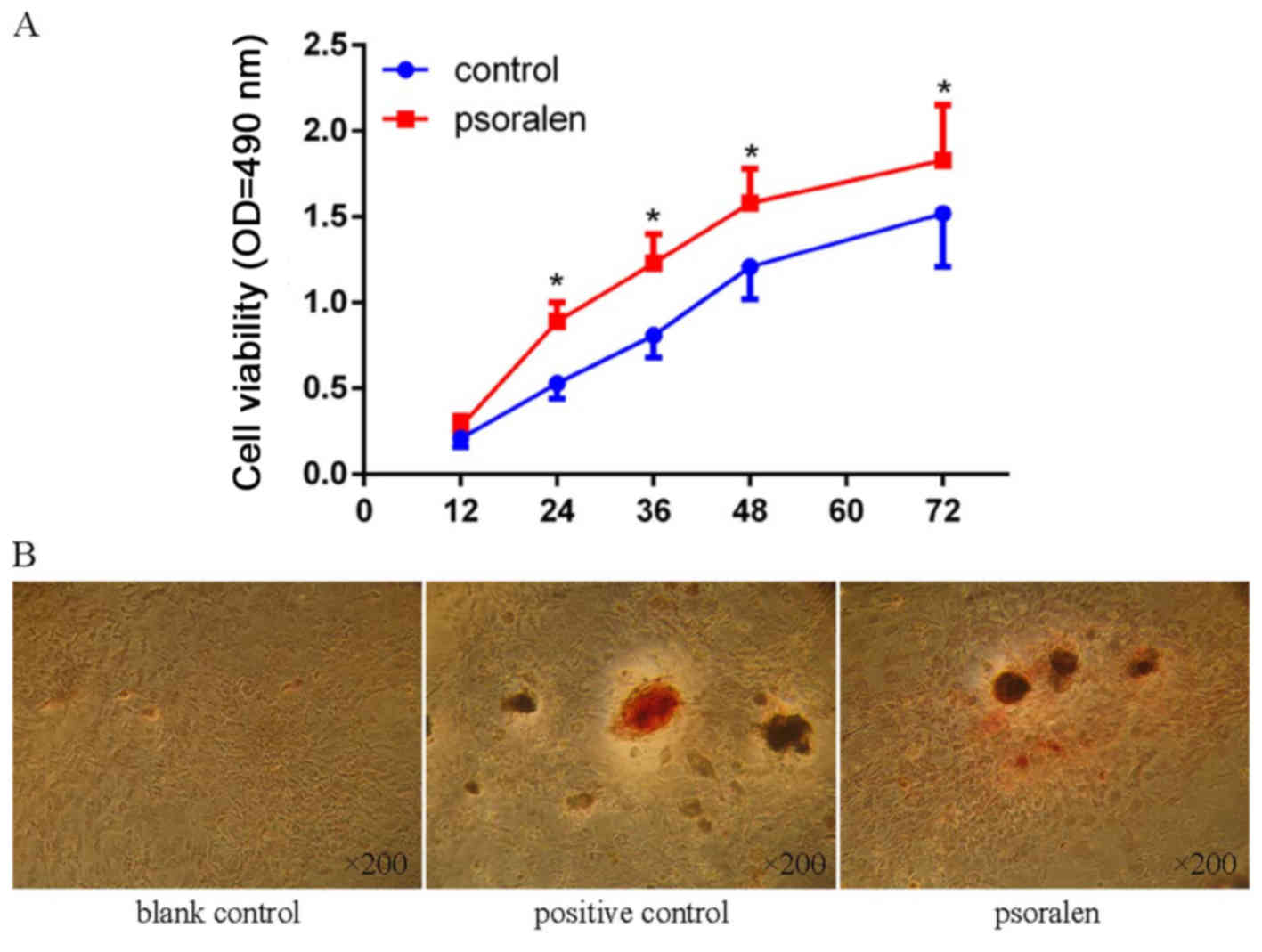
The CCK-8 assay showed that cell viability was
significantly elevated in BMSCs treated with psoralen at all time
points, with the exception of 12 h (Fig. 1A). The AR-S showed negligible
calcium mineral deposition in the blank control group, while
similar calcium mineral deposition was observed in the psoralen and
positive control groups (Fig.
1B).
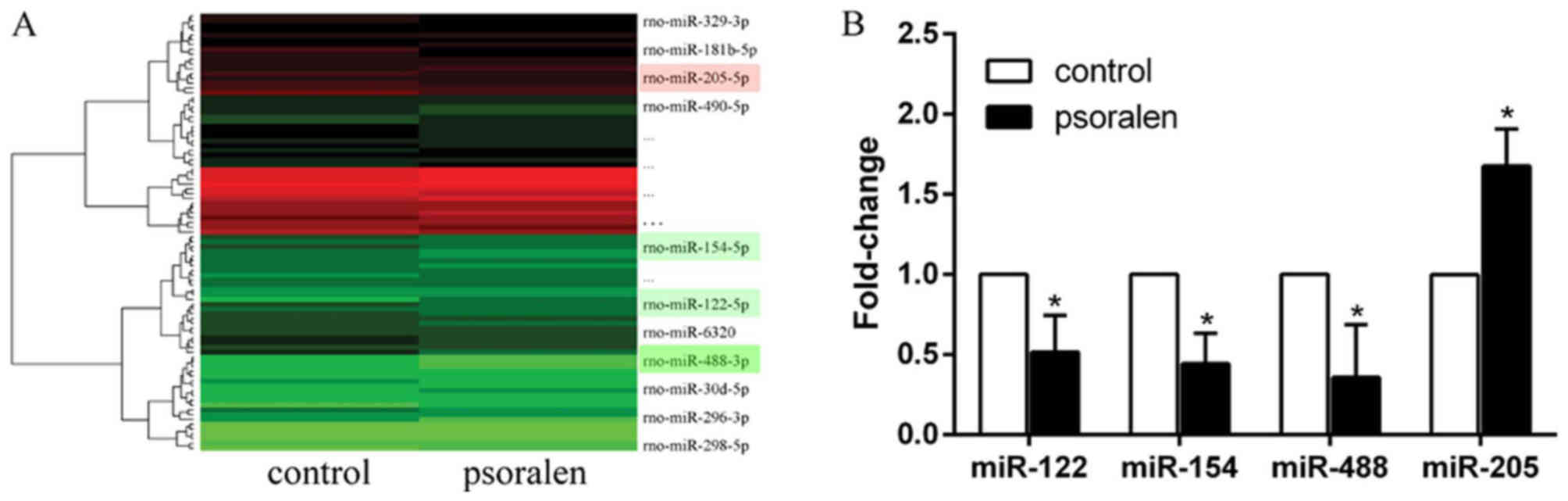
miR-488 is downregulated in BMSCs
during psoralen-induced osteogenic differentiation
To study the different expression levels of miRNAs
during psoralen-induced osteogenic differentiation, BMSCs were
treated with psoralen for 3 days and then the cells were subjected
to miRNA microarray analysis. In total, 91 miRNAs were
differentially expressed between the control and psoralen groups.
Compared with the control group, 55 miRNAs were upregulated and 36
were downregulated in the psoralen group. The data were analysed
and illustrated in a heat map (Fig.
2A). Among the miRNAs, miR-122, miR-154, miR-488 and miR-205
were randomly selected to confirm the results of the miRNA
microarray analysis via RT-qPCR. Compared with the control group,
miR-205 was upregulated, while miR-122, miR-154 and miR-488 were
downregulated, and miR-488 was the most downregulated (Fig. 2B), which was consistent with the
results of the miRNA microarray analysis.
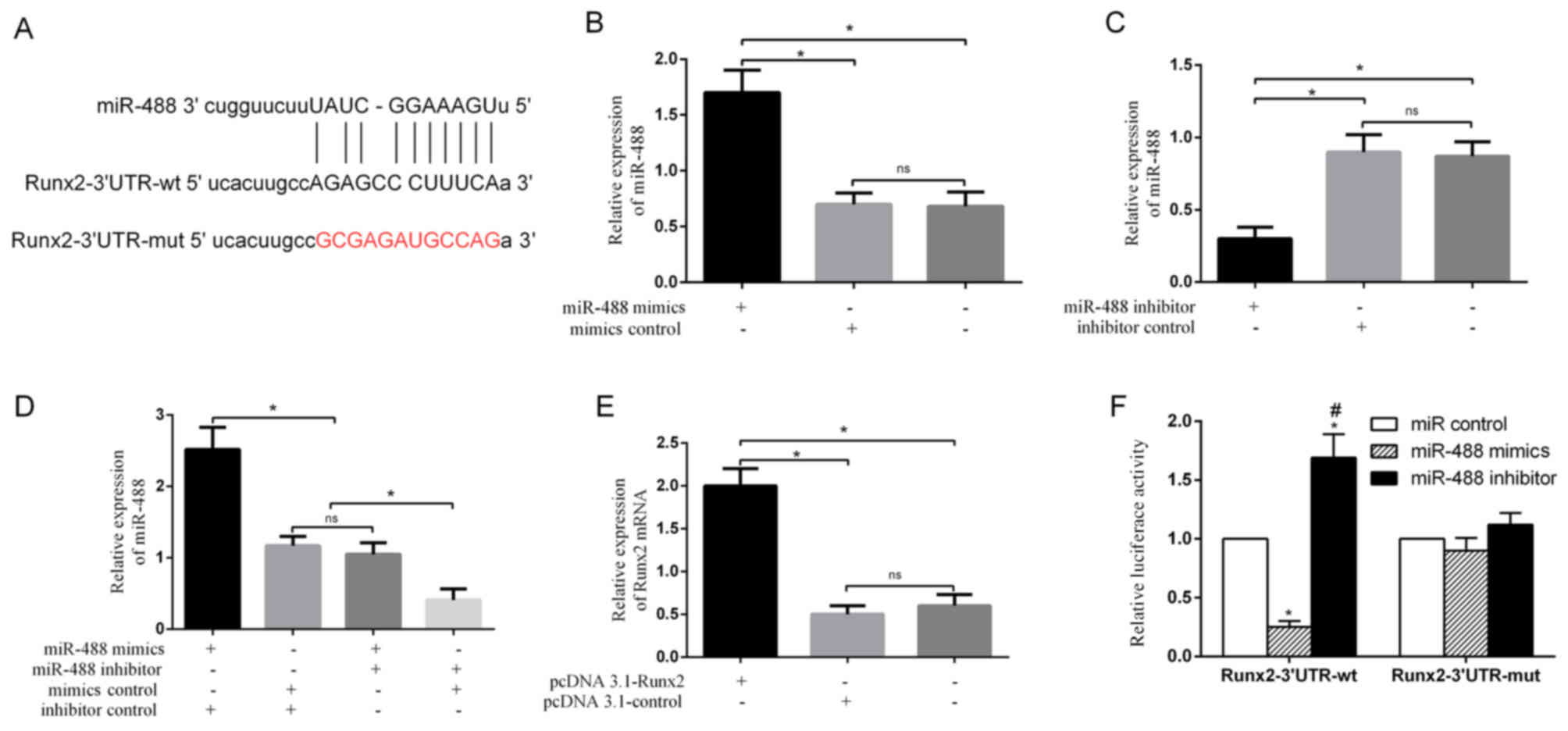
Runx2 is a potential target of
miR-488
To gain insight into the molecular mechanisms by
which miRNAs regulate the osteogenic differentiation of BMSCs, the
potential miRNAs were predicted using TargetScan and it was
identified that the osteogenic transcription factor Runx2 has
miR-488 binding sites in its 3′UTR (Fig. 3A). None of the other miRNAs
regulate the expression of Runx2, thus miR-488 was further
investigated. RT-qPCR showed that miR-488 was significantly
upregulated in BMSCs transfected with miR-488 mimics (Fig. 3B), and was downregulated in BMSCs
transfected with miR-488 inhibitor (Fig. 3C and D). Runx2 was significantly
upregulated in BMSCs transfected with pcDNA 3.1-Runx2 (Fig. 3E). These results indicated that
miR-488 mimics, miR-488 inhibitor and pcDNA 3.1-Runx2 were
successfully transfected. To further examine whether miR-488
directly targets Runx2, a luciferase reporter assay was performed
and it was demonstrated that the miR-488 mimics significantly
inhibited the luciferase activity of the wild-type Runx2 3′UTR but
not that of the mutated 3′UTR (Fig.
3F). Inversely, the miR-488 inhibitor significantly enhanced
the luciferase activity of the wild-type Runx2 3′UTR but not that
of the mutated 3′UTR (Fig.
3F).
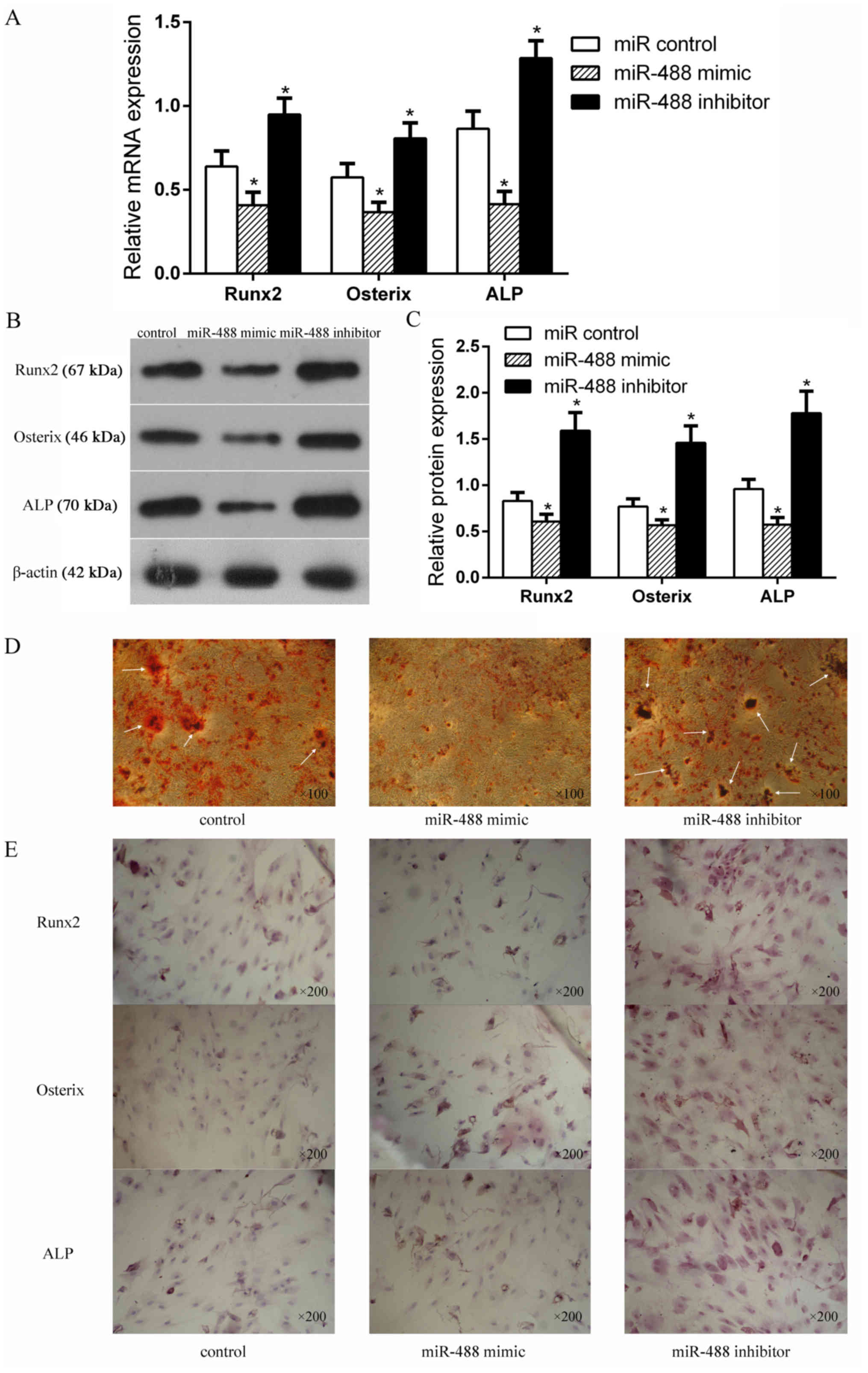
miR-488 negatively regulates the
osteogenic differentiation of BMSCs
To better understand the functional role of miR-488
in the osteogenic differentiation of BMSCs, the expressions of
Runx2, Osterix and ALP were detected after the BMSCs were
transfected with the miR-488 mimics or inhibitor. As shown in
Fig. 4A-C, the mRNA and protein
expression levels of Runx2, Osterix and ALP were significantly
decreased in the miR-488 mimics group but were significantly
increased in the miR-488 inhibitor group compared with the control
group. The AR-S result also showed that more calcium mineral
deposition was found in the miR-488 inhibitor group than in the
control and miR-488 mimic groups (Fig.
4D). Immunocytochemistry detected the expression levels of
Runx2, Osterix and ALP expressed in BMSCs, where the cytoplasm was
stained (Fig. 4E). The expression
levels of Runx2, Osterix and ALP in the miR-488 mimic group were
notably lower than those in the miR-488 inhibitor group, indicating
that miR-488 suppressed osteogenic differentiation of the
BMSCs.
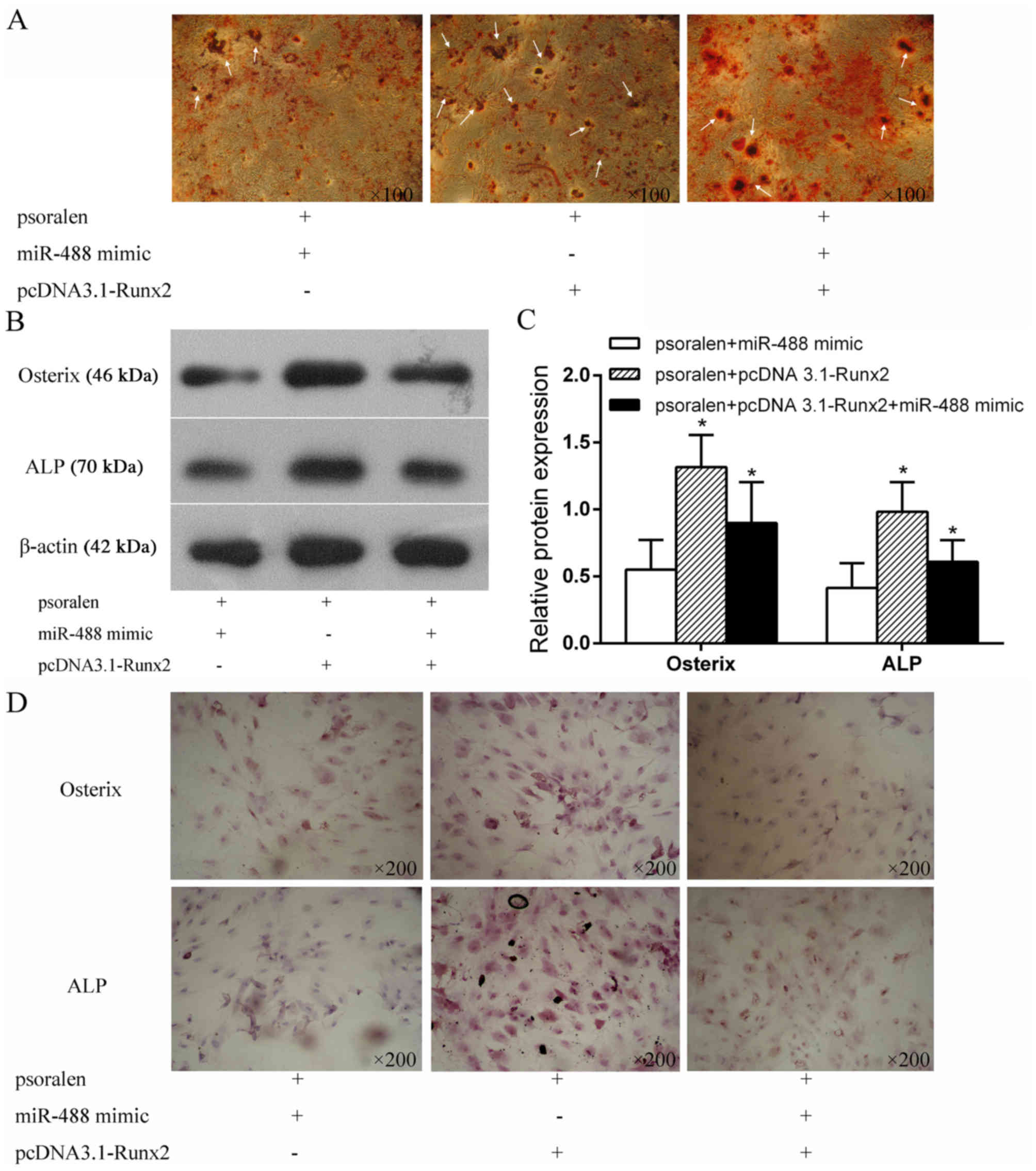
Runx2 overexpression partially rescues
the inhibitory effect of miR-488 on osteogenic differentiation
To further explore the underlying mechanism of
miR-488/Runx2 in the osteogenic differentiation of BMSCs, BMSCs
were transfected with miR-488 mimics and pcDNA 3.1-Runx2 under
treatment with psoralen. As shown in Fig. 5A, AR-S indicated that more calcium
mineral deposition was found in the psoralen+pcDNA
3.1-Runx2+miR-488 mimics group than in the psoralen+miR-488 mimics
group, while the calcium mineral deposition in the psoralen+pcDNA
3.1-Runx2+miR-488 mimics group was less than that in the
psoralen+pcDNA 3.1-Runx2 group. Runx2 partially rescued the
inhibitory effect of miR-488 during the psoralen-induced osteogenic
differentiation of BMSCs. Western blotting (Fig. 5B and C) and immunocytochemistry
(Fig. 5D) detected the protein
expression of osteogenic specific factors in BMSCs transfected with
miR-488 mimics and pcDNA 3.1-Runx2, and the results were similar to
the AR-S results.
Discussion
A previous study has shown that psoralen has a
smooth muscle diastolic effect, stimulating bone formation and
inducing osteogenic differentiation without affecting cell growth
(9), consistent with the present
findings. However, little is understood regarding the roles of
miRNAs in BMSCs induced by psoralen. In the present study, miRNA
microarray analysis was conducted to identify the differently
expressed miRNAs during psoralen-induced osteogenic differentiation
of BMSCs, indicating that miR-488 was significantly decreased.
However, the underlying mechanism of miR-488 in regulating BMSCs to
stimulate osteogenic differentiation under psoralen induction
remains largely unknown.
Each miRNA can bind to hundreds of different mRNAs
and assemble with Argonaute proteins into miRNA-induced silencing
complexes to direct post-transcriptional silencing of complementary
mRNA targets (17). For example,
miR-488 has been identified as a cancer-associated miRNA that
participates in various diseases, such as hepatocellular carcinoma
(18), ovarian cancer (19) and colorectal cancer (20). This is the first time, to the best
of the authors' knowledge, that miR-488 has been reported as a key
regulator in the osteogenic differentiation of BMSCs. An important
mechanism revealed by the present study is that Runx2 is directly
targeted by miR-488. Runx2 is a transcription factor that is
indispensable for skeletal development and controls bone formation
by acting as a signalling hub and transcriptional regulator to
coordinate target gene expression (13,14).
According to existing literature, miR-690, a Runx2-targeted miRNA,
regulated the osteogenic differentiation of C2C12 myogenic
progenitor cells by targeting NF-κB p65 (21). Heparin-binding EGF-like growth
factor and miR-1192 exert opposite effects on Runx2-induced
osteogenic differentiation (22).
However, to the best of the authors' knowledge, functional evidence
of miR-488 targeting Runx2 in the regulation of BMSCs to undergo
osteogenic differentiation has not been previously documented.
Therefore, in the present study, Runx2 was examined,
and the hypothesis that the osteogenic differentiation of BMSCs may
be regulated by miR-488 targeting Runx2 was evaluated. The
pLUC-Runx2 plasmid construct, which contained the miR-488 target
site in Runx2-3′UTR-wild-type, was constructed to be co-transfected
with the miR-488 mimics or inhibitor. The results showed that the
miR-488 mimics significantly reduced the luciferase activity of
pLUC-Runx2, while the miR-488 inhibitor significantly increased the
luciferase activity of pLUC-Runx2.
To further confirm the hypothesis, the mimics and
inhibitor of miR-488 were transfected into BMSCs, and the mRNA and
protein expression levels of Runx2 and a downstream signalling
mediator (Osterix) (14,23) and osteogenic-specific marker (ALP)
(24) were detected via RT-qPCR,
western blotting and immunocytochemistry. The Runx2, Osterix and
ALP mRNA and protein expression levels were reduced in the miR-488
mimics group and were enhanced in the miR-488 inhibitor group
compared with the control group. The AR-S result also showed that
more calcium mineral deposition was found in the miR-488 inhibitor
group than the control and miR-488 mimic groups. Further rescue
assays demonstrated that Runx2 overexpression partially rescued the
inhibitory effect of miR-488 on osteogenic differentiation under
psoralen induction. It should be noted that the present study only
observed calcium mineral deposition by AR-S and lacked precise
quantification. However, despite this limitation, the present study
does support the hypothesis that Runx2 is targeted by miR-488 and
could be directly responsible for the osteogenic differentiation of
BMSCs under psoralen induction.
The results of the present study have two important
clinical implications. Using the miRNA-target gene network to
examine the mechanism of psoralen promoting the osteogenic
differentiation of BMSCs, the present study provided insight for
the development and application of psoralen in osteoporosis
treatment. Additionally, the present study identified a potential
molecular approach to promote the osteogenic differentiation of
BMSCs, and miR-488/Runx2 could be therapeutic targets for
osteoporosis. miR-488/Runx2 constitutes a system that has not yet
been used in osteoporosis treatment. Inhibition of miR-488 or
overexpression of Runx2 appears to be an interesting novel
therapeutic option for osteoporosis.
The present study suggested that miR-488 is involved
in the osteogenic differentiation of BMSCs by targeting Runx2 under
psoralen induction. Altering miR-488/Runx2 may be a targeted and
mechanism-based therapeutic strategy against osteoporosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation (grant nos. 81473699 and 81804047),
Natural Science Foundation of Guangdong Province (grant nos.
2018A030313694 and 2017A030312009), the Honk Kong Scholar Program
(grant no. XJ2018059) and Guangdong Provincial Traditional Chinese
Medicine Research Project (grant nos. 20181095 and 20182043).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author on reasonable request.
Authors' contributions
YH, TJ and YL designed the research. YH, QH, TJ and
DC performed the experiments. YH, HS and DC analysed the data. YH
and QH wrote the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All animals received humane care in accordance with
the guidelines set by the Care of Experimental Animals Committee of
Guangzhou University of Chinese Medicine. Additionally, the present
study was approved by the Ethics Committee of Guangzhou University
of Chinese Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Collison J: Osteoporosis: Teriparatide
preferable for fracture prevention. Nat Rev Rheumatol. 14:42018.
View Article : Google Scholar
|
|
2
|
Chrischilles EA, Butler CD, Davis CS and
Wallace RB: A model of lifetime osteoporosis impact. Arch Intern
Med. 151:2026–2032. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pepe J, Cipriani C, Cecchetti V, et al:
Patients' reasons for adhering to long-term alendronate therapy.
Osteoporosis international. 2019 May 14;[Epub ahead of print] doi:
10.1007/s00198-019-05010-w. View Article : Google Scholar
|
|
4
|
Wang C, Wang J, Li J, Hu G, Shan S, Li Q
and Zhang X: KDM5A controls bone morphogenic protein 2-induced
osteogenic differentiation of bone mesenchymal stem cells during
osteoporosis. Cell Death Dis. 7:e23352016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Elango J, Robinson J, Zhang J, Bao B, Ma
N, de Val JEMS and Wu W: Collagen peptide upregulates
osteoblastogenesis from bone marrow mesenchymal stem cells through
MAPK-Runx2. Cells. 8:2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qi M, Zhang L, Ma Y, Shuai Y, Li L, Luo K,
Liu W and Jin Y: Autophagy maintains the function of bone marrow
mesenchymal stem cells to prevent estrogen deficiency-induced
osteoporosis. Theranostics. 7:4498–4516. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jing H, Liao L, An Y, Su X, Liu S, Shuai
Y, Zhang X and Jin Y: Suppression of EZH2 prevents the shift of
osteoporotic MSC fate to adipocyte and enhances bone formation
during osteoporosis. Mol Ther. 24:217–229. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
An J, Yang H, Zhang Q, Liu C, Zhao J,
Zhang L and Chen B: Natural products for treatment of osteoporosis:
The effects and mechanisms on promoting osteoblast-mediated bone
formation. Life Sci. 147:46–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang DZ, Yang F, Yang Z, Huang J, Shi Q,
Chen D and Wang YJ: Psoralen stimulates osteoblast differentiation
through activation of BMP signaling. Biochem Biophys Res Commun.
405:256–261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Czech MP: MicroRNAs as therapeutic
targets. N Engl J Med. 354:1194–1195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Laxman N, Mallmin H, Nilsson O and
Kindmark A: miR-203 and miR-320 regulate bone morphogenetic
protein-2-induced osteoblast differentiation by targeting
distal-less homeobox 5 (Dlx5). Genes (Basel). 8:2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chao C, Li F, Tan Z, Zhang W, Yang Y and
Luo C: miR-195 inhibited abnormal activation of osteoblast
differentiation in MC3T3-E1 cells via targeting RAF-1. Exp Cell
Res. 362:293–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tian Z, Zhou H, Xu Y and Bai J:
MicroRNA-495 inhibits new bone regeneration via targeting high
mobility group AT-Hook 2 (HMGA2). Med Sci Monit. 23:4689–4698.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Almalki SG and Agrawal DK: Key
transcription factors in the differentiation of mesenchymal stem
cells. Differentiation. 92:41–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stephens AS and Morrison NA: Novel target
genes of RUNX2 transcription factor and 1,25-dihydroxyvitamin D3. J
Cell Biochem. 115:1594–1608. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meister G: Argonaute proteins: Functional
insights and emerging roles. Nat Rev Genet. 14:447–459. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu D, Shen D, Zhang M, Jiang N, Sun F,
Yuan S and Wan K: MiR-488 suppresses cell proliferation and
invasion by targeting ADAM9 and lncRNA HULC in hepatocellular
carcinoma. Am J Cancer Res. 7:2070–2080. 2017.PubMed/NCBI
|
|
19
|
Yang Z, Feng Z, Gu J, Li X, Dong Q, Liu K,
Li Y and OuYang L: microRNA-488 inhibits chemoresistance of ovarian
cancer cells by targeting Six1 and mitochondrial function.
Oncotarget. 8:80981–80993. 2017.PubMed/NCBI
|
|
20
|
Lv Y, Shi Y, Han Q and Dai G: Histone
demethylase PHF8 accelerates the progression of colorectal cancer
and can be regulated by miR-488 in vitro. Mol Med Rep.
16:4437–4444. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu S, Geng Q, Pan Q, Liu Z, Ding S, Xiang
Q, Sun F, Wang C, Huang Y and Hong A: MiR-690, a Runx2-targeted
miRNA, regulates osteogenic differentiation of C2C12 myogenic
progenitor cells by targeting NF-kappaB p65. Cell Biosci. 6:102016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu S, Geng Q, Ma J, Sun F, Yu Y, Pan Q and
Hong A: Heparin-binding EGF-like growth factor and miR-1192 exert
opposite effect on Runx2-induced osteogenic differentiation. Cell
Death Dis. 4:e8682013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Z, Wang W, Xu H, Ning Y, Fang W, Liao
W, Zou J, Yang Y and Shao N: Effects of altered CXCL12/CXCR4 axis
on BMP2/Smad/Runx2/Osterix axis and osteogenic gene expressions
during osteogenic differentiation of MSCs. Am J Transl Res.
9:1680–1693. 2017.PubMed/NCBI
|
|
24
|
Kim HK, Cho SG, Kim JH, Doan TK, Hu QS,
Ulhaq R, Song EK and Yoon TR: Mevinolin enhances osteogenic genes
(ALP, type I collagen and osteocalcin), CD44, CD47 and CD51
expression during osteogenic differentiation. Life Sci. 84:290–295.
2009. View Article : Google Scholar : PubMed/NCBI
|