Introduction
Oral squamous cell carcinoma (OSCC) is one of the
most aggressive neoplasms among head and neck malignant tumors
(1). Smoking and alcohol
consumption are the main environmental factors affecting the
development of OSCC (2). Each year
~540,000 novel cases are diagnosed and mortality rates have not
been lowered significantly in recent years. Although several
advances have been obtained in its treatment, the prognosis of OSCC
remains poor, with a 5-year survival ratio of nearly 50% (3). Better understanding of the genetic
and molecular disorders of the disease is the key to early
diagnosis, appropriate treatment and improved prognosis of patients
with OSCC.
Circular RNAs (circRNAs) are another class of
non-coding RNAs that are widely expressed in mammals besides
microRNAs and long non-coding (lnc) RNAs (4,5).
They are formed by exon skipping or back-splicing events with
neither 5′ to 3′ polarity nor a polyadenylated tail. Nowadays,
circRNAs have been extensively identified among a variety of
transcriptomes. Previously, they were mostly misinterpreted as
splicing errors. Only in recent years, circRNAs were rediscovered
from RNA sequencing (RNA-seq) data and demonstrated to be
widespread and diverse in eukaryotic cells (6,7).
This discovery has permanently altered perspectives toward cancer,
especially in carcinogenesis and cancer progression (8,9).
However, the current understanding of the molecular characteristics
and functional role of circRNAs in carcinogenesis remains
limited.
CircPVT1, alternatively named hsa_circ_0001821, is
derived from exon 3 within the oncogene PVT1 locus, which is
located on chr8:128902834-128903244, a cancer susceptibility locus
(10). Previously, PVT1 has been
reported as an oncogene during tumor growth and metastasis
(11), however, the expression and
functional role of circPVT1 are not well known, including in OSCC.
Previously, Chen et al (12) demonstrated that circPVT1 is a
proliferative factor in gastric cancer by serving as a competing
endogenous (ce)RNA. More recently, circPVT1 was identified to be
upregulated in head and neck squamous carcinoma and mediated by the
mutant p53/YAP/TEAD transcription-competent complex (13). Therefore, the role of circPVT1 in
OSCC progression may be important.
In this study, it was hypothesized that circPVT1 is
upregulated in OSCC and promotes cell proliferation by acting as a
ceRNA. To test this hypothesis, circPVT1 levels were measured in
OSCC cell lines. In addition, the oncogenic role of circPVT1 was
also verified by performing several gain-or loss-functional
assays.
Materials and methods
Clinical tissue samples
The present study included 50 patients with OSCC who
underwent partial or total surgical resection at the Department of
Otorhinolaryngology Head and Neck Surgery of the China-Japan
Friendship Hospital between January 2015 and June 2017. The
patients were enrolled between the ages of 28 and 79 years old and
the median age at the time of diagnosis was 53 years. The male to
female ratio was 37:13. Tissues were obtained from the tongue (21
cases), gingiva (12 cases), bucca (9 cases) and floor of the mouth
(8 cases). Primary cancer tissues and adjacent non-tumor tissues
(>2 cm distal from cancer area to avoid the encroachment of
cancer cells) were collected. No patients received radiotherapy
prior to the surgical resection. The obtained tissue samples were
immediately snap-frozen in liquid nitrogen upon resection and then
stored at −80°C until use. The present study was approved by the
Research Scientific Ethics Committee of the China-Japan Friendship
Hospital. All participants signed informed consent prior to using
the tissues for scientific research.
Cell culture
A total of two OSCC cell lines (SCC-9 and CAL-27)
were purchased from the Chinese Academy of Sciences (Shanghai,
China) and normal oral epithelial keratinocytes (HOK) from
ScienCell Research Laboratories (San Diego, CA, USA). The source of
the OSCC cell lines was derived from tongue cancer and has been
authenticated in oral cancer research (14). CAL-27 and SCC-9 cells were cultured
in Dulbecco's modified Eagle's medium: Nutrient Mixture F-12
(DMEM/F12, Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal
bovine serum (FBS; Interpath Services Pty, Ltd.). HOK was cultured
in DMEM supplemented with 10% FBS and penicillin-streptomycin (100
unit/ml-100 µg/ml). The culture condition is 37°C with a 5%
CO2 containing air.
Vector construction and cell
transfection
The specific silencing RNAs against circPVT1 small
interfering (si)-circPVT1, si-signal transducer and activator of
transcription 3 (si-STAT3) or negative control and miRNA mimics
(miR-125b and negative control) were purchased from Sangon Biotech
Co., Ltd., (Shanghai, China). The full sequences of circPVT1 and
negative control sequences were amplified and synthesized in
plasmids between two frames. In total, 5×105 cells were
cultured in each well of a six-well plate and were transfected with
above-mentioned vectors using Lipofectamine 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The nucleic acids were transfected at a concentration of
100 nM. Cells were harvested following 48 h for validation and
subsequent functional analysis. The sequences of the
oligonucleotides are presented in Table I.
 | Table I.Information of the RT-qPCR primer
sequences and oligonucleotides sequences. |
Table I.
Information of the RT-qPCR primer
sequences and oligonucleotides sequences.
| RT-qPCR primer
name | Sequence
(5′-3′) |
|---|
| CircPVT1
Forward |
ATCGGTGCCTCAGCGTTCGG |
| CircPVT1
Reverse |
CTGTCCTCGCCGTCACACCG |
| GAPDH Forward |
GCACCGTCAAGGCTGAGAAC |
| GAPDH Reverse |
ATGGTGGTGAAGACGCCAGT |
| U6 Forward |
CTCGCTTCGGCAGCACA |
| U6 Reverse |
AACGCTTCACGAATTTGCGT |
| U1 Forward |
GGGAGATACCATGATCACGAA |
| U1 Reverse |
CCACAAATTATGCAGTCGAGTT |
| Oligonucleotide
name |
|
| si-PVT1 (1) |
UGGGCUUGAGGCCUGAUCU |
| si-PVT1 (2) |
GCUUGAGGCCUGAUCUUUU |
| si-STAT3 |
GCAGCAGCTGAACAACATG |
| si-NC |
UUCUCCGAACGUGUCACGUTT |
| miR-125b |
UCCCUGAGACCCUAACUUGUGA |
| miR-NC |
CGAAUUAACGGGCCUAUCCGAU |
| Oligos for plasmid
construction |
|
|
circPVT1-exp-vector-F |
gcagaattcAGAGGCAGCTCTCACCTGAA |
|
circPVT1-exp-vector-R |
gcaggtaccTCAGAAAATGGTGACAGGTA |
|
LUC-UTRcricPVT1-F |
gcagaattcGCCTGATCTTTTGGCCAGAA |
|
LUC-UTRcricPVT1-R |
gcaggatcCAGCCTCACCTCAAGCCCAG |
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA extraction from cell fraction and tissue samples
was performed using a TRIzol regent (Invitrogen; Thermo Fisher
Scientific, Inc.). RT and qPCR kits were used to evaluate the
expression of target RNAs. The 20 µl RT reactions were performed
using a PrimeScript® RT reagent kit (Takara Bio, Inc.)
and incubated for 30 min at 37°C, 5 sec at 85°C. For assaying
circRNAs, mRNAs and miRNAs expression levels, the RT-qPCR analyses
were performed using SYBR Premix Ex Taq™ kit (Takara) and a
StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The values of 2−ΔΔCq relative to one
of the samples was calculated to analyze relative expression levels
of the genes (15). GAPDH and U6
were used to normalize the relative expression levels of circRNAs
and miRNAs, respectively. The relative expression level of
different genes was calculated using the ΔΔCq method. The primer
sequences were presented in Table
I.
Cell Counting kit-8 (CCK-8) assay
The proliferation of transfected cells was detected
by CCK-8 (Dojindo Molecular Technologies, Inc.). A total of
~2×103 OSCC cells were seeded in 96-well plates in
triplicate. CCK-8 solution (10 µl) was added into each well and
incubated at 37°C for 2.5 h. The optical density value at 450 nm
was measured using the SpectraMax M5 microplate reader (Molecular
Devices, LLC).
Cell cycle assay
A total of >1×104 transfected
cells were harvested and fixed with 75% ethanol at 4°C for 24 h.
The fixed cells were washed with cold PBS twice and mixed with
propidium iodide (PI; cat. no. 550825; BD Biosciences) for 20 min
at room temperature. Cell cycle analysis was performed by flow
cytometry analysis using a BD FACSCalibur (BD Biosciences) flow
cytometer and the data were analyzed using the ModFit LT software
(version 4.1; Verity Software House, Inc.).
Bioinformatics analysis
The putative binding sites of miR-125b in the
circPVT1 sequence were predicted using miRBase (http://www.mirbase.org/) and TargetScan (http://www.targetscan.org).
Nucleocytoplasmic separation
The PARIS™ kit (Ambion; Thermo Fisher Scientific,
Inc.) was used for the nucleic-cytoplasmic separation experiment.
Briefly, 5×106 cells were resuspended in 0.6 ml
resuspension buffer for 15 min's incubation at room temperature,
followed by homogenization. The pellet consisted the nuclear
fraction following a 1×105 × g centrifugation at 4°C for
10 min. CircPVT1 expression was determined by qPCR with GAPDH as
cytoplasmic control and U1 as nuclear control.
Fluorescence in situ hybridization
(FISH) analysis
Cy3-labeled circPVT1 probes were obtained from
Guangzhou RiboBio Co., Ltd. Hybridizations were carried out using a
FISH kit (Guangzhou RiboBio Co., Ltd.) according to the
manufacturer's protocol. All fluorescence images were captured
using Nikon A1Si Laser Scanning Confocal Microscope (Nikon
Corporation).
Immunofluorescence
Cell were permeabilized with 0.3% Triton X-100
(Beyotime Institute of Biotechnology) at room temperature for 15
min after being fixed with 4% paraformaldehyde for 30 min at room
temperature. Cells were blocked using 5% goat serum (Beyotime
Institute of Biotechnology) for 30 min at room temperature, and
then were incubated with an anti-Ki67 antibody (1:100; cat. no.
ab15580; Abcam) overnight at 4°C. The slides were then incubated
with an anti-rabbit Alexa Fluor 488-labeled secondary antibody
(1:500; cat. no. 016-540-084; Jackson ImmunoResearch Laboratories,
Inc.) for 1 h at room temperature. DAPI was used for nuclear
counterstaining at 4°C for 10 min. The samples were observed under
a fluorescence microscope (IX73; Olympus Corporation;
magnification, ×20).
RNA immunoprecipitation (RIP)
The RIP assay was performed using the Magna RIP™
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore).
Briefly, the cells were harvested and resuspended in the NP-40
lysis buffer (1 mM; Beyotime Institute of Biotechnology) for 5 min
at room temperature following 48 h of transfection. A total of 10
µl of the cell lysates were sampled for input and the remaining
lysates were incubated with anti-STAT3 antibody (1:100; cat. no.
ab119352; Abcam) or negative control immunoglobulin G (1:100; cat.
no. 12-370; EMD Millipore) at 4°C overnight. Next, the beads were
washed with wash buffer and the RNAs captured by beads were
extracted using the RNeasy Mini kit (Qiagen, Inc.). Finally,
RT-qPCR was performed to detect the enrichment and analyze. The
magnetic bead bound complexes were immobilized with a magnet and
unbound materials were washed off. Then, RNAs were extracted and
analyzed by qPCR.
Luciferase reporter assay
OSCC cells were seeded in 24-well plates at a
density of 5×103 cells per well and maintained for 24 h.
A total of 250 ng Firefly Luciferase (FL) reporter, 500 ng
Renilla luciferase reporter vectors and miR-125b mimics/NC
mimics were co-transfected into the cells, and the cells were
harvested in 48 h. The luciferase activity was measured with a dual
luciferase reporter assay detection kit (Promega Corporation) on an
Omega device (Omega Bio-Tek, Inc.). The Firefly luciferase activity
was normalized to Renilla luciferase activity.
Western blotting and antibodies
The total proteins were extracted using RIPA buffer
(Sigma-Aldrich; Merck KGaA), and their concentrations were detected
using Total Protein Extraction kit (Beijing Solarbio Science &
Technology Co., Ltd.). Cell lysates (25 µg protein in each well)
were separated by 10% SDS-PAGE and transferred to nitrocellulose
membranes (GE Healthcare). Then, 5% concentration non-fat milk in
TBST buffer (containing 0.1% Tween-20) was used for the blockage of
membranes containing protein for 2 h at room temperature. The
antibodies used included primary antibodies anti-STAT3 (1:1,000;
Abcam; cat. no. ab65222), anti-GAPDH (1:1,000; cat. no. 5174; Cell
Signaling Technology, Inc.), which were incubated with membranes at
4°C overnight; horseradish peroxidase-conjugated secondary goat
anti-mouse (1:5,000; cat. no. SA00001-1) or goat anti-rabbit
(1:5,000; cat. no. SA00001-2; both ProteinTech Group, Inc.)
antibodies (2 h at room temperature). The relative grey values of
immunoreactive bands were calculated based on GAPDH.
Statistical analysis
All experiments were performed in triplicate. Data
are presented as the mean ± standard error of the mean and analyzed
using GraphPad Prism 6.0 (GraphPad Software, Inc.). Parametric
pared t-test was performed to compare the difference between OSCC
tissues and paired normal tissues. A Chi-squared test was used to
analyze the correlation between clinicopathological features of
OSCC patients and circPVT1 expression. One-way ANOVA was performed
to evaluate difference among multiple groups. Fisher's Least
Significant Difference test was used as post-hoc test. Receiver
operation characteristic (ROC) analysis was performed using MedCalc
version 11.4 (MedCalc Software bvba) to evaluate the diagnostic
performance of circPVT1. P<0.05 was considered to indicate a
statistically significant difference.
Results
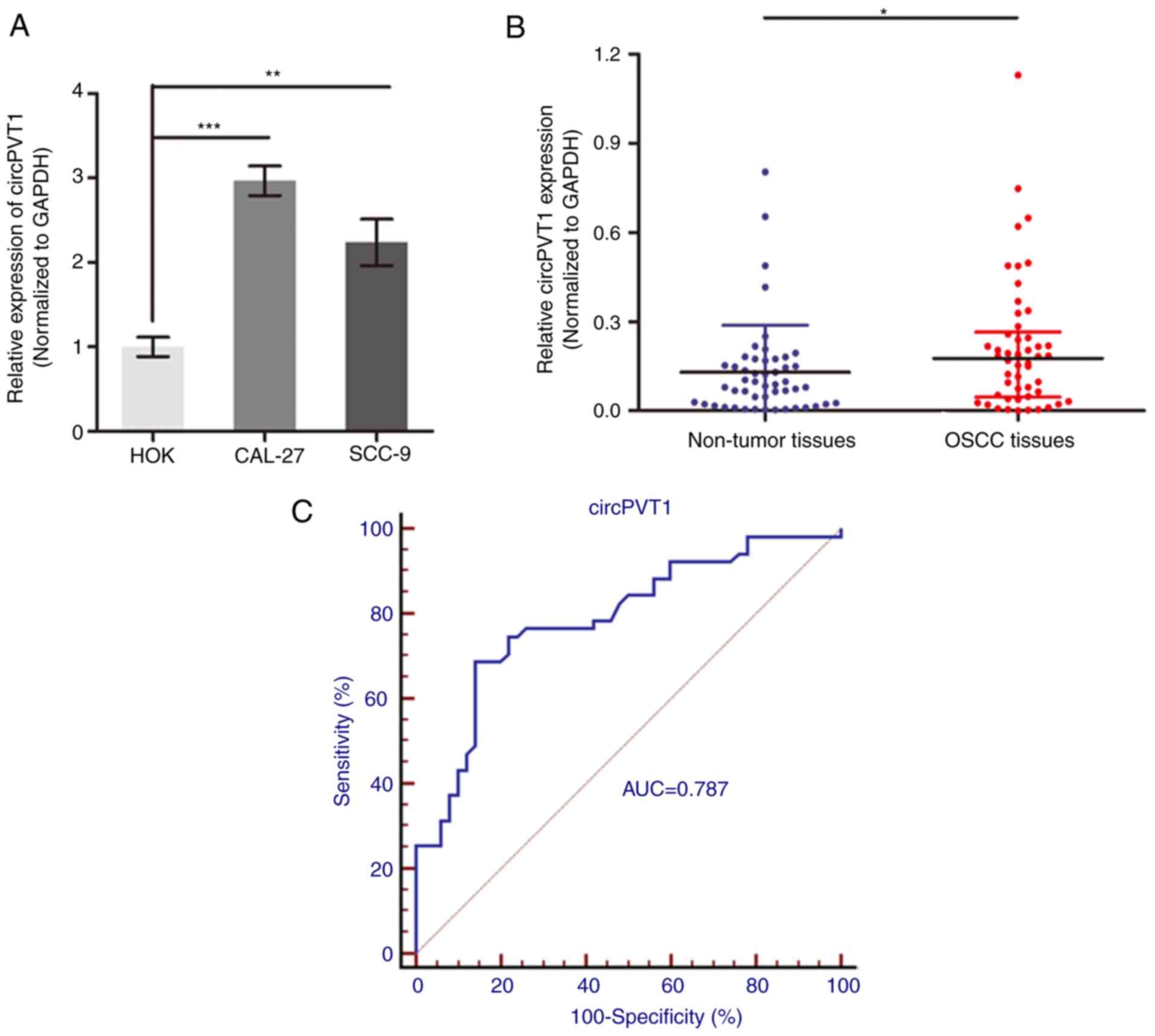
CircPTV1 is upregulated in OSCC
To determine the potential role of circPVT1 in OSCC,
its expression level was measured in OSCC by RT-qPCR. As presented
in Fig. 1A, circPVT1 was
significantly upregulated in two OSCC cell lines, CAL-27 and SCC-9,
when compared to the normal oral epithelial keratinocytes, HOK
cells (P<0.01). In addition, circPVT1 level was also
significantly increased in 50 OSCC tissues compared with
precancerous tissues (P<0.05; Fig.
1B). Following validation of circPVT1 upregulation in OSCC
patients, the diagnostic role of circPVT1 was investigated by
establishing ROC curves using the 50 paired OSCC tissues (Fig. 1C). The area under the curve of the
ROC curve was 0.787 and the detective sensitivity, and specificity
were 68.6 and 86.0% respectively. Analysis of association between
circPVT1 expression and clinical pathological factors of revealed
that circPVT1 expression was positively correlated with tumor size
and tumor node and metastasis, but not with other factors,
including sex, age and histological grade (Table II). This indicates a potential
prognostic significance of circPVT1 for OSCC patients.
 | Table II.Clinical characteristics of 50
patients and the expression of circPVT1 in primary oral squamous
cell carcinoma tissues. |
Table II.
Clinical characteristics of 50
patients and the expression of circPVT1 in primary oral squamous
cell carcinoma tissues.
|
Characteristics | Case | circPVT1 median
(range) | P-value |
|---|
| Age (years) |
|
| 0.776 |
|
<55 | 25 | 0.15
(0.02–0.67) |
|
|
≥55 | 25 | 0.17
(0.06–0.82) |
|
| Sex |
|
| 0.275 |
|
Male | 34 | 0.18
(0.02–0.82) |
|
|
Female | 16 | 0.11
(0.04–0.67) |
|
| Tumor size |
|
| 0.021 |
| <5
cm | 33 | 0.10
(0.04–0.74) |
|
| ≥5
cm | 17 | 0.28
(0.01–0.82) |
|
| Histological
grade |
|
| 0.106 |
|
Well | 11 | 0.13
(0.01–0.64) |
|
|
Moderate | 27 | 0.17
(0.05–0.61) |
|
|
Poor | 12 | 0.21
(0.08–0.82) |
|
| TNM stage |
|
| 0.002 |
|
I–II | 23 | 0.09
(0.01–0.56) |
|
|
III–IV | 27 | 0.26
(0.06–0.82) |
|
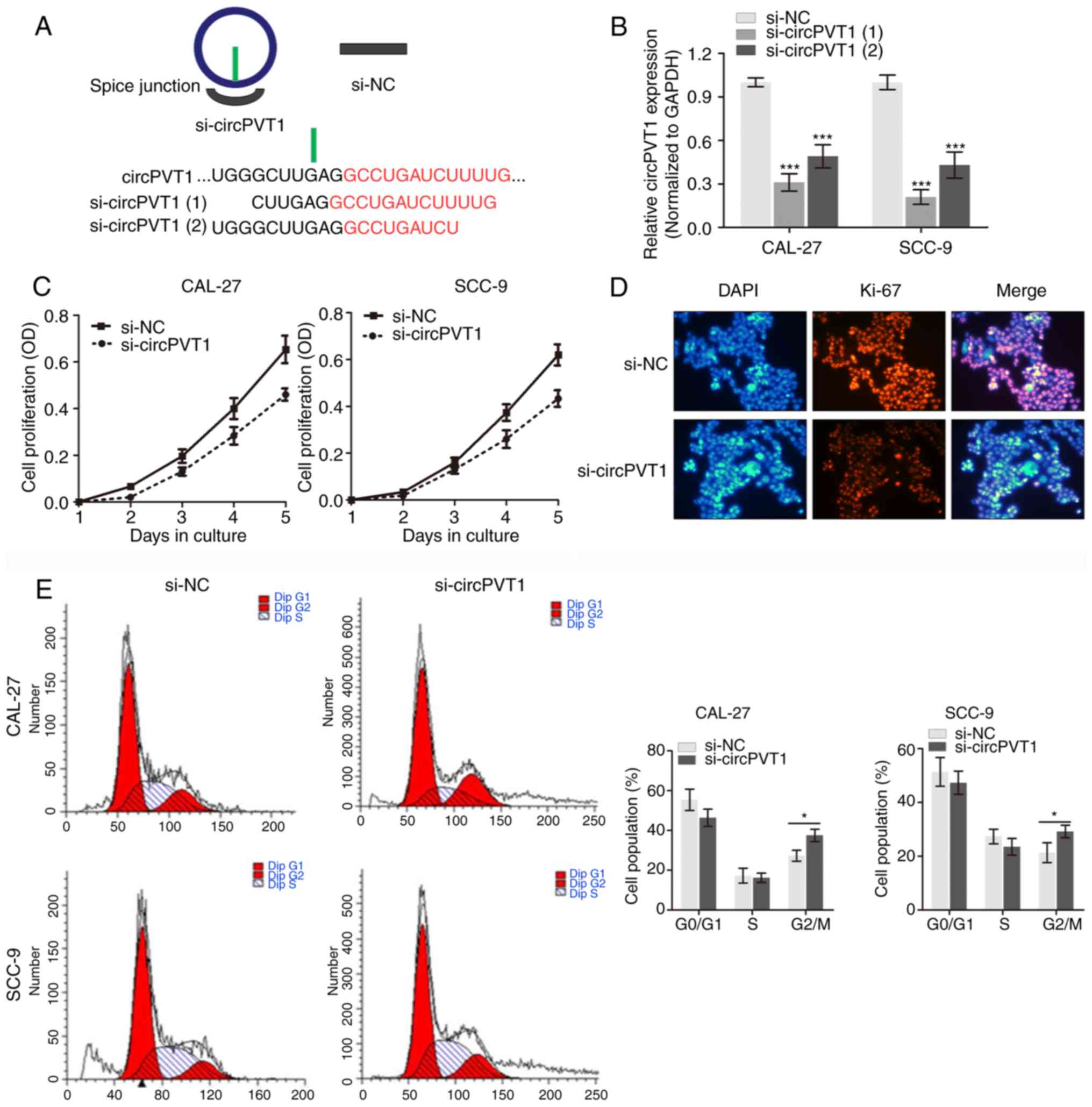
Knockdown of circPVT1 inhibits cell
proliferation in OSCC
For the upregulated expression levels of circPVT1 in
OSCC cells, the present study hypothesized that circPVT1 may serve
an oncogenic role in biological processes of OSCC. To verify the
hypothesis, specific siRNAs targeting circPVT1 were constructed
(Fig. 2A) to silence its
expression in cell lines CAL-27 and SCC-9 cells and perform several
experiments. As si-circPVT1 (1)
demonstrated a better silencing efficiency, it was selected for the
following experimental assays (Fig.
2B). Cell proliferation was determined by performing a CCK-8
assay and the results demonstrated that silencing of circPVT1
suppressed OSCC cell growth in contrast to the negative controls
(Fig. 2C). Immunofluorescence
analysis using Ki-67 antibody demonstrated that silencing circPVT1
decreased Ki-67 expression level in (Fig. 2D), which is a well-known
proliferative protein marker. Meanwhile, the cell cycle assay
indicated that knockdown of circPVT1 induced cell cycle arrest at
the G2/M phase in CAL-27 and SCC-9 cells compared with
controls. Therefore, the results of the present study indicate
circPVT1 promoted proliferation of OSCC cells.
CircPVT1 serves as a ceRNA via
sponging miR-125b in OSCC
To test whether circPVT1 functions as a ceRNA in
OSCC, the subcellular location of circPVT1 in OSCC cells was first
determined. A nucleocytoplasmic separation assay demonstrated that
circPVT1 may be distributed mainly in the cell cytoplasm (Fig. 3A). The FISH assay with specific
probe verified circPVT1 was located in the cytoplasm (Fig. 3B). Subsequently, whether circPVT1
influenced the biofunctions of OSCC cells was investigated through
sponging of miRNAs. As expected, miR-125b was verified as a
potential sponged miRNA with two targeting sites within circPVT1
according to miRBase and TargetScan (Fig. 3C). Using two OSCC cell lines, it
was identified that knockdown of circPVT1 significantly promoted
miR-125b expression (P<0.01; Fig.
3D). Then, a luciferase reporter assay was performed by
generating miR-125b mimics (Fig.
3E). As expected, the results demonstrated that miR-125b mimics
significantly silenced the luciferase activity of circPVT1
(Fig. 3F) but exhibited no effect
on mutant controls. Taken together, the sponging of miR-125b by
circPVT1 in OSCC cells was verified.
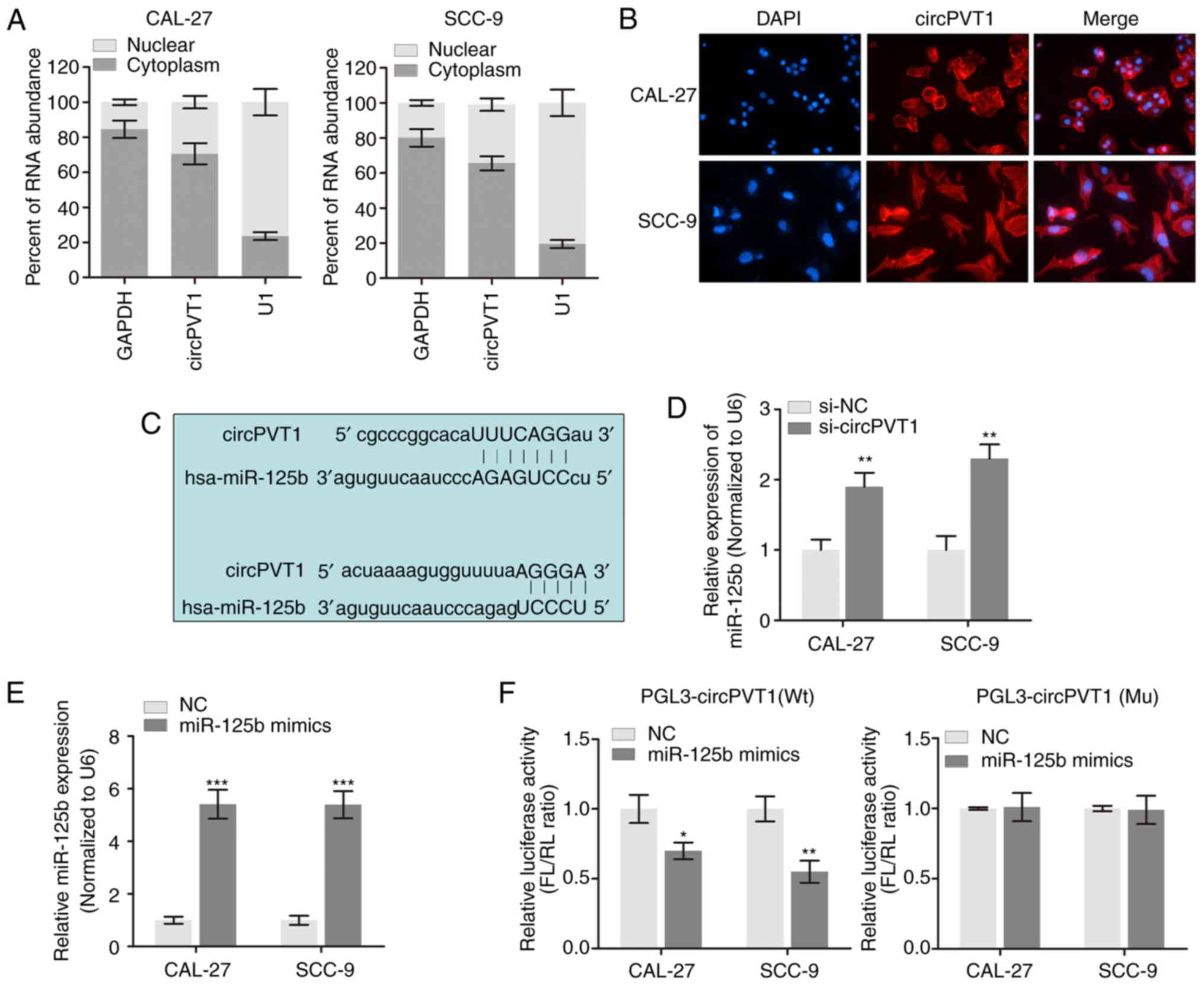 | Figure 3.circPVT1 serve as a competing
endogenous RNA of miR-125b. (A) The expression level of circPVT1 in
nuclear and cytoplasm of OSCC cells were verified via reverse
transcription-qPCR. (B) The expression density of circPVT1 in OSCC
cells was determined using fluorescence in situ hybridization
assay. Magnification, ×40. (C) The putative sequences of miR-125b
and circPVT1 with two binding sites. (D) miR-125b expression was
increased in OSCC cells following knockdown of circPVT1.
**P<0.01 vs. the si-NC group. (E) qPCR validation of
overexpression of miR-125b by transfection of miR-125b mimics.
***P<0.001 vs. the NC group. (F) Firefly luciferase activity
normalized to Renilla luciferase activity (FL/RL) in OSCC
cells co-transfected with luciferase reporters with Wt or Mu
transcripts of circPVT1. *P<0.05 and **P<0.01 vs. the NC
group. qPCR, quantitative polymerase chain reaction; NC, negative
control; OSCC, oral squamous cell carcinoma; circ, circular; miR,
microRNA; Wt, wild type; Mu, mutant; si, small interfering; NC,
negative control; miR, microRNA; FL/RL, firefly
luciferase/Renilla luciferase. |
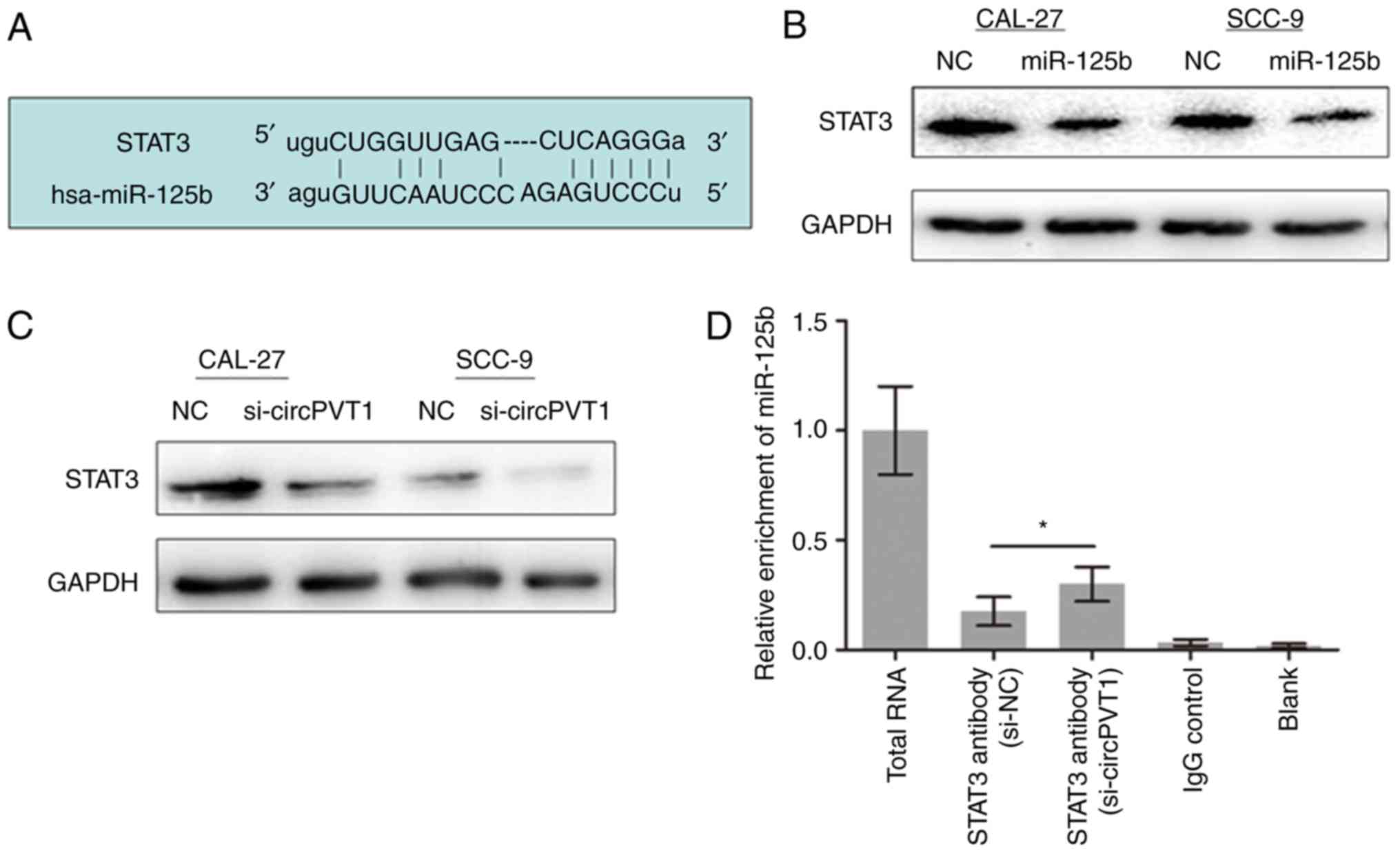
CircPVT1 affects the STAT3 expression
level through sponging miR-125b
Previously, it was reported that STAT3 was directly
targeted by miR-125b (16,17). Furthermore, STAT3 is known to serve
a key role in a number of cellular processes including cell growth
(18). Therefore, whether STAT3 is
essential for the OSCC proliferation change mediated by the
circPVT1/miR-125b pathway was investigated. Using bioinformatics
analysis, STAT3 was predicted to be targeted by miR-125b (Fig. 4A). miR-125b mimics suppressed STAT3
expression level in OSCC cells (Fig.
4B). Meanwhile, knockdown of circPVT1 decreased STAT3 levels
(Fig. 4C). RIP experiment revealed
that miR-125b was pulled down by STAT3 antibody. Furthermore, the
enrichment between miR-125b and STAT3 was increased when circPVT1
was silenced in CAL-27 cells (Fig.
4D). These indicate that circPVT1 released STAT3 by binding to
miR-125b in OSCC.
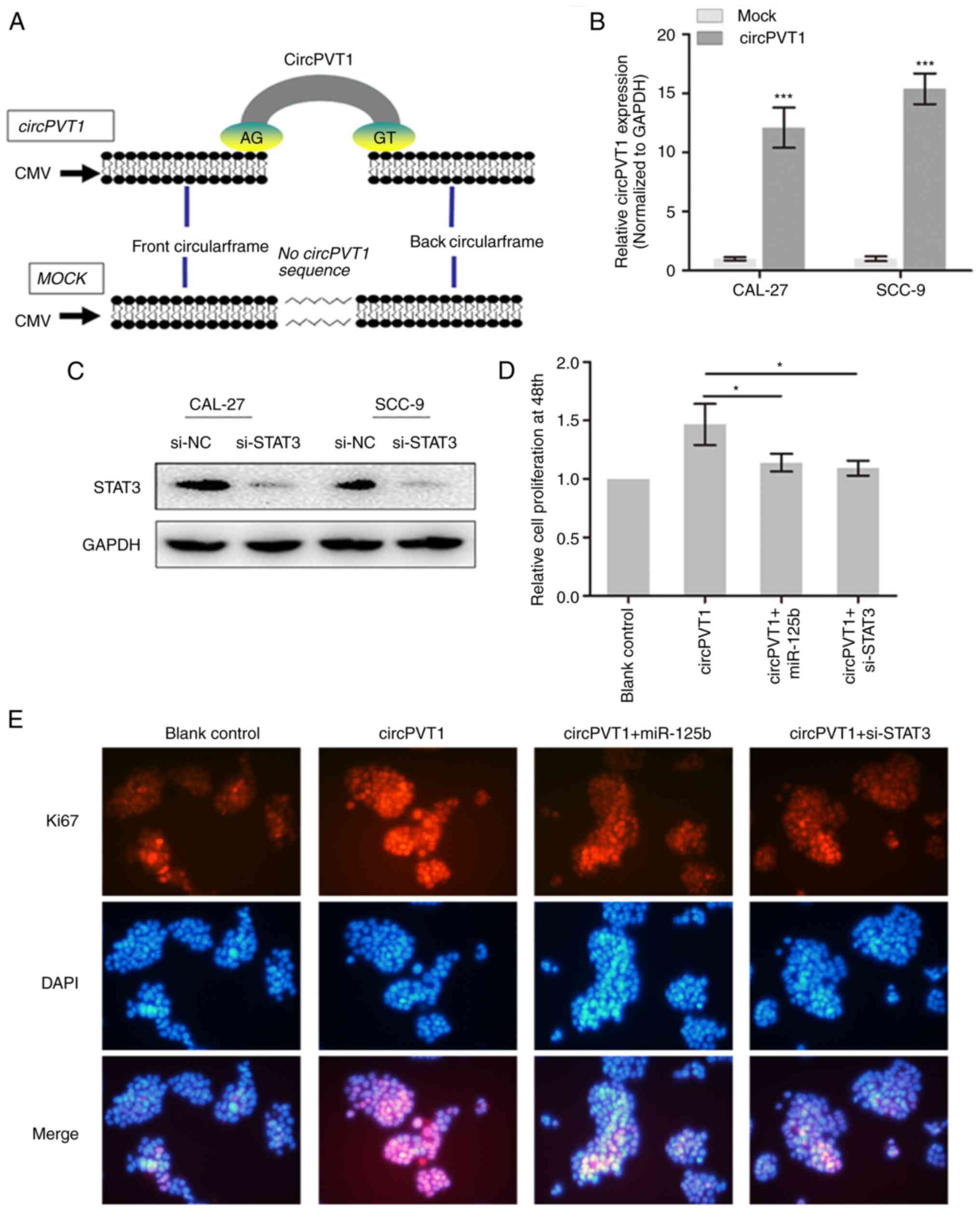
CircPVT1 promotes cell proliferation
via sponging miR-125b and releasing STAT3
Full-length cDNA of circPVT1 from CAL-27 cells was
generated and loaded into the plasmid vectors (Fig. 5A). The upregulation of circPVT1 in
OSCC cells was also confirmed following transfection of the vector
by qPCR (Fig. 5B). In addition,
STAT3 was silenced by transfection of specific siRNA (Fig. 5C). Next, the CCK-8 assay
demonstrated that circPVT1 promoted the cell growth of OSCC cells,
however, the enhanced cell growth was partially reversed by
knockdown of STAT3 or overexpression of miR-125b (Fig. 5D). Furthermore, immunofluorescence
demonstrated an elevated expression of Ki-67 by overexpression of
circPVT1 and this elevation was reversed by co-expression of
miR-125b or si-STAT3 (Fig. 5E). To
conclude, circPVT1 may promote cell growth by sponging miR-125b and
therefore increasing STAT3 expression.
Discussion
Numerous studies have been performed to get a
thorough understanding of biological characteristics and functional
roles of noncoding RNAs in cancer progression; however, their
specific roles are still largely unknown in cancer, including OSCC
(19,20). More recently, circRNA have been
increasingly recognized as oncogenes or tumor-suppressor genes in
different types of cancers, including gastric cancer (21), breast cancer (22) and colorectal cancer (23). However, the role of circRNAs in
OSCC is largely unknown. In this study, the expression and function
of circPVT1 in OSCC was investigated. The results of the present
study demonstrated that increased expression of circPVT1 in OSCC
could enhance cell proliferation via sponging miR-125b and
upregulating the STAT3 expression level.
During the past years, miRNAs and lncRNAs have been
investigated to be used as potential diagnostic predictors or
therapeutic targets (24,25). However, the outcome is not
satisfactory and the translational studies have grown stagnant. In
the present study, the novel findings of circRNAs open up a new
area. As non-classical products of transcription, circRNAs are
thought to be synthesized co-transcriptionally in the nucleus and
are modified in the cytoplasm (26). CircRNAs can selectively bind to
target miRNAs in a sequence specific manner to form the
circRNA-miRNA complexes that block the functions of miRNAs
(27). Unlike known unidimensional
lncRNAs, circRNAs in general are produced from pre-mRNAs
back-spicing events, which covalently linked the 3′ and 5′ ends of
exons or introns together and formed single-stranded continuous
loop structures with neither a 5′ cap nor a 3′ polyadenylated tail.
Compared to linear RNAs, circRNAs are much more resistant to RNase
R and more stable probably due to the absence of 5′ and 3′ ends
(28,29). Importantly, it was identified that
one circRNA named circPVT1 was upregulated in OSCC tissues and
cells, which indicates that circPVT1 has the potential to serve as
useful biological marker and may be critical in OSCC
progression.
Several studies have demonstrated that the
expression profiles of circRNAs are aberrant in diverse cancer
types and certain ones serve very important roles in oncogenesis
and cancer development, including as circPVT1 (13,30,31).
The specific role of circPVT1 in OSCC is still unknown. In this
study, it was identified that circPVT1 was downregulated in OSCC
compared to noncancerous tissues and predominantly localized in the
cytoplasm. Importantly, circPVT1 could directly sponge miR-125b to
release STAT3 and subsequently promotes proliferation of OSCC by
upregulating STAT3 expression. Therefore, the aforementioned
studies suggest that circPVT1 serves quite important roles in
neoplasia and cancer development. In addition, it was reported that
G2/M arrest was involved in cellular dormancy.
Therefore, the role of circPVT1 in dormancy still needs to be
further investigated.
It was demonstrated that circPVT1 was enriched in
the cytoplasm section and revealed that it acts as a miRNA sponge
in OSCC to exert the regulatory function. An increasing amount of
evidence suggests that miRNAs are abnormally expressed in diverse
human cancer and have important roles in the tumorigenesis,
progression and metastasis of these cancers (32). The first well-known circRNA serving
as miRNA sponge is ciRS-7. Expression of ciRS-7 efficiently tethers
miR-7, resulting in reduced miR-7 activity and increased levels of
miR-7-targeted transcripts (33).
After validating the direct interaction between circPVT1 and
miR-125b, STAT3 mRNA was verified as a downstream target of
miR-125b. STAT3 is a central regulator of numerous cellular
processes, particularly cell proliferation (34,35).
The oncogenic property of STAT3 has been observed in multiple types
of cancer due to its effect on promoting proliferation progression.
However, positive and negative associations between STAT3
activation and patient survival or tumor progression have been
reported (36–38). Therefore, the role of STAT3 in
tumorigenesis seems complex and requires a careful and repeated
validation. The current study suggested that STAT3 promoted cell
growth and was affected by the circPVT1/miR-125b axis. Furthermore,
STAT3 and miR-125b are essential for the circPVT1-induced cell
proliferation, indicating the oncogenic role of STAT3.
The present study has limitations. First, the
investigation of the prognostic significance of circPVT1 expression
in OSCC patient survival is needed. A follow-up study will be
conducted with the auhors' subsequent research work. Second,
although the in vitro experimental study was repeated using
two OSCC cell lines, the functional role of circPVT1 in OSCC needs
a further validation with other OSCC cell lines. In the future, the
functional role of circPVT1 in OSCC cell proliferation will be
comprehensively validated.
In conclusion, the present study's findings revealed
that overexpression of circPVT1 promoted cell proliferation via
serving as a sponge for miR-125b. Therefore, circPVT1 may be a
promising predictive biomarker and therapeutic target for OSCC
patients.
Acknowledgements
Not applicable.
Funding
The present study was funded by The National Natural
Science Foundation of China (grant no. 86722321).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TH and LT designed the study and performed the
experiments; XL and DX performed the statistical work and wrote the
manuscript; TH constructed the figures and tables and LT approved
the data and the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Clinical
Research Ethics Committee of China-Japan Friendship Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu
CP, Yang WE, Su CW, Chuang CY, Li WH, et al: Exome sequencing of
oral squamous cell carcinoma reveals molecular subgroups and novel
therapeutic opportunities. Theranostics. 7:1088–1099. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yan L, Chen F, Liu F, Qiu Y, Wang J, Wu J,
Bao X, Hu Z, Peng X, Lin X, et al: Differences in modifiable
factors of oral squamous cell carcinoma in the upper and lower of
oral fissure. Oncotarget. 8:75094–75101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Glazer CA, Chang SS, Ha PK and Califano
JA: Applying the molecular biology and epigenetics of head and neck
cancer in everyday clinical practice. Oral Oncol. 45:440–446. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsiao KY, Sun HS and Tsai SJ: Circular
RNA-New member of noncoding RNA with novel functions. Exp Biol Med
(Maywood). 242:1136–1141. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular roles and function of circular RNAs in eukaryotic cells.
Cell Mol Life Sci. 75:1071–1098. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao D, Zhang X, Liu B, Meng D, Fang K, Guo
Z and Li L: Screening circular RNA related to chemotherapeutic
resistance in breast cancer. Epigenomics. 9:1175–1188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kristensen LS, Hansen TB, Veno MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meyer KB, Maia AT, O'Reilly M, Ghoussaini
M, Prathalingam R, Porter-Gill P, Ambs S, Prokunina-Olsson L,
Carroll J and Ponder BA: A functional variant at a prostate cancer
predisposition locus at 8q24 is associated with PVT1 expression.
PLoS Genet. 7:e10021652011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu D, Luo P, Wang Q, Ye Y and Wang B:
lncRNA PVT1 in cancer: A review and meta-analysis. Clin Chim Acta.
474:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Verduci L, Ferraiuolo M, Sacconi A, Ganci
F, Vitale J, Colombo T, Paci P, Strano S, Macino G, Rajewsky N and
Blandino G: The oncogenic role of circPVT1 in head and neck
squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD
transcription-competent complex. Genome Biol. 18:2372017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao M, Sano D, Pickering CR, Jasser SA,
Henderson YC, Clayman GL, Sturgis EM, Ow TJ, Lotan R, Carey TE, et
al: Assembly and initial characterization of a panel of 85
genomically validated cell lines from diverse head and neck tumor
sites. Clin Cancer Res. 17:7248–7264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dai CY, Tsai YS, Chou WW, Liu T, Huang CF,
Wang SC, Tsai PC, Yeh ML, Hsieh MY, Huang CI, et al: The IL-6/STAT3
pathway upregulates microRNA-125b expression in hepatitis C virus
infection. Oncotarget. 9:11291–11302. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gorczynski RM, Alexander C, Brandenburg K,
Chen Z, Heini A, Neumann D, Mach JP, Rietschel ET, Terskikh A,
Ulmer AJ, et al: Corrigendum to ‘An altered REDOX environment,
assisted by over-expression of fetal hemoglobins, protects from
inflammatory colitis and reduces inflammatory cytokine expression’
[Int. Immunopharmacol. 50 (2017) 69–76]. Int Immunopharmacol.
59:4142018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
MacPherson S, Horkoff M, Gravel C,
Hoffmann T, Zuber J and Lum JJ: STAT3 regulation of citrate
synthase is essential during the initiation of lymphocyte cell
growth. Cell Rep. 19:910–918. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Momen-Heravi F and Bala S: Emerging role
of non-coding RNA in oral cancer. Cell Signal. 42:134–143. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemoresistance through EZH2. Mol Cancer Ther. 16:739–751.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang J, Meng X, Pan J, Jiang N, Zhou C, Wu
Z and Gong Z: CRISPR/Cas9-mediated noncoding RNA editing in human
cancers. RNA Biology. 15:35–43. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hamam R, Hamam D, Alsaleh KA, Kassem M,
Zaher W, Alfayez M, Aldahmash A and Alajez NM: Circulating
microRNAs in breast cancer: Novel diagnostic and prognostic
biomarkers. Cell Death Dis. 8:e30452017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanwal R, Plaga AR, Liu X, Shukla GC and
Gupta S: MicroRNAs in prostate cancer: Functional role as
biomarkers. Cancer Lett. 407:9–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ungerleider N, Jain V, Wang Y, Maness NJ,
Blair RV, Alvarez X, Midkiff C, Kolson D, Bai S, Roberts C, et al:
Comparative analysis of gammaherpesvirus circRNA repertoires:
Conserved and unique viral circular RNAs. J Virol. 93(pii):
e01952–18. 2019.PubMed/NCBI
|
|
28
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Greene J, Baird AM, Brady L, Lim M, Gray
SG, McDermott R and Finn SP: Circular RNAs: Biogenesis, Function
and Role in Human Diseases. Front Mol Biosci. 4:382017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu J, Han Q, Gu Y, Ma J, McGrath M, Qiao
F, Chen B, Song C and Ge Z: Circular RNA PVT1 expression and its
roles in acute lymphoblastic leukemia. Epigenomics. 10:723–732.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mali SB: Review of STAT3 (Signal
Transducers and Activators of Transcription) in head and neck
cancer. Oral Oncol. 51:565–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Sepich DS and Solnica-Krezel L:
Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis
extension during zebrafish gastrulation. PLoS Genet.
13:e10065642017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Cui J, Zhang J, Liu Y, Han L, Jia C,
Deng J and Liang H: PIAS3, an inhibitor of STAT3, has intensively
negative association with the survival of gastric cancer. Int J
Clin Exp Med. 8:682–689. 2015.PubMed/NCBI
|
|
37
|
Kanda N, Seno H, Konda Y, Marusawa H,
Kanai M, Nakajima T, Kawashima T, Nanakin A, Sawabu T, Uenoyama Y,
et al: STAT3 is constitutively activated and supports cell survival
in association with survivin expression in gastric cancer cells.
Oncogene. 23:4921–4929. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|