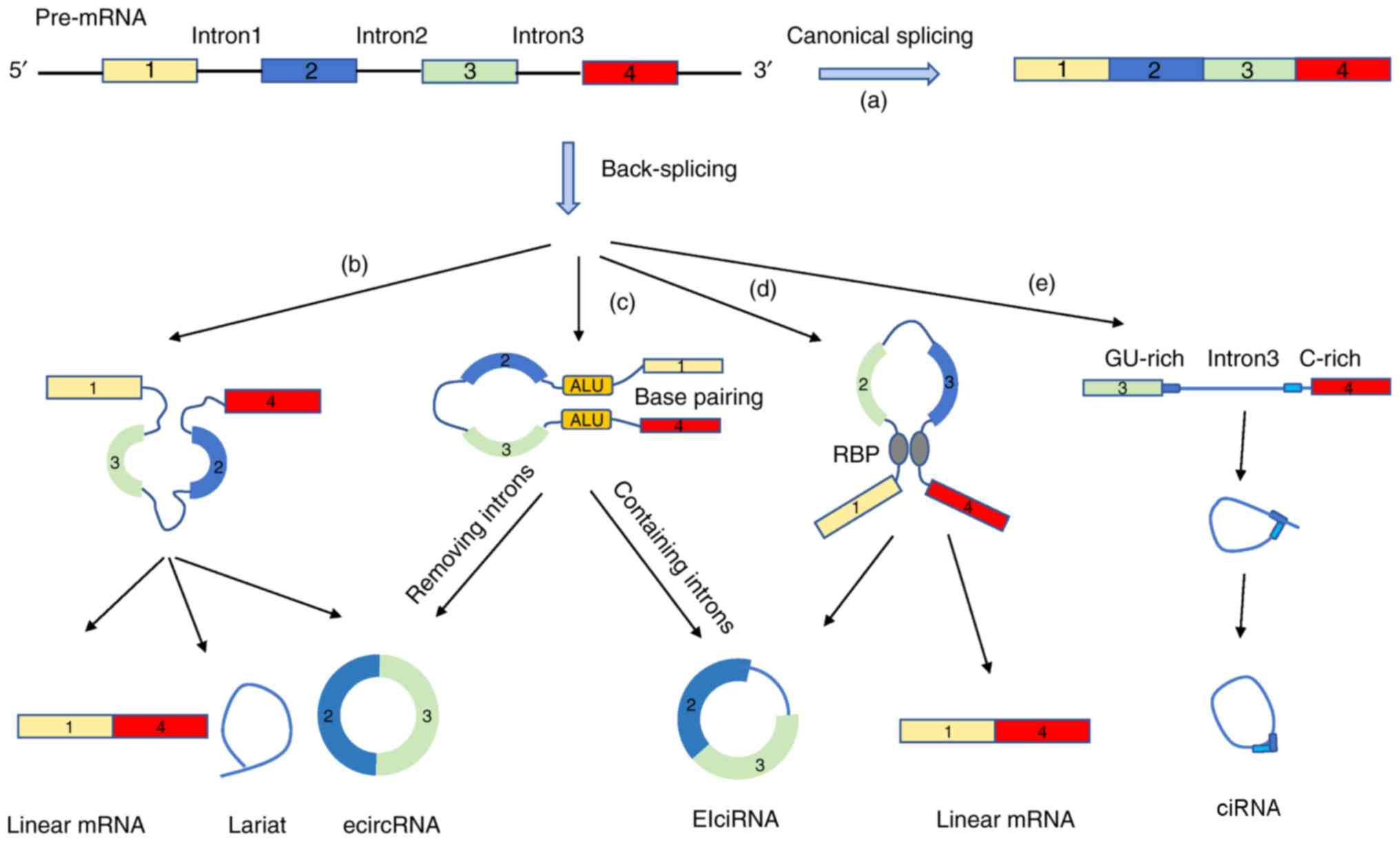
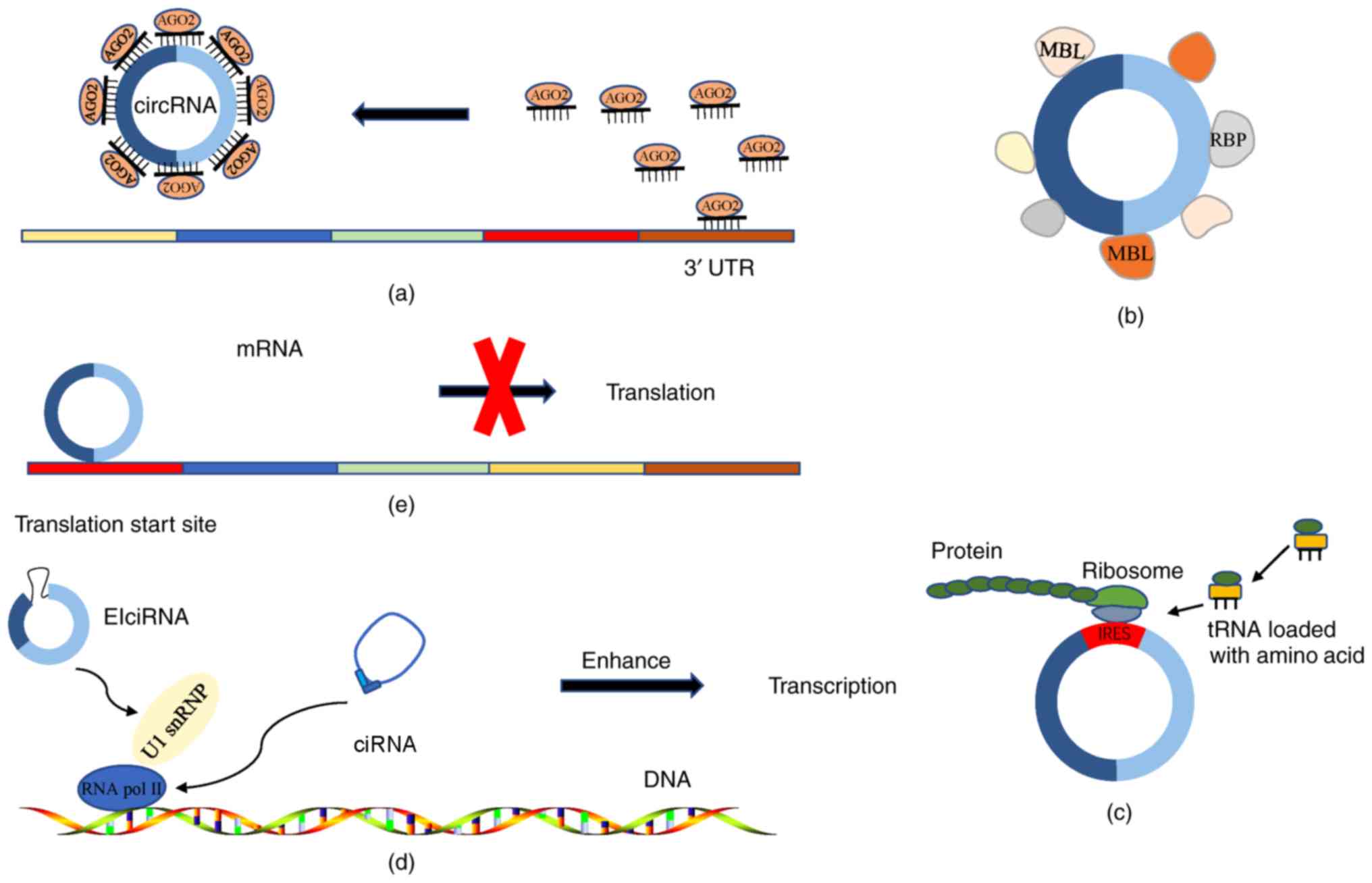
|
1
|
Dong Y, Xu S, Liu J, Ponnusamy M, Zhao Y,
Zhang Y, Wang Q, Li P and Wang K: Non-coding RNA-linked epigenetic
regulation in cardiac hypertrophy. Int J Biol Sci. 14:1133–1141.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abu N and Jamal R: Circular RNAs as
promising biomarkers: A mini-review. Front Physiol. 7:3552016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pasman Z, Been MD and Garcia-Blanco MA:
Exon circularization in mammalian nuclear extracts. RNA. 2:603–610.
1996.PubMed/NCBI
|
|
11
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cocquet J, Chong A, Zhang G and Veitia RA:
Reverse transcriptase template switching and false alternative
transcripts. Genomics. 88:127–131. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lopez-Jimenez E, Rojas AM and Andrés-León
E: RNA sequencing and prediction tools for circular RNAs analysis.
Adv Exp Med Biol. 1087:17–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong R, Ma XK, Li GW and Yang L: CIRCpedia
v2: An updated database for comprehensive circular RNA annotation
and expression comparison. Genomics Proteomics Bioinformatics.
16:226–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Glazar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Figueiredo C, Camargo MC, Leite M,
Fuentes-Pananá EM, Rabkin CS and Machado JC: Pathogenesis of
gastric cancer: Genetics and molecular classification. Curr Top
Microbiol Immunol. 400:277–304. 2017.PubMed/NCBI
|
|
17
|
Wu Z, Shi W and Jiang C: Overexpressing
circular RNA hsa_circ_0002052 impairs osteosarcoma progression via
inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis.
Biochem Biophys Res Commun. 502:465–471. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu
K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted
from adipocytes promotes the growth of hepatocellular carcinoma by
targeting deubiquitination-related USP7. Oncogene. 38:2844–2859.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rong D, Tang W, Li Z, Zhou J, Shi J, Wang
H and Cao H: Novel insights into circular RNAs in clinical
application of carcinomas. Onco Targets Ther. 10:2183–2188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi Y, Guo Z, Fang N, Jiang W, Fan Y, He
Y, Ma Z and Chen Y: hsa_circ_0006168 sponges miR-100 and regulates
mTOR to promote the proliferation, migration and invasion of
esophageal squamous cell carcinoma. Biomed Pharmacother.
117:1091512019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen
W, Jiang B, Qin H, Guo X, Liu M, et al: Circular RNA circSLC8A1
acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer
progression via regulating PTEN. Mol Cancer. 18:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang C, Wei Y, Yu L and Xiao Y:
Identification of altered circular RNA expression in serum exosomes
from patients with papillary thyroid carcinoma by high-throughput
sequencing. Med Sci Monit. 25:2785–2791. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiong S, Peng H, Ding X, Wang X, Wang L,
Wu C, Wang S, Xu H and Liu Y: Circular RNA expression profiling and
the potential role of hsa_circ_0089172 in Hashimoto's thyroiditis
via sponging miR125a-3p. Mol Ther Nucleic Acids. 17:38–48. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guarnerio J, Zhang Y, Cheloni G, Panella
R, Mae Katon J, Simpson M, Matsumoto A, Papa A, Loretelli C, Petri
A, et al: Intragenic antagonistic roles of protein and circRNA in
tumorigenesis. Cell Res. Jun 17–2019.doi: 10.1038/s41422-019-0192-1
(Epub ahead of print). View Article : Google Scholar
|
|
27
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Muller S and Appel B: In vitro
circularization of RNA. RNA Biol. 14:1018–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Dong R, Zhang Y, Zhang JL, Luo Z,
Zhang J, Chen LL and Yang L: Diverse alternative back-splicing and
alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang M and Xin Y: Circular RNAs: A new
frontier for cancer diagnosis and therapy. J Hematol Oncol.
11:212018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Z: Not just a sponge: New functions
of circular RNAs discovered. Sci China Life Sci. 58:407–408. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kwiatkowski TJ Jr, Bosco DA, Leclerc AL,
Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis
EJ, Munsat T, et al: Mutations in the FUS/TLS gene on chromosome 16
cause familial amyotrophic lateral sclerosis. Science.
323:1205–1208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vance C, Rogelj B, Hortobágyi T, De Vos
KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P,
et al: Mutations in FUS, an RNA processing protein, cause familial
amyotrophic lateral sclerosis type 6. Science. 323:1208–1211. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Errichelli L, Dini Modigliani S, Laneve P,
Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R,
Peruzzi G, et al: FUS affects circular RNA expression in murine
embryonic stem cell-derived motor neurons. Nat Commun. 8:147412017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Borchardt EK, Meganck RM, Vincent HA, Ball
CB, Ramos SBV, Moorman NJ, Marzluff WF and Asokan A: Inducing
circular RNA formation using the CRISPR endoribonuclease Csy4. RNA.
23:619–627. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liao Q, Wang B, Li X and Jiang G: miRNAs
in acute myeloid leukemia. Oncotarget. 8:3666–3682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dragomir M, Mafra ACP, Dias SMG, Vasilescu
C and Calin GA: Using microRNA networks to understand cancer. Int J
Mol Sci. 19(pii): E18712018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fumagalli MR, Zapperi S and La Porta CAM:
Impact of the cross-talk between circular and messenger RNAs on
cell regulation. J Theor Biol. 454:386–395. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Piwecka M, Glazar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357(pii): eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen H, Liu T, Liu J, Feng Y, Wang B, Wang
J, Bai J, Zhao W, Shen Y, Wang X, et al: Circ-ANAPC7 is upregulated
in acute myeloid leukemia and appears to target the MiR-181 family.
Cell Physiol Biochem. 47:1998–2007. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gartel A and Radhakrishnan SK: Lost in
transcription: p21 repression, mechanisms, and consequences. Cancer
Res. 65:3980–3985. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Abdelmohsen K, Panda A, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang Y and Wang Z: Efficient backsplicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen CY and Science S: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 Is a Circular RNA that Can Be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel Role of FBXW7
Circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 1102018.doi: 10.1093/jnci/djx166.
|
|
60
|
Zhao J, Wu J, Xu T, Yang Q, He J and Song
X: IRESfinder: Identifying RNA internal ribosome entry site in
eukaryotic cell using framed k-mer features. J Genet Genomics.
45:403–406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kwek K, Murphy S, Furger A, Thomas B,
O'Gorman W, Kimura H, Proudfoot NJ and Akoulitchev A: U1 snRNA
associates with TFIIH and regulates transcriptional initiation. Nat
Struct Biol. 9:800–805. 2002.PubMed/NCBI
|
|
63
|
Huang C and Shan G: What happens at or
after transcription: Insights into circRNA biogenesis and function.
Transcription. 6:61–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R
and Li H: The emerging functions and roles of circular RNAs in
cancer. Cancer Lett. 414:301–309. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Somervaille TC and Cleary ML: Grist for
the MLL: How do MLL oncogenic fusion proteins generate leukemia
stem cells? Int J Hematol. 91:735–741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of Fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 165:289–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K,
Liu Y, Liu L, Wei YQ and Peng Y: Circular RNA F-circEA produced
from EML4-ALK fusion gene as a novel liquid biopsy biomarker for
non-small cell lung cancer. Cell Res. 28:693–695. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
69
|
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M,
Liu X, Sun H, Pan Y and Wang S: The pro-metastasis effect of
circANKS1B in breast cancer. Mol Cancer. 17:1602018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhang XL, Xu LL and Wang F:
Hsa_circ_0020397 regulates colorectal cancer cell viability,
apoptosis and invasion by promoting the expression of the miR-138
targets TERT and PD-L1. Cell Biol Int. 41:1056–1064. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Huang Y, Yu P, Li W, Ren G, Roberts AI,
Cao W, Zhang X, Su J, Chen X, Chen Q, et al: p53 regulates
mesenchymal stem cell-mediated tumor suppression in a tumor
microenvironment through immune modulation. Oncogene. 33:3830–3838.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang J, Zhang Q, Zhou S, Xu H, Wang D,
Feng J, Zhao J and Zhong S: Circular RNA expression in exosomes
derived from breast cancer cells and patients. Epigenomics.
11:411–421. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou
J, Jin H, Zhao A, Tang WW and Cao XF: Down-regulation of circPVRL3
promotes the proliferation and migration of gastric cancer cells.
Sci Rep. 8:101112018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li P, Chen H, Chen S, Mo X, Li T, Xiao B,
Yu R and Guo J: Circular RNA 0000096 affects cell growth and
migration in gastric cancer. Br J Cancer. 116:626–633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Montagner A, Le Cam L and Guillou H:
β-catenin oncogenic activation rewires fatty acid catabolism to
fuel hepatocellular carcinoma. Gut. 68:183–185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhong L, Wang Y, Cheng Y, Wang W, Lu B,
Zhu L and Ma Y: Circular RNA circC3P1 suppresses hepatocellular
carcinoma growth and metastasis through miR-4641/PCK1 pathway.
Biochem Biophys Res Commun. 499:1044–1049. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jiang W, Wen D, Gong L, Wang Y, Liu Z and
Yin F: Circular RNA hsa_circ_0000673 promotes hepatocellular
carcinoma malignance by decreasing miR-767-3p targeting SET.
Biochem Biophys Res Commun. 500:211–216. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Seneviratne S, Lawrenson R, Scott N, Kim
B, Shirley R and Campbell I: Breast cancer biology and ethnic
disparities in breast cancer mortality in new zealand: A cohort
study. PLoS One. 10:e01235232015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R,
Yang SY, Yang DC and Wang XL: Circular RNA hsa_circ_0001982
promotes breast cancer cell carcinogenesis through decreasing
miR-143. DNA Cell Biol. 36:901–908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Du WW, Yang W, Li X, Awan FM, Yang Z, Fang
L, Lyu J, Li F, Peng C, Krylov SN, et al: A circular RNA circ-DNMT1
enhances breast cancer progression by activating autophagy.
Oncogene. 37:5829–5842. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH suppresses lung cancer proliferation
via inhibiting the Wnt/β-catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ma X, Yang X, Bao W, Li S, Liang S, Sun Y,
Zhao Y, Wang J and Zhao C: Circular RNA circMAN2B2 facilitates lung
cancer cell proliferation and invasion via miR-1275/FOXK1 axis.
Biochem Biophys Res Commun. 498:1009–1015. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S
and Yuan H: Microarray profile of circular RNAs identifies
hsa_circ_0014130 as a new circular RNA biomarker in non-small cell
lung cancer. Sci Rep. 8:28782018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Jin P, Huang Y, Zhu P, Zou Y, Shao T and
Wang O: CircRNA circHIPK3 serves as a prognostic marker to promote
glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem
Biophys Res Commun. 503:1570–1574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang
J, Xu Y, Hu J, Dong G, Xu PL and Yin R: Circular RNA
has_circ_0067934 is upregulated in esophageal squamous cell
carcinoma and promoted proliferation. Sci Rep. 6:355762016.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG,
Wang WF and Fang MH: CiRS-7 promotes growth and metastasis of
esophageal squamous cell carcinoma via regulation of miR-7/HOXB13.
Cell Death Dis. 9:8382018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Xiao-Long M, Kun-Peng Z and Chun-Lin Z:
Circular RNA circ_HIPK3 is down-regulated and suppresses cell
proliferation, migration and invasion in osteosarcoma. J Cancer.
9:1856–1862. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Huang L, Chen M, Pan J and Yu W: Circular
RNA circNASP modulates the malignant behaviors in osteosarcoma via
miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 500:511–517.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Song Y and Li J: Circular RNA
hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis
by acting miRNA sponge. Biochem Biophys Res Commun. 495:2369–2375.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Xia L, Wu L, Bao J, Li Q, Chen X, Xia H
and Xia R: Circular RNA circ-CBFB promotes proliferation and
inhibits apoptosis in chronic lymphocytic leukemia through
regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem Biophys Res
Commun. 503:385–390. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Li S, Ma Y, Tan Y, Ma X, Zhao M, Chen B,
Zhang R, Chen Z and Wang K: Profiling and functional analysis of
circular RNAs in acute promyelocytic leukemia and their dynamic
regulation during all-trans retinoic acid treatment. Cell Death
Dis. 9:6512018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wu DM, Wen X, Han XR, Wang S, Wang YJ,
Shen M, Fan SH, Zhang ZF, Shan Q, Li MQ, et al: Role of Circular
RNA DLEU2 in Human Acute Myeloid Leukemia. Mol Cell Biol. 38(pii):
e00259–18. 2018.PubMed/NCBI
|
|
95
|
Ma HB, Yao YN, Yu JJ, Chen XX and Li HF:
Extensive profiling of circular RNAs and the potential regulatory
role of circRNA-000284 in cell proliferation and invasion of
cervical cancer via sponging miR-506. Am J Transl Res. 10:592–604.
2018.PubMed/NCBI
|
|
96
|
Liu J, Wang D, Long Z, Liu J and Li W:
CircRNA8924 promotes cervical cancer cell proliferation, migration
and invasion by competitively binding to MiR-518d-5p /519-5p family
and modulating the expression of CBX8. Cell Physiol Biochem.
173–184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhong Z, Huang M, Lv M, He Y, Duan C,
Zhang L and Chen J: Circular RNA MYLK as a competing endogenous RNA
promotes bladder cancer progression through modulating VEGFA/VEGFR2
signaling pathway. Cancer Lett. 403:305–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang X, Yuan W, Tao J, Li P, Yang C, Deng
X, Zhang X, Tang J, Han J, Wang J, et al: Identification of
circular RNA signature in bladder cancer. J Cancer. 8:3456–3463.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Huang G, Li S, Yang N, Zou Y, Zheng D and
Xiao T: Recent progress in circular RNAs in human cancers. Cancer
Lett. 404:8–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Alhasan AA, Izuogu OG, Al-Balool HH, Steyn
JS, Evans A, Colzani M, Ghevaert C, Mountford JC, Marenah L,
Elliott DJ, et al: Circular RNA enrichment in platelets is a
signature of transcriptome degradation. Blood. 127:e1–e11. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Nicolet BP, Engels S, Aglialoro F, van den
Akker E, von Lindern M and Wolkers MC: Circular RNA expression in
human hematopoietic cells is widespread and cell-type specific.
Nucleic Acids Res. 46:8168–8180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Geng Y, Jiang J and Wu C: Function and
clinical significance of circRNAs in solid tumors. J Hematol Oncol.
11:982018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang T, Shigdar S, Shamaileh HA, Gantier
MP, Yin W, Xiang D, Wang L, Zhou SF, Hou Y, Wang P, et al:
Challenges and opportunities for siRNA-based cancer treatment.
Cancer Lett. 387:77–83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Frazier KS: Antisense oligonucleotide
therapies: The promise and the challenges from a toxicologic
pathologist's perspective. Toxicol Pathol. 43:78–89. 2015.
View Article : Google Scholar : PubMed/NCBI
|