Introduction
Cholangiocarcinoma (CCA) is a malignant tumor
derived from the intrahepatic and extrahepatic bile duct (1,2). At
present, CCA is the most common hepatic and bile duct-associated
malignancy with poor prognosis besides hepatocellular carcinoma
(3–6). The rates of all types of CCA have
been increasing in the last few decades (7). However, the molecular mechanism of
CCA remains to be elucidated, and efficient therapeutic options are
lacking, which leads to the urgency of exploring specific
biomarkers and effective therapies for CCA.
Among all possible molecular events involved in the
tumorigenesis and development of CCA, microRNAs (miRNAs/miRs) have
been documented to serve essential roles. As small noncoding
single-stranded RNA molecules, miRNAs participate in biological
processes by functioning as post-transcriptional regulatory factors
(8–12). In recent years, miRNAs have been
reported to be closely associated with numerous human diseases,
including CCA (13). Previous
studies have reported that several miRNAs may regulate the
proliferation, invasion and metastasis of CCA, and may be potential
biomarkers used to evaluate the progression and prognosis of
patients with CCA (5–7,14–16).
However, the number of known miRNAs involved in CCA is limited and
the majority of studies conducted to date have focused on a small
sample size of clinical specimens. Therefore, it is necessary to
identify novel miRNA candidates and explore the clinical value of
these miRNAs in a larger sample size.
miR-132-3p has been reported to be differentially
expressed in several cancer types, and to participate in the
formation and metastasis of several malignant tumors. miR-132-3p
has been associated with the post-transcriptional regulation of
BCRP/ABCG2 in renal cell carcinoma (8,17).
Overexpressed miR-132-3p can regulate executioner caspase-7 in
pancreatic ductal adenocarcinoma and may contribute to malignant
progression (18). Furthermore,
miR-132-3p is significantly decreased in gastric cancer tissues
compared with in peripheral non-cancer gastric tissues, and its
alteration may serve as a novel molecular marker for gastric cancer
(19). However, the association
between miR-132-3p and CCA, and its clinical significance, remain
to be investigated. A limited number of genes have been identified
as targets of miR-132-3p in various diseases. For instance, Cai
et al (20) reported that
miR-132-3p regulates BCL2L11 in tuberous sclerosis complex
angiomyolipoma, whereas Zhou et al (21) reported that miR-132-3p regulates
ADAMTS-5 expression and promotes chondrogenic differentiation in
rat mesenchymal stem cells. It is well known that a single miRNA
can have multiple targets. However, the targets and signaling
pathways of miR-132-3p in CCA remain unknown.
Our preliminary work mining miRNA microarray and
miRNA-sequencing data of CCA revealed several aberrantly expressed
miRNAs (Wu et al, unpublished data), and miR-132-3p was
among them. Therefore, to identify the role of miR-132-3p in CCA,
the present study reprocessed the expression profiles of miR-132-3p
in CCA and non-tumor tissues from microarray and miRNA-sequencing
data. Furthermore, meta-analyses were performed to evaluate the
expression levels of miR-132-3p in CCA based on all available
cases. Subsequently, the potential role of miR-132-3p as a
biomarker for predicting prognosis in CCA was investigated. In
order to explore the molecular function of miR-132-3p in CCA, Gene
Ontology (GO) and WikiPathway analyses were performed to
investigate the prospective targets of miR-132-3p. Finally, the
mRNA and protein expression levels of hub target genes were
verified.
Materials and methods
Collection of clinical samples and
reverse transcription-quantitative PCR (RT-qPCR)
Samples were collected from patients with CCA from
the First Affiliated Hospital, Guangxi Medical University between
December 2010 and September 2017. In total, 25 CCA and 22
paracarcinoma bile duct tissues were collected, which included 16
males and 9 females with the ages ranging from 34 to 65 years old.
In three patients, paracarcinoma bile duct tissue was not
collected. This study was approved by the Ethics Committee of First
Affiliated Hospital, Guangxi Medical University, China. Informed
written consent was obtained from all patients participating in the
study. Total RNA from fixed (overnight with 10% neutral-buffered
formalin at 25°C) and paraffin embedded samples was extracted from
sections using the miRNeasy FFPE kit (Qiagen GmbH). For RT into
cDNA, the All-in-One™ miRNA First-Strand cDNA Synthesis kit (QP013,
GeneCopoeia, Inc.) was used to transcribe 10 µl purified RNA. The
prepared reaction mixture was gently mixed, incubated at 37°C for
60 min after brief centrifugation, followed by incubation at 85°C
for 5 min for RT. qPCR, using miR-specific primers and universal
adaptor PCR primers purchased from GeneCopoeia, Inc. with
confidential sequences, was performed using the Applied Biosystems
7500 Real-Time PCR system (cat. no. QP010; Applied Biosystems;
Thermo Fisher Scientific, Inc.). The All-in-one miRNA qPCR kit was
used for the qPCR (cat. no. QP013; GeneCopoeia, Inc.). The
reactions were incubated in a 96-well plate at 95°C for 10 min,
followed by 40 cycles at 95°C for 10 sec, 60°C for 20 sec and 72°C
for 34 sec, fluorescent signals were measured during the extension
phase. The results were normalized to the reference of U6RNA and
calculated using the 2−ΔΔCq method (22). All reactions were run in
triplicate.
Collection and analysis of
miRNA-microarray and miRNA-sequencing data of CCA
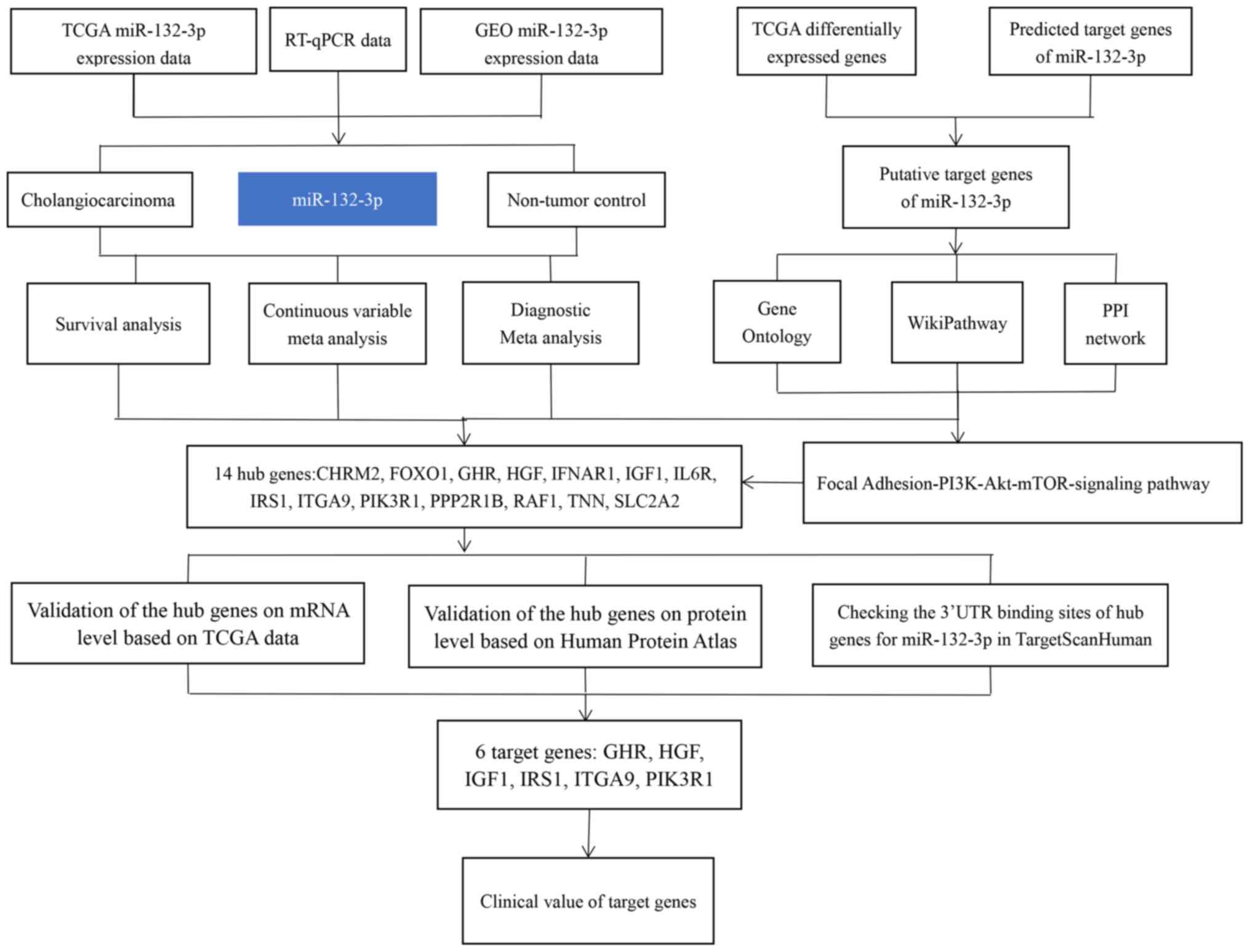
The flowchart of the present study is shown in
Fig. 1. To find potential datasets
of interest, CCA-associated Gene Expression Omnibus (GEO;
http://www.ncbi.nlm.nih.gov/geo/),
ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) and Sequence Read
Archive (SRA; http://www.ncbi.nlm.nih.gov/sra/) (23–25)
microarray or miRNA-sequencing data were used with the following
search strategies: (‘bile duct’ OR ‘cholangiocellular’) AND
(‘cancer’ OR ‘carcinoma’ OR ‘tumor’ OR ‘neoplas*’ OR ‘malignan*’ OR
‘adenocarcinoma’ OR ‘cholangiocarcinoma’). The screening criteria
were as follows: Firstly, the samples in each dataset must contain
CCA and non-tumor control groups; both the serum and tissue data of
patients with CCA were included in this study, but cell line data
were excluded. Secondly, in terms of sample size, each dataset
should contain at least three samples. Thirdly, the raw expression
data of mature or precursor miRNAs should be available and
reprocessable. Finally, all miRNA-microarray and miRNA-sequencing
data with other intervening factors were removed, such as
experiments with gene knockdown and other treatments. In addition,
The Cancer Genome Atlas (TCGA) cholangiocarcinoma miRNA mature
strand expression RNA-sequencing data were downloaded from UCSC
Xena (xena.ucsc.edu), which included 36 CCA
samples and nine normal samples. The corresponding
clinicopathological parameters (sex, age, smoking status, Tumor,
Node and Metastasis stage, pathological stage and grade) of CCA
samples were also investigated, but the clinical parameters of some
samples were not available on TCGA. Transcripts Per Kilobase
Million (TPM) was used to present RNA-sequencing results. For each
sample, isoform expression for the same miRNA mature strand was
combined after being log2 (total_TPM +1) transformed.
For the statistical analysis, the present study
first explored the association between miR-132-3p and
clinicopathological features of CCA, and the miR-132-3p expression
profile data acquired from each public database and RT-qPCR data
were analyzed with independent sample Student's t-test or a paired
samples t-test in SPSS 22.0 (IBM Corp.). The data were presented as
the means ± standard deviation. P<0.05 was considered to
indicate a statistically significant difference. In addition, the
possibility of miR-132-3p being used as a biomarker for
distinguishing CCA tumor tissues from normal tissues was evaluated
using a receiver operating characteristic (ROC) curve analysis. The
present study also performed a continuous variable meta-analysis in
Stata12.0 (StataCorp LP) to calculate the standardized mean
difference (SMD). This meta-analysis initially used a fixed effect
model; if heterogeneity was present, a random effect model was
selected instead. The heterogeneity was evaluated with a
χ2 test. The result would be regarded as heterogeneous
if P<0.05 or I2>50%. A Begg's test was performed
to evaluate potential publication bias. Subsequently, a summary ROC
(sROC) was conducted to combine the effect of single datasets.
Diagnostic odds ratio, as well as negative and positive likelihood
ratios, were also analyzed. In addition, the present study
performed sROC analysis to further appraise the distinguishing
ability of miR-132-3p (26–28).
Thirdly, a Kaplan-Meier analysis was conducted to reveal the
prognostic ability of miR-132-3p in CCA based on TCGA data, and a
log-rank test was performed to compare the survival between high
and low miR-132-3p expression groups. Furthermore, to improve the
accuracy of the present study, survival analysis was also conducted
on the GSE53870 dataset. The GSE53870 dataset is a miRNA-related
GEO dataset, which includes the survival data of 63 patients with
CCA.
Analysis of differentially expressed
genes based on TCGA CCA mRNA expression data
The TCGA gene expression profile data of CCA and
non-tumor samples were downloaded from UCSC Xena, and
differentially expressed genes [defined as those with a log (fold
change) equal to 1 and P<0.05] were analyzed using the edgeR
package (29).
Analysis of target genes of miR-132-3p
in CCA
Predicted target genes of miR-132-3p were obtained
from miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/),
which included 12 online tools [TargetScan version 6.2; MicroT4;
miRanda (release date; November 1st, 2010); miRBridge (release
date; April 9th, 2010); PITA version 6.0; miRMap (release date;
January 9th, 2013); miRNAMap version 2; PICTAR version 2; miRDB
version 4.0; RNA22 version 2; RNAhybrid version 2.1 and miRWalk
version 2.0] (30–33). Genes predicted by at least three
databases were selected for subsequent analysis. To increase the
reliability of the predicted targets, downregulated differentially
expressed genes from RNA-sequencing data were considered as
possible targets, and the present study only focused on this group
of genes whose mRNA expression could be influenced by miR-132-3p.
The overlapped genes from differential expression and prediction
findings were selected for further analysis.
Signaling pathway analysis of
miR-132-3p in CCA
Gene Ontology (GO) annotation of the aforementioned
overlapped genes was performed in WebGestalt 2017 (http://www.webgestalt.org/) (34). Pathway analysis of the
aforementioned overlapped genes was also performed in WebGestalt
using the WikiPathway functional database. Visualization of GO
annotation was conducted using the BiNGO app of Cytoscape version
3.5.0, and protein-protein interaction (PPI) analyses were
conducted in STRING (35–39).
Validation of the hub target genes at
the mRNA and protein levels
TCGA mRNA expression profiles were used to validate
the expression levels of hub genes at the mRNA level. The Human
Protein Atlas (https://www.proteinatlas.org/) (40) was used to evaluate the protein
expression levels of hub genes in CCA tissues and non-tumor
intrahepatic biliary epithelium. Additionally, the binding site of
miR-132-3p in hub genes was investigated with TargetScanHuman
(www.targetscan.org/vert_72), and only
those genes that contained 3′-untranslated region (UTR) binding
sites for miR-132-3p were selected. Finally, the clinical
significance of hub genes was explored by plotting ROC and survival
curves.
Results
Expression of miR-132-3p in CCA
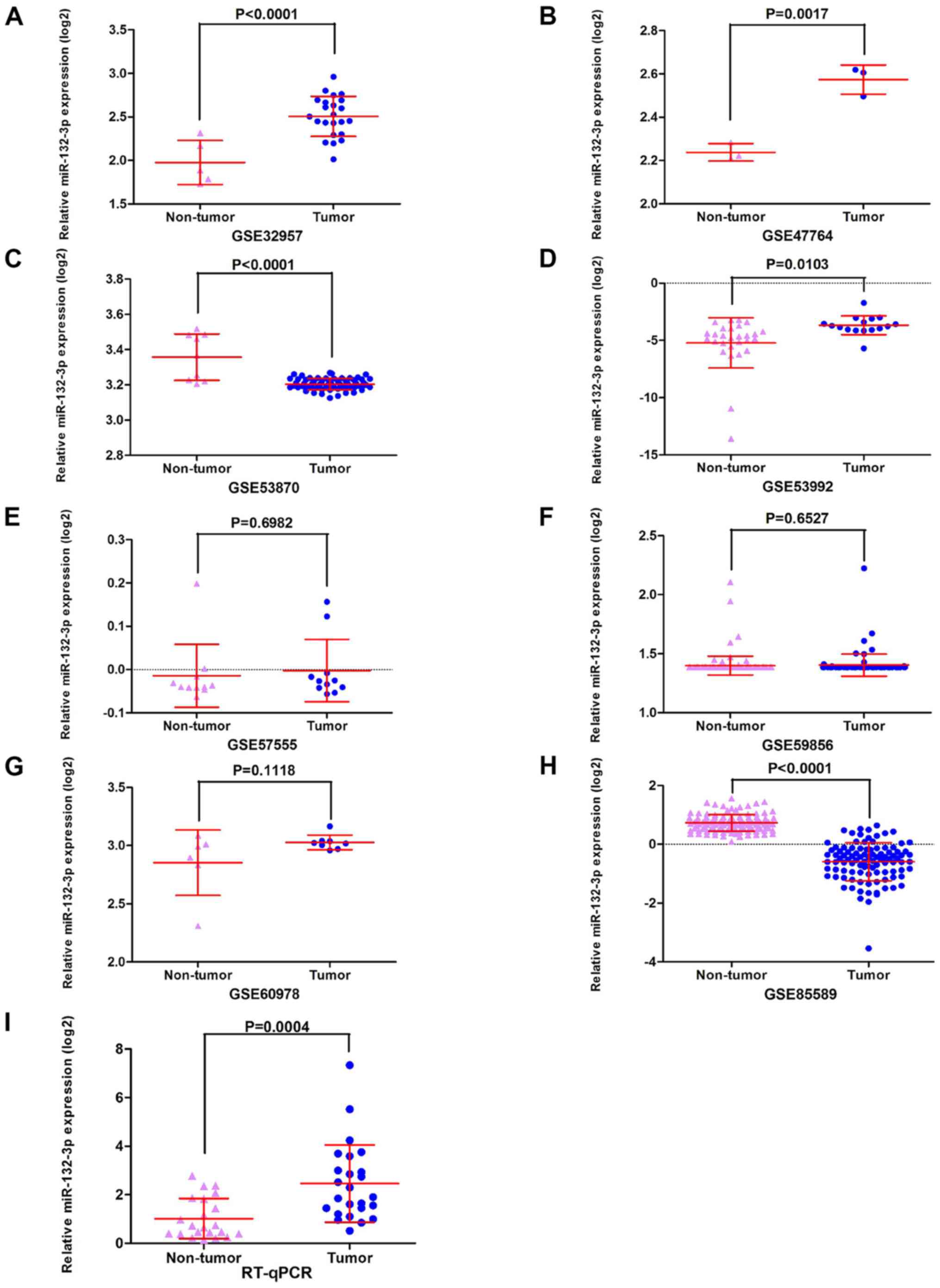
The GEO datasets (GSE32957, GSE47764 and GSE53992)
indicated that miR-132-3p was upregulated in CCA (Fig. 2A, B and D). Besides, according to
RT-qPCR, the relative expression levels of miR-132-3p were higher
in the CCA group (2.4634±1.59019) compared with in non-tumor
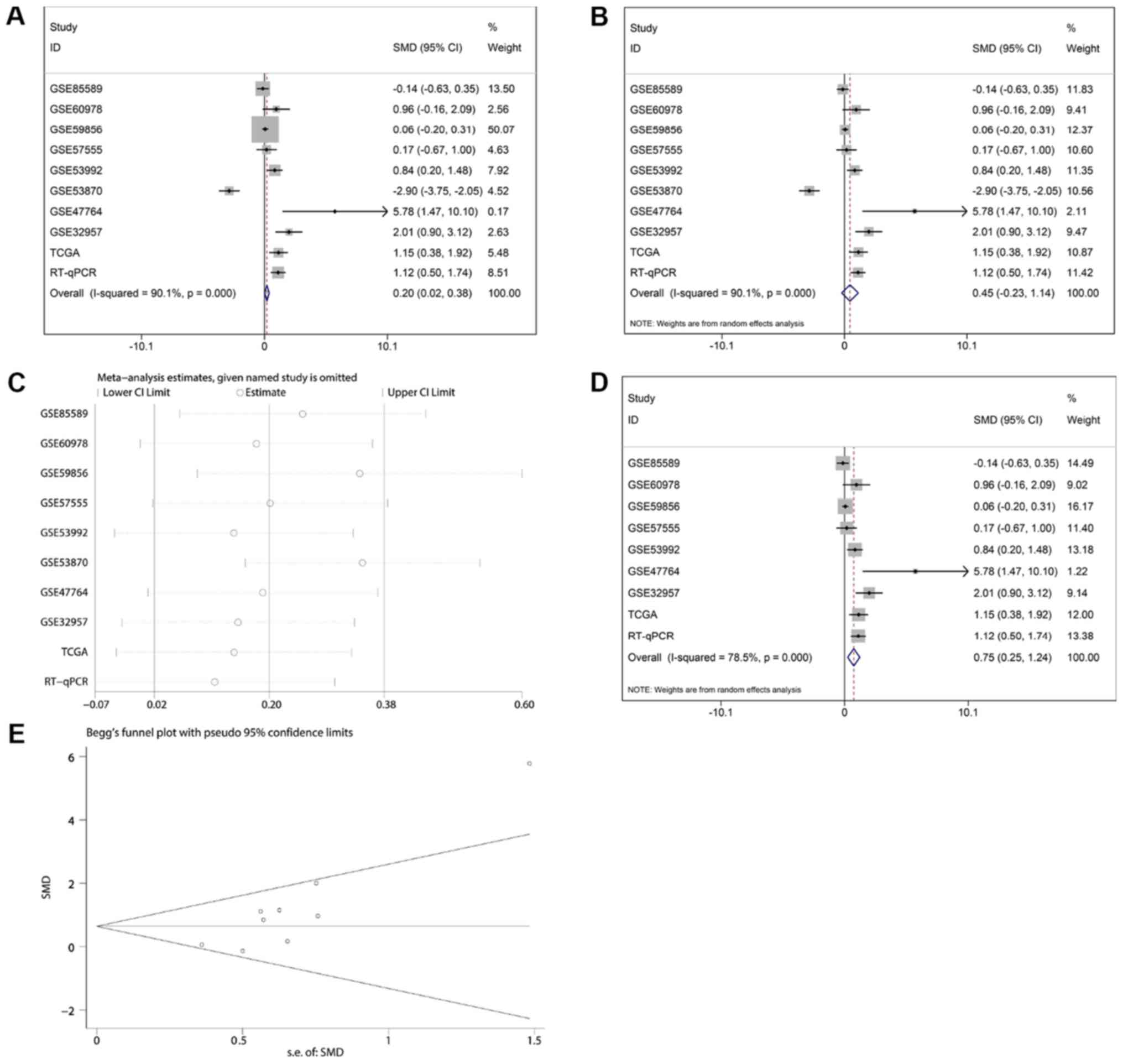
control tissues (1.0190±0.83004; P<0.001; Fig. 2I). The meta-analysis contained 10
groups of data, including in-house RT-qPCR data, eight GEO
miRNA-microarray datasets and one TCGA miRNA-sequencing dataset;
the results revealed consistent upregulation of miR-132-3p in CCA
using a random effect model (Fig. 3A
and B). Regarding the SMD of miR-132-3p in CCA, heterogeneity
existed with an I2 of 78.5%, and GSE53870 had the
greatest impact on the results of the meta-analysis based on the
sensitivity analysis conducted (Fig.
3C). Therefore, a forest plot was generated following exclusion
of GSE53870, which revealed an SMD of 0.75 (95% CI: 0.25, 1.24)
(Fig. 3D and E), indicating that
miR-132-3p had a general increasing trend in CCA tissues based on
multiple detecting methods.
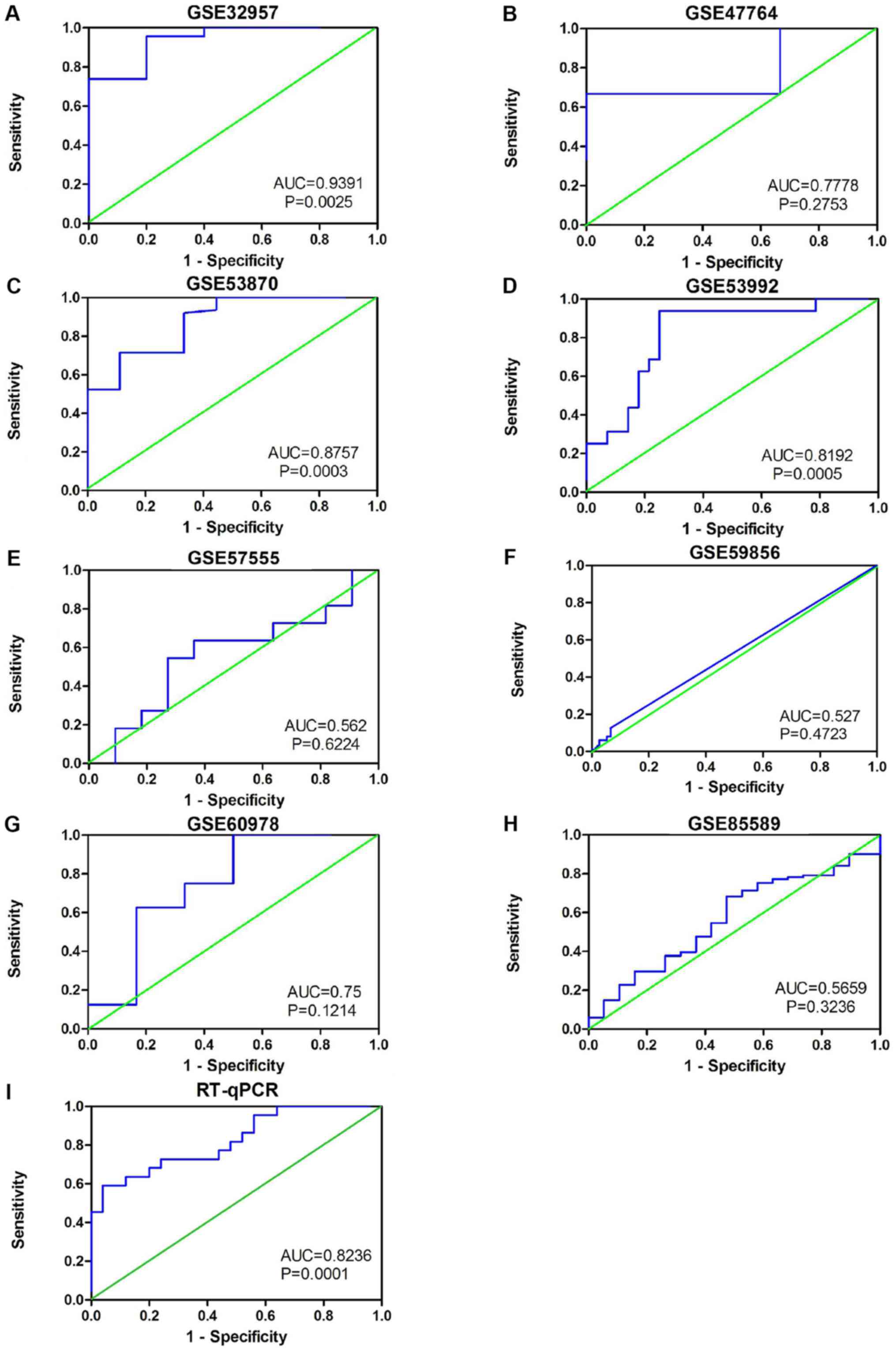
The results also indicated that miR-132-3p may have
potential as a biomarker to distinguish CCA tissues from non-cancer
tissues based on ROC curves of RT-qPCR, miRNA-microarray and
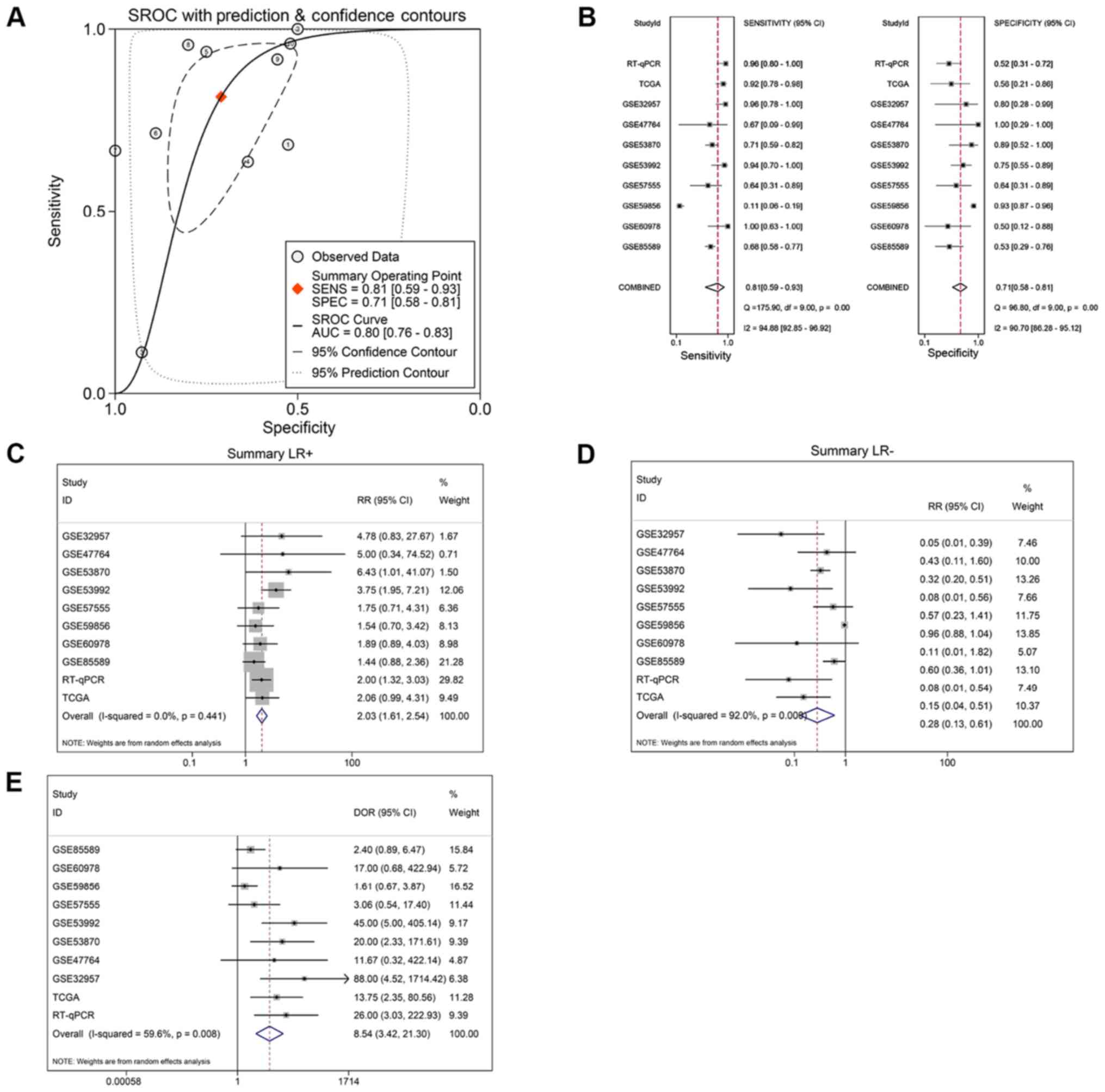
RNA-sequencing data (Fig. 4).
Moreover, the sROC revealed a consistent result with an AUC of 0.80
(95% CI: 0.76, 0.83). The pooled sensitivity, specificity, positive
likelihood ratio, negative likelihood ratio and diagnostic odds
ratio of miR-132-3p in CCA tissues were 0.81 (95% CI: 0.59, 0.93),
0.71 (95% CI: 0.58, 0.81), 2.03 (95% CI: 1.61, 2.54), 0.28 (95% CI:
0.13, 0.61) and 8.54 (95% CI: 3.42, 21.30), respectively (Fig. 5B-E).
Association between miR-132-3p and
clinicopathological features of CCA
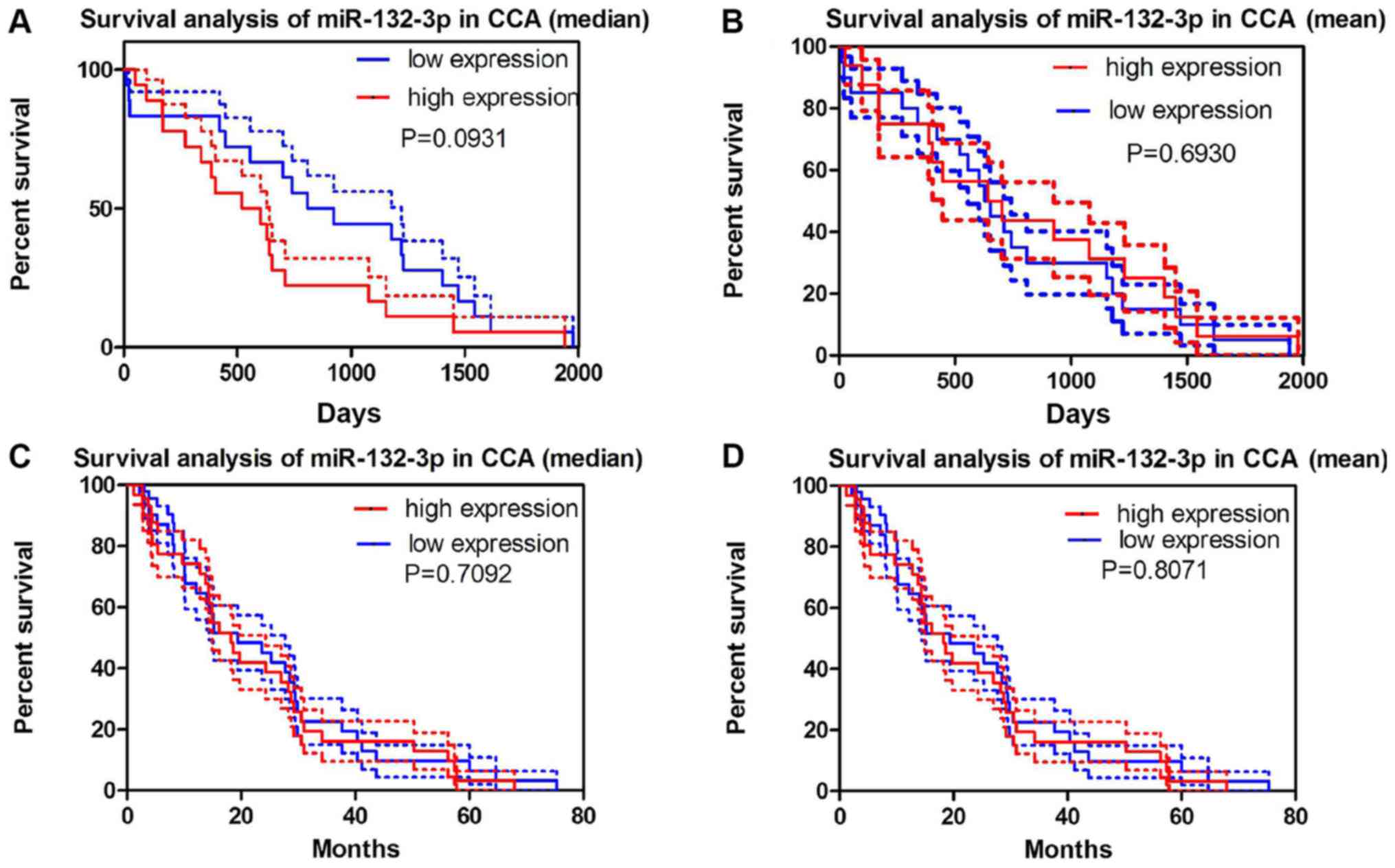
No significant association between miR-132-3p levels
and survival was observed either in the miRNA-sequencing data from
TCGA or GEO GSE53870 data (Fig.
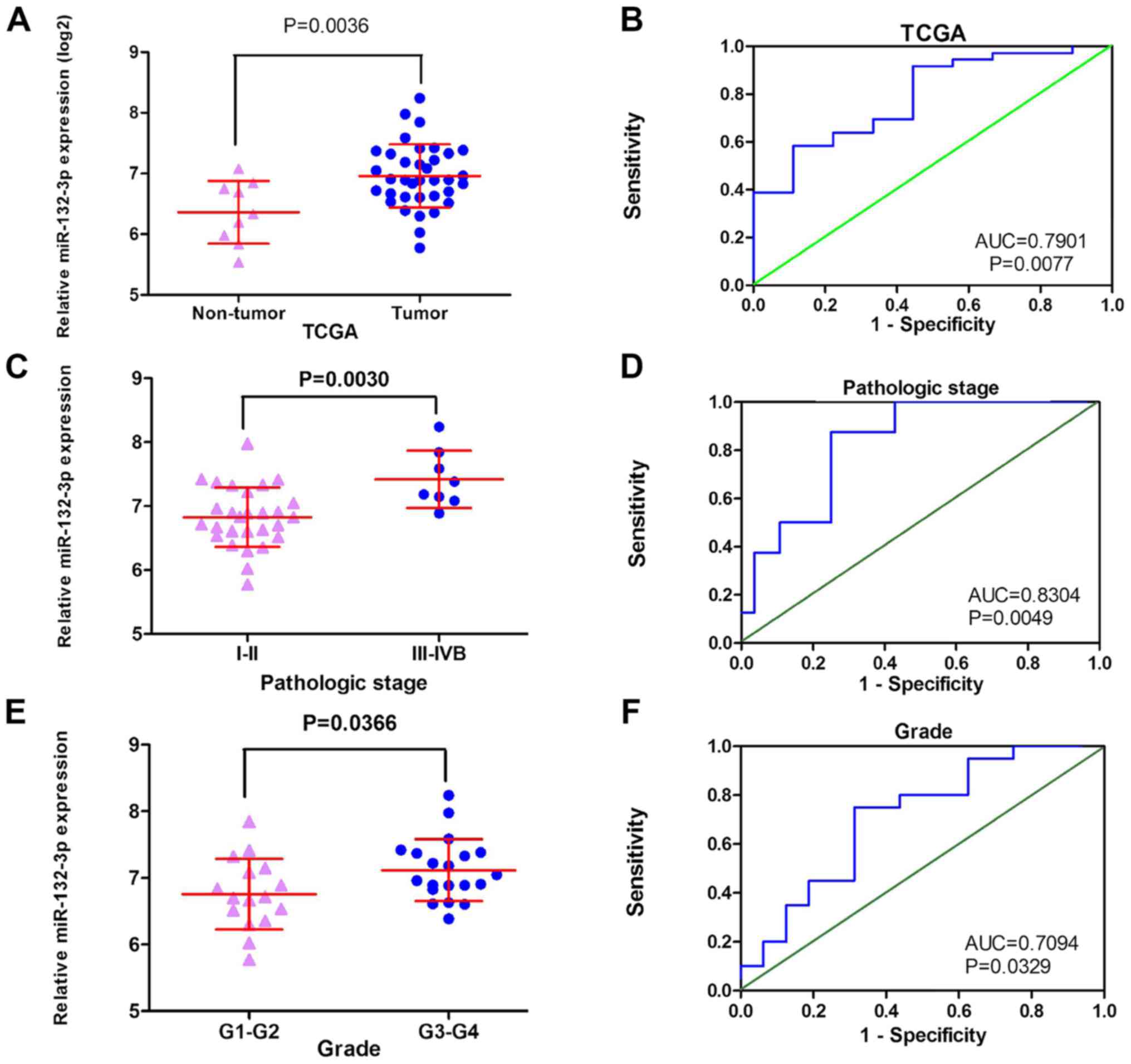
6). TCGA data exhibited a similar tendency, with increased
miR-132-3p expression detected in cancer tissues (6.9586±0.52035)
compared with in non-cancer tissues (6.3613±0.51683; P=0.004;
Fig. 7A and B). Notably, the
expression of miR-132-3p was significantly different in each
pathological stage and grade of CCA. The expression levels of
miR-132-3p in the early stage of CCA (I–II) were markedly lower
than those observed in patients with advanced stage (III–IVB)
(6.8754±0.5279 vs. 7.3034±0.3267; P=0.003) (Fig. 7C and D). Consistently, miR-132-3p
expression in low-grade CCA (G1-G2) was 6.7581±0.5297, whereas in
patients with high-grade CCA (G3-G4) it was 7.1191±0.4651 (P=0.037;
Fig. 7E and F; Table II). We have attempted to collect
more data concerning the relationships between miR-132-3p level and
the progression of CCA; however, no sufficient data were available
from the literature or GEO/ArrayExpress/SRA datasets.
 | Table II.Relationship between miR-132-3p
expression and clinicopathological features in patients with CCA
based on The Cancer Genome Atlas data. |
Table II.
Relationship between miR-132-3p
expression and clinicopathological features in patients with CCA
based on The Cancer Genome Atlas data.
| Clinicopathological
feature | n | miR-132-3p
expression log2 (total_TPM +1), mean ± standard deviation | t | P-value |
|---|
| Tissue |
|
| −3.084 | 0.004 |
|
CCA | 36 | 6.9586±0.5204 |
|
|
|
Non-tumor | 9 | 6.3613±0.5168 |
|
|
| Sex |
|
| −0.356 | 0.724 |
|
Male | 16 | 6.9236±0.5276 |
|
|
|
Female | 20 | 6.9866±0.1177 |
|
|
| Age, years |
|
| −1.504 | 0.142 |
|
<60 | 12 | 7.1398±0.4039 |
|
|
|
≥60 | 24 | 6.8680±0.5553 |
|
|
| Smoking status |
|
| −0.054 | 0.958 |
| No | 22 | 6.9617±0.6113 |
|
|
|
Yes | 12 | 6.9719±0.3962 |
|
|
| T stage |
|
| −1.462 | 0.153 |
| T1 | 19 | 6.8406±0.3793 |
|
|
|
T2-T3 | 17 | 7.0905±0.6289 |
|
|
| N stage |
|
| −1.411 | 0.167 |
| N0 | 26 | 6.8838±0.5470 |
|
|
|
N1-NX | 10 | 7.1532±0.4042 |
|
|
| M stage |
|
| −0.738 | 0.465 |
| M0 | 28 | 6.9242±0.5332 |
|
|
|
M1-MX | 8 | 7.0792±0.4856 |
|
|
| Pathological
stage |
|
| −2.039 | 0.003 |
|
I–II | 29 | 6.8754±0.5279 |
|
|
|
III–IVB | 7 | 7.3034±0.3267 |
|
|
| Grade |
|
| −2.176 | 0.037 |
|
G1-G2 | 16 | 6.7581±0.5297 |
|
|
|
G3-G4 | 20 | 7.1191±0.4651 |
|
|
GO and WikiPathway analysis of
miR-132-3p in CCA
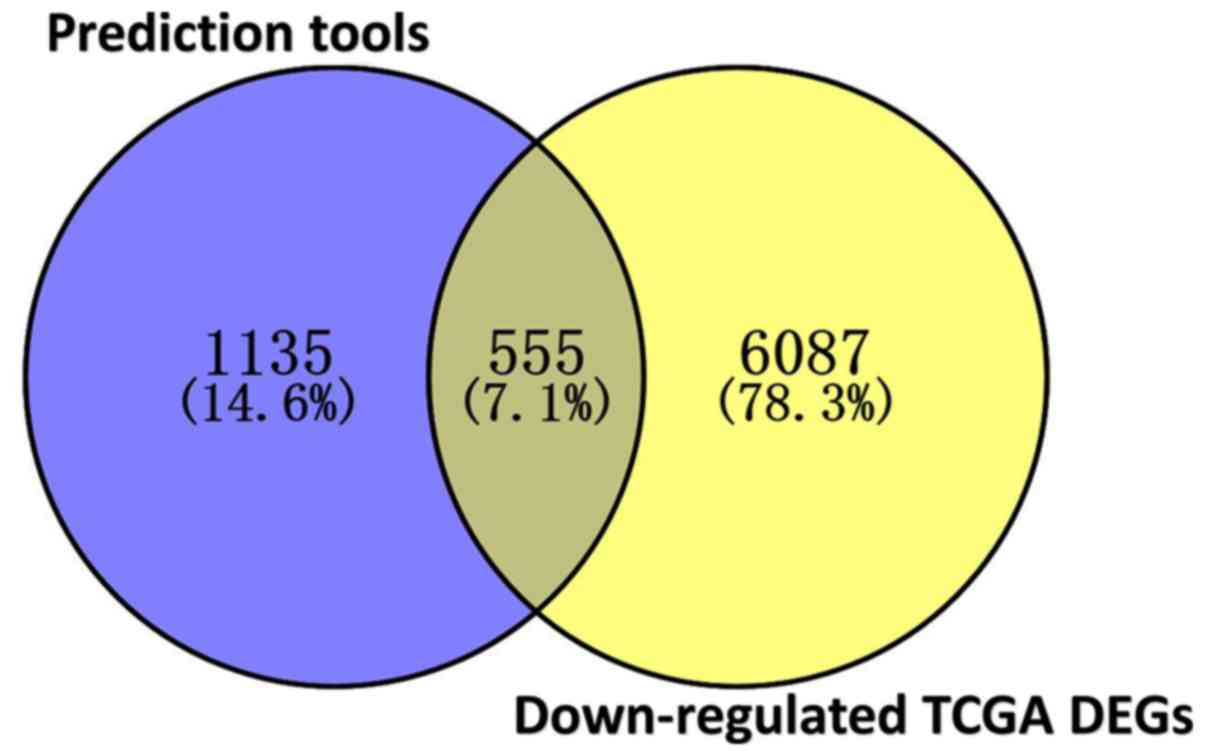
In total, 3,309 differentially expressed genes in
CCA were identified, including 1,619 upregulated and 1,690
downregulated genes. Since miR-132-3p is highly expressed in CCA,
downregulated genes are more likely to be direct targets of
miR-132-3p. Due to the absence of data on the protein levels of the
predicted genes, the current study focused on genes influenced by
miR-132-3p at the mRNA level. In total, 555 overlapped genes were
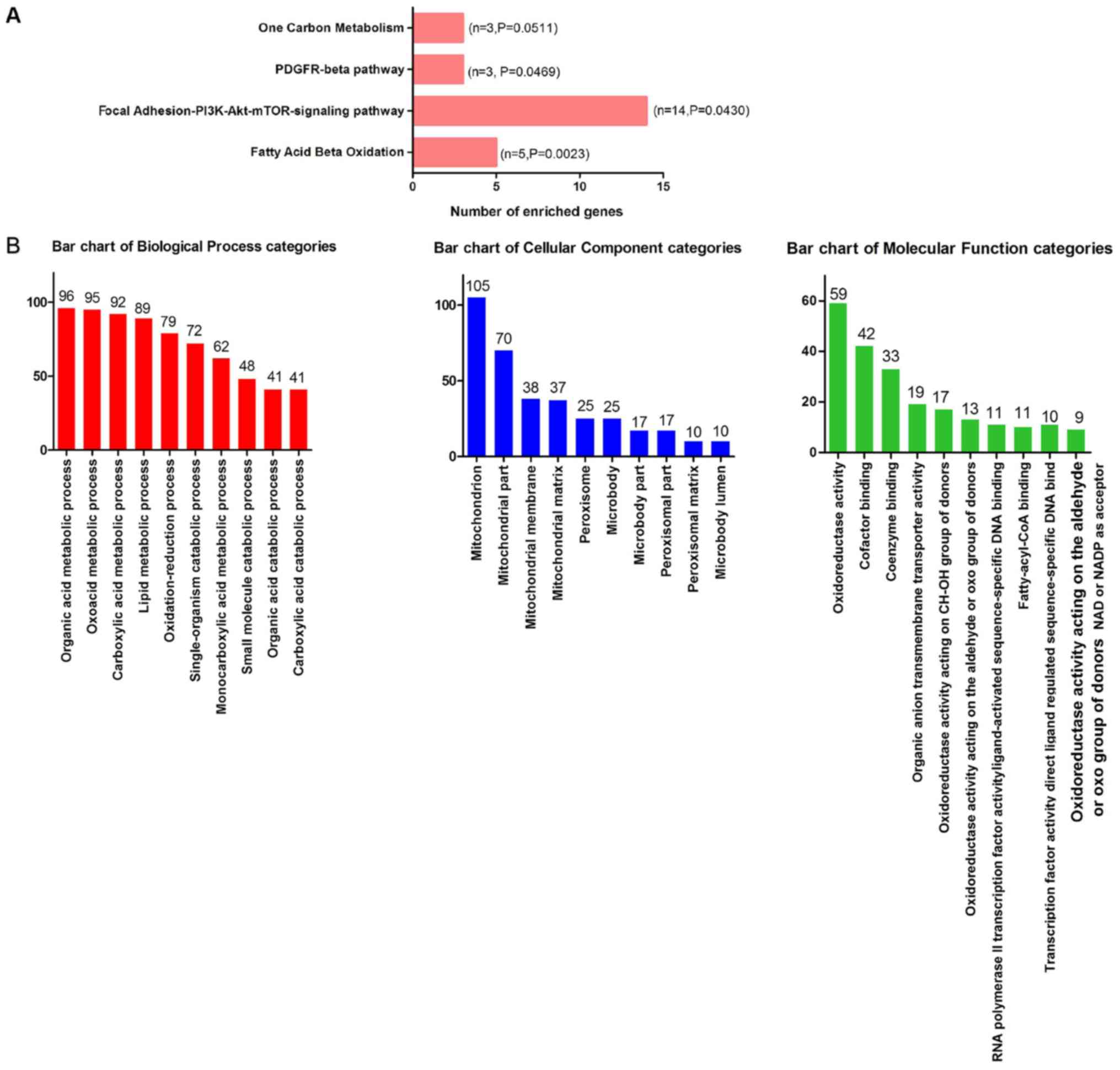
obtained, including downregulated and predicted genes (Fig. 8). The outcome of WikiPathway
analysis revealed that the targets of miR-132-3p were mainly
enriched in the ‘Focal Adhesion-PI3K-Akt-mTOR-signaling pathway’,
which is a cancer-associated pathway, and a total of 14 target
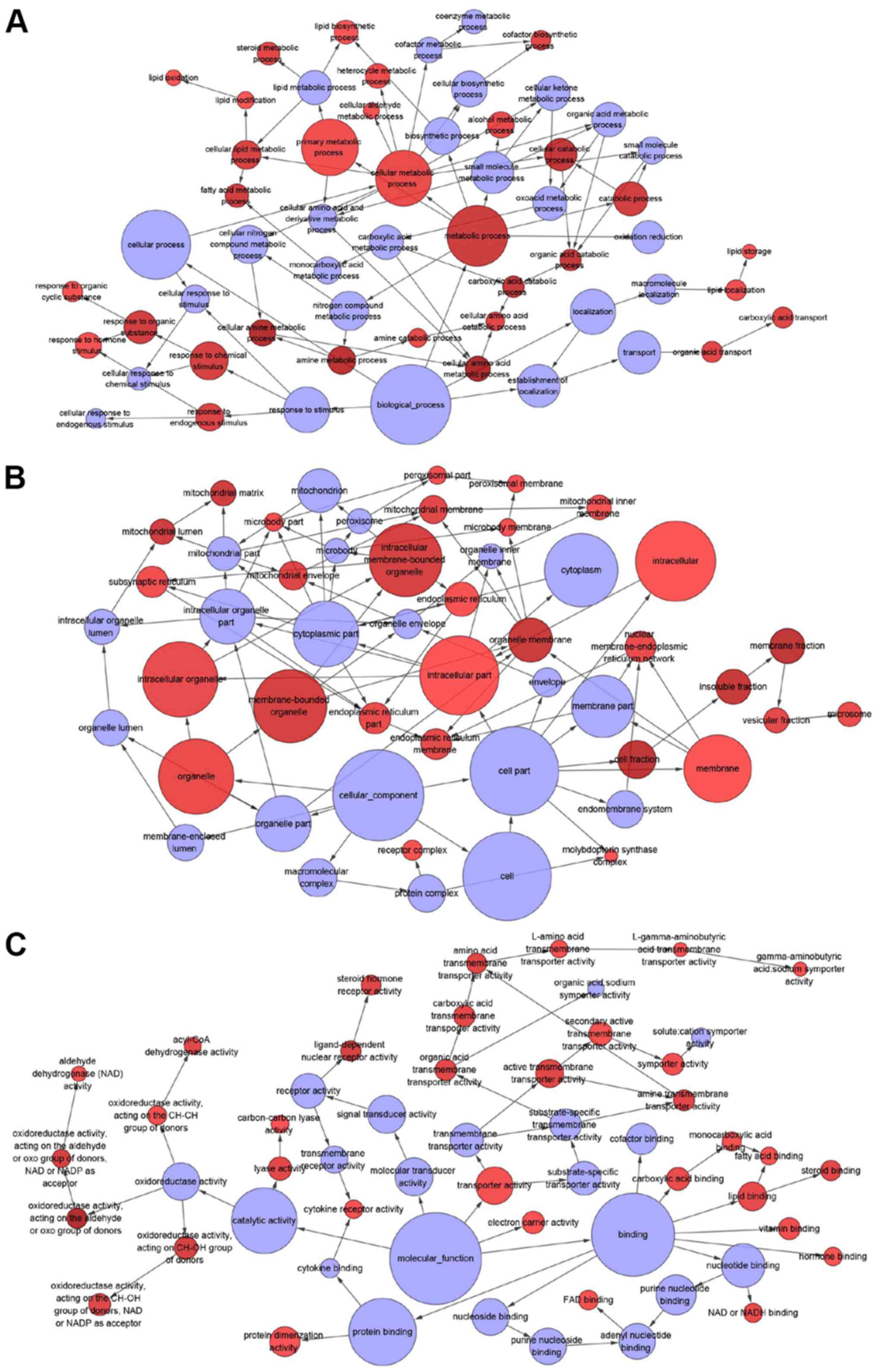
genes were enriched in this pathway (Figs. 9 and 10A). In terms of GO annotation, the
target genes of miR-132-3p were most significantly enriched in
‘organic acid metabolic process’, ‘mitochondrion’ and ‘cofactor
binding’ (Figs. 10B and 11; Table
III).
 | Table III.Pathways related to microRNA-132-3p
in cholangiocarcinoma. |
Table III.
Pathways related to microRNA-132-3p
in cholangiocarcinoma.
| Geneset | Description | Count | P-value |
|---|
| WikiPathway |
|
|
|
|
WP143 | Fatty Acid β
Oxidation | 5 | 0.002 |
|
WP3932 | Focal
Adhesion-PI3K-Akt-mTOR-signaling pathway | 14 | 0.043 |
|
WP3972 | PDGFR-β
pathway | 3 | 0.046 |
| Biological
process |
|
|
|
|
GO:0006082 | Organic acid
metabolic process | 96 | <0.001 |
|
GO:0016054 | Organic acid
catabolic process | 41 | <0.001 |
|
GO:0019752 | Carboxylic acid
metabolic process | 92 | <0.001 |
|
GO:0032787 | Monocarboxylic acid
metabolic process | 62 | <0.001 |
|
GO:0043436 | Oxoacid metabolic
process | 95 | <0.001 |
|
GO:0044282 | Small molecule
catabolic process | 48 | <0.001 |
|
GO:0046395 | Carboxylic acid
catabolic process | 41 | <0.001 |
|
GO:0055114 | Oxidation-reduction
process | 79 | <0.001 |
|
GO:0044712 | Single-organism
catabolic process | 72 | <0.001 |
|
GO:0006629 | Lipid metabolic
process | 89 | <0.001 |
| Cellular
component |
|
|
|
|
GO:0005739 | Mitochondrion | 105 | <0.001 |
|
GO:0044429 | Mitochondrial
part | 70 | <0.001 |
|
GO:0005777 | Peroxisome | 25 | <0.001 |
|
GO:0042579 | Microbody | 25 | <0.001 |
|
GO:0005759 | Mitochondrial
matrix | 37 | <0.001 |
|
GO:0044438 | Microbody part | 17 | <0.001 |
|
GO:0044439 | Peroxisomal
part | 17 | <0.001 |
|
GO:0005782 | Peroxisomal
matrix | 10 | <0.001 |
|
GO:0031907 | Microbody
lumen | 10 | <0.001 |
|
GO:0031966 | Mitochondrial
membrane | 38 | <0.001 |
| Molecular
function |
|
|
|
|
GO:0048037 | Cofactor
binding | 42 | <0.001 |
|
GO:0050662 | Coenzyme
binding | 33 | <0.001 |
|
GO:0016491 | Oxidoreductase
activity | 59 | <0.001 |
|
GO:0016903 | Oxidoreductase
activity, acting on the aldehyde or oxo group of donors | 13 | <0.001 |
|
GO:0000062 | Fatty-acyl-CoA
binding | 10 | <0.001 |
|
GO:0004879 | RNA polymerase II
transcription factor activity, ligand-activated sequence-specific
DNA binding | 11 | <0.001 |
|
GO:0098531 | Transcription
factor activity, directligand regulated sequence-specific DNA
binding | 11 | <0.001 |
|
GO:0016614 | Oxidoreductase
activity, acting on CH-OH group of donors | 17 | <0.001 |
|
GO:0016620 | Oxidoreductase
activity, acting on the aldehyde or oxo group of donors, NAD or
NADP as acceptor | 9 | <0.001 |
|
GO:0008514 | Organic anion
transmembrane transporter activity | 19 | <0.001 |
PPI network and validation of hub
genes
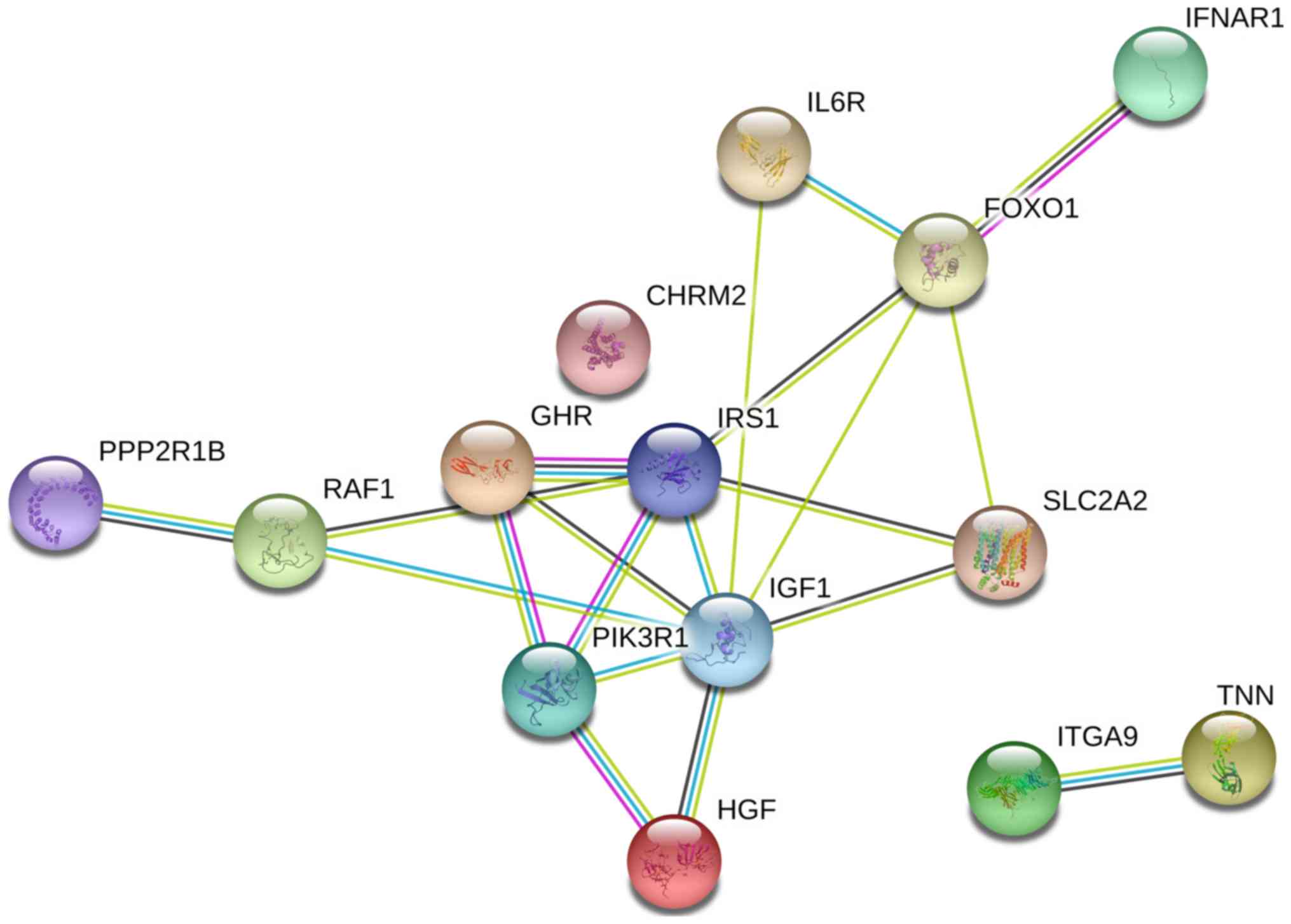
The PPI network constructed in the present study is
shown in Fig. 12. Genes that were
enriched in the ‘Focal Adhesion-PI3K-Akt-mTOR-signaling pathway’
were selected as hub genes for further analysis. In total, 14 hub
genes were selected, including CHRM2, FOXO1, GHR, HGF, IFNAR1,
IGF1, IL6R, IRS1, ITGA9, PIK3R1, PPP2R1B, RAF1, TNN and SLC2A2. Of
these, 13 genes (CHRM2, FOXO1, GHR, HGF, IGF1, IL6R, IRS1, ITGA9,
PIK3R1, PPP2R1B, RAF1, TNN and SLC2A) were downregulated in CCA
tissues compared with non-tumor tissues based on the TCGA and GTEx
mRNA expression data (Fig. 13).
These 14 hub genes were further validated at the protein expression
level in The Human Protein Atlas. The results indicated that the
expression levels of 10 hub genes were markedly reduced in CCA
tissues compared with in normal tissues, whereas three hub genes
(FOXO1, IFNAR1 and PPP2R1B) were upregulated or not differentially
expressed in CCA tissues. However, the protein expression levels of
IL6R in CCA tissues were not available in The Human Protein Atlas
(Fig. 14). Furthermore, since the
sample size was not sufficient to perform further statistical
analyses, additional samples were required for verification of the
present findings in future studies. In the present study, only
genes with low expression levels were considered potential targets
of miR-132-3p in CCA. Upon evaluating the binding site of
miR-132-3p in the hub genes, it was observed that miR-132-3p binds
to the 3′-UTR of six hub target genes (GHR, HGF, IGF1, IRS1, ITGA9
and PIK3R1), whereas four genes (CHRM2, RAF1, TNN and SLC2A2) are
not considered targets of miR-132-3p due to lacking the binding
site for miR-132-3p in their 3′-UTR (Table IV). Therefore, these six hub genes
with low expression levels were selected for further analysis. The
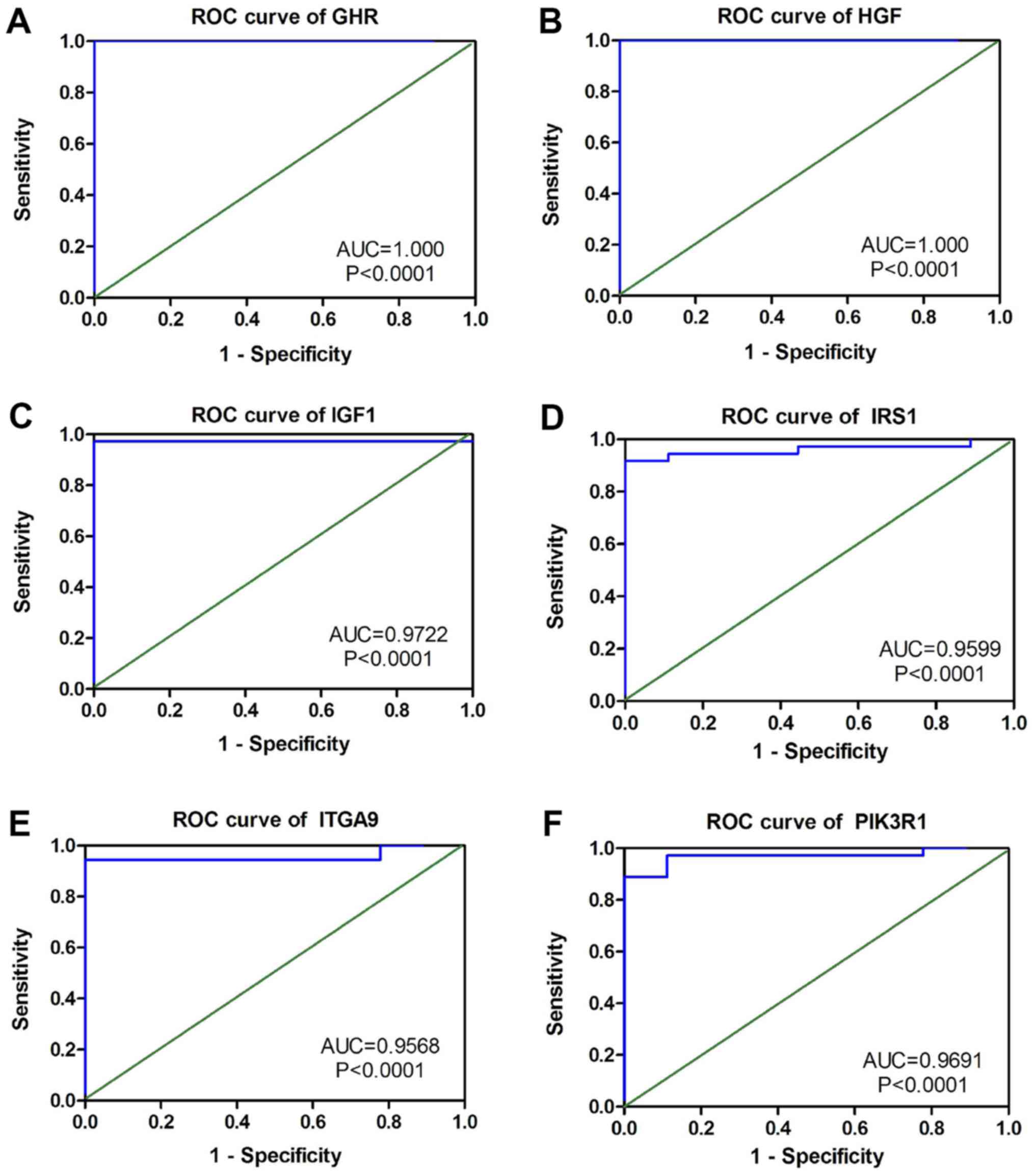
ROC of these six hub genes revealed that all could serve as
biomarkers to distinguish CCA tissues from normal tissues with high
sensitivity and specificity (Fig.
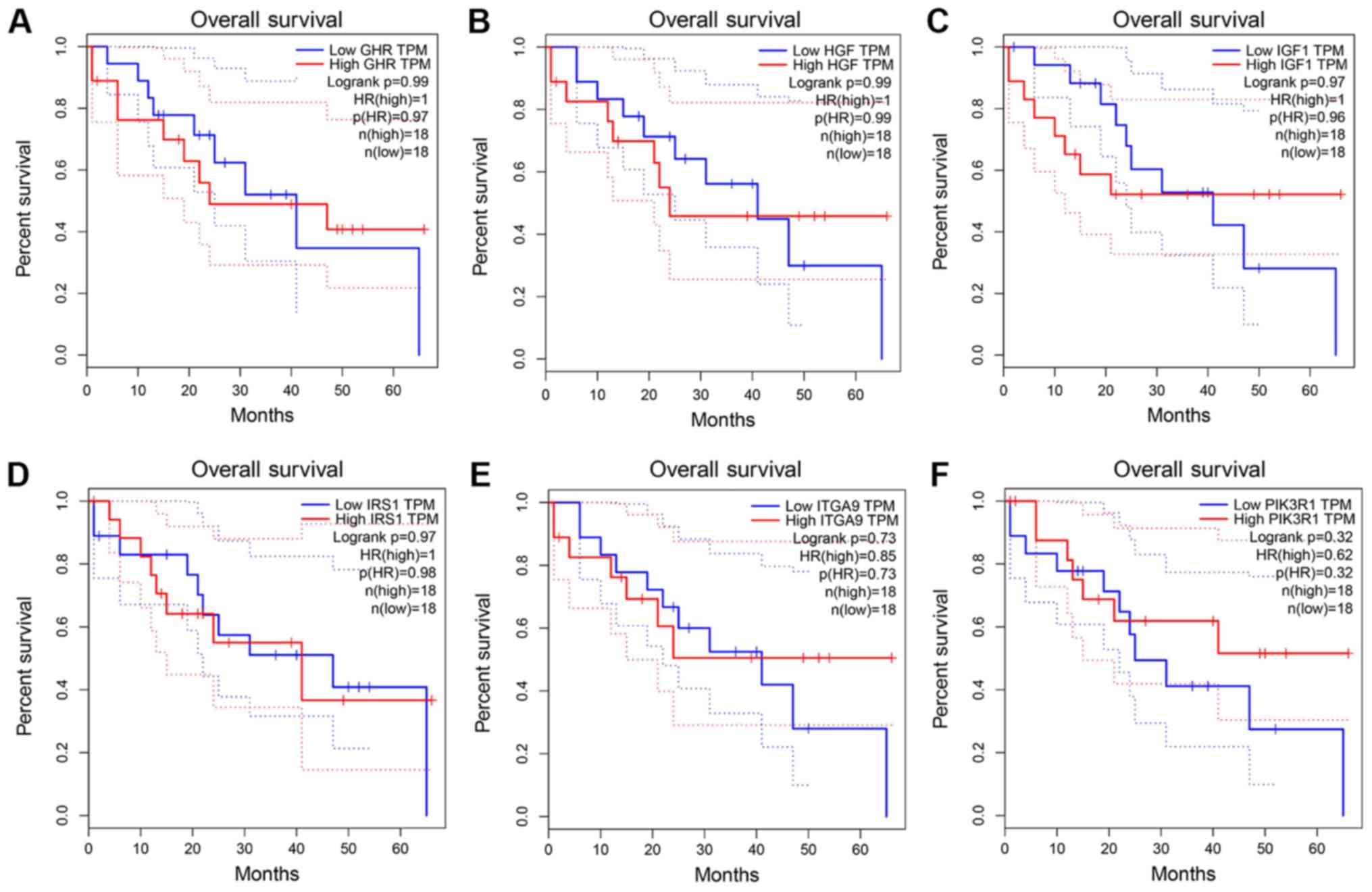
15). However, there were no statistically significant
differences in the survival analysis of these genes (Fig. 16).
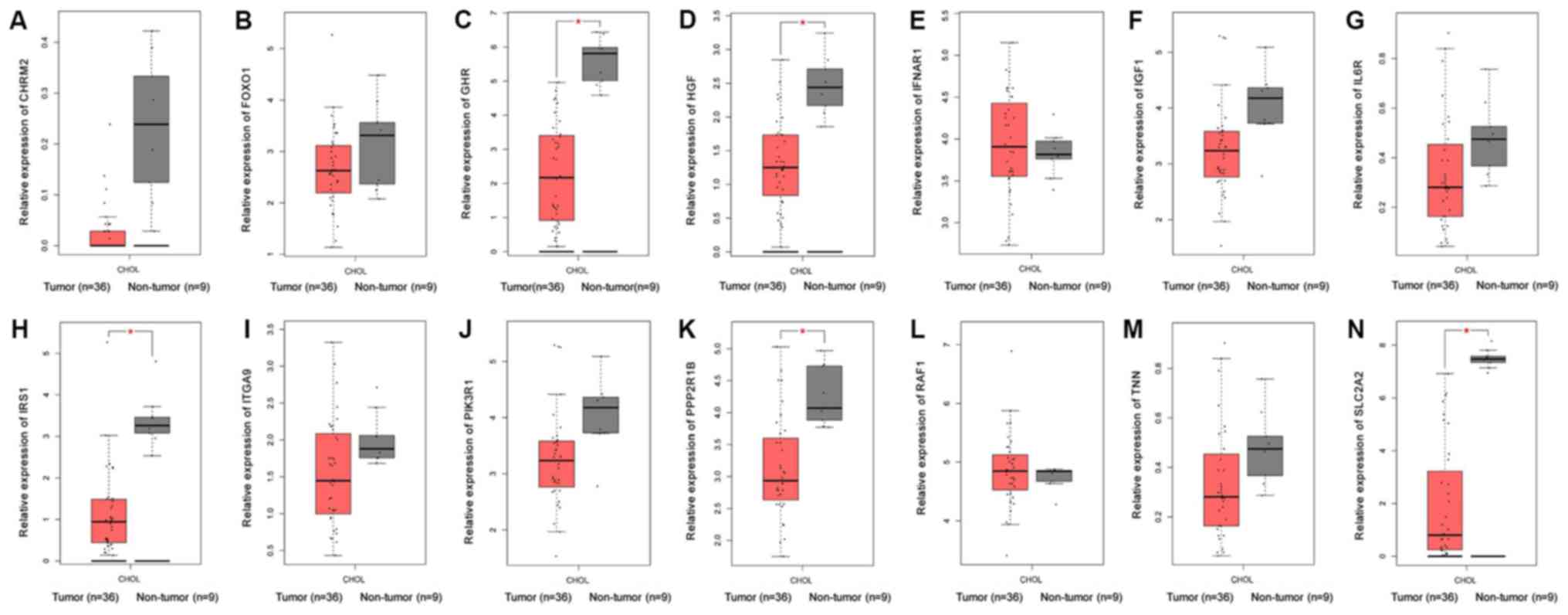 | Figure 13.Validation of 14 hub genes at the
mRNA level based on The Cancer Genome Atlas data. (A) CHRM2, (B)
FOXO1, (C) GHR, (D) HGF, (E) IFNAR1, (F) IGF1, (G) IL6R, (H) IRS1,
(I) ITGA9, (J) PIK3R1, (K) PPP2R1B, (L) RAF1, (M) TNN and (N)
SLC2A2. |
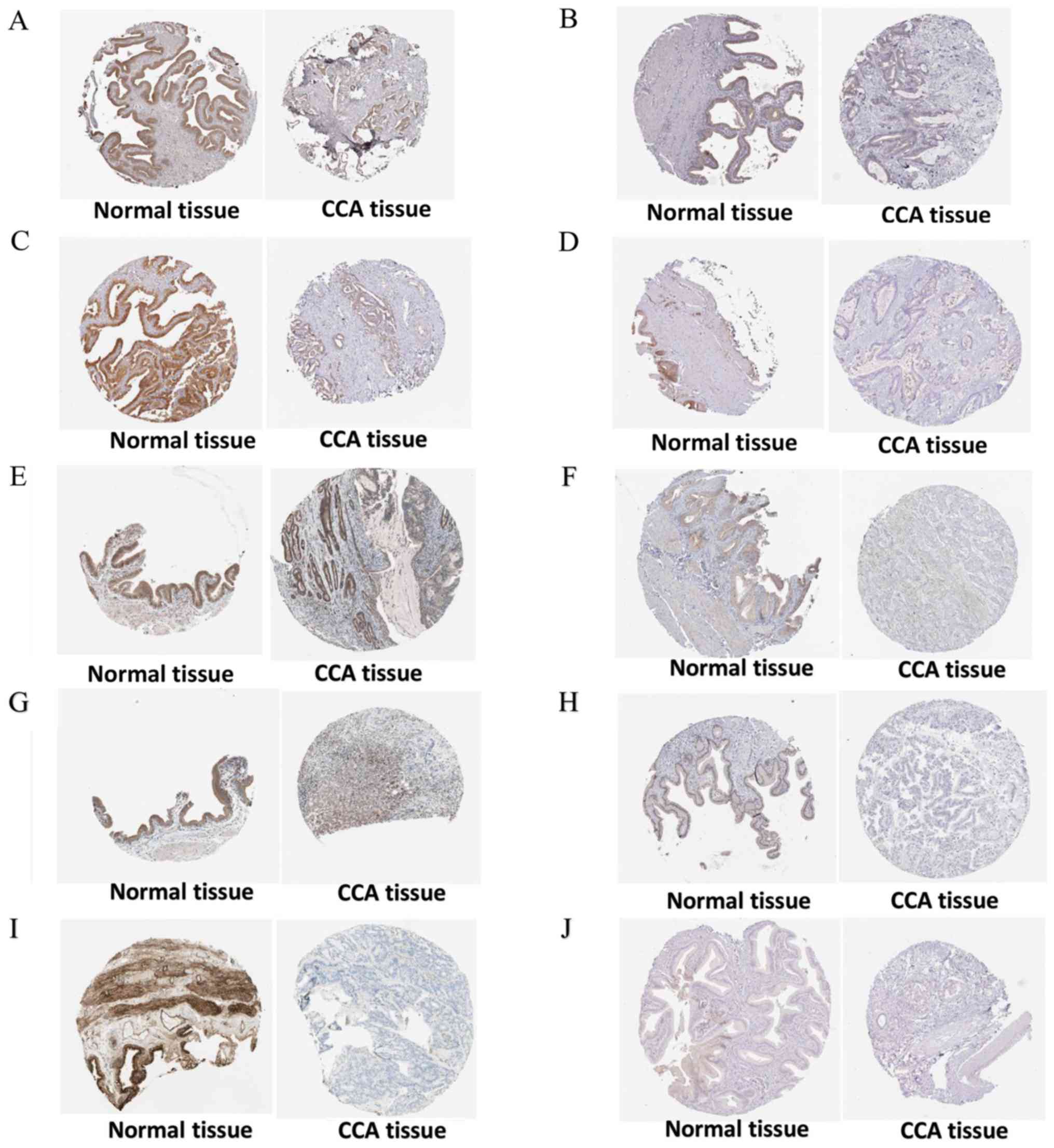 | Figure 14.Validation of 10 hub genes at the
protein level based on The Human Protein Atlas data. (A) CHRM2,
available from: https://www.proteinatlas.org/ENSG00000181072-CHRM2/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000181072-CHRM2/tissue/gallbladder.
(B) GHR, available from: https://www.proteinatlas.org/ENSG00000112964-GHR/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000112964-GHR/tissue/gallbladder.
(C) HGF, available from: https://www.proteinatlas.org/ENSG00000019991-HGF/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000019991-HGF/tissue/gallbladder.
(D) IGF1, available from: https://www.proteinatlas.org/ENSG00000017427-IGF1/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000017427-IGF1/tissue/gallbladder.
(E) IRS1, available from: https://www.proteinatlas.org/ENSG00000169047-IRS1/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000169047-IRS1/tissue/gallbladder.
(F) ITGA9, available from: https://www.proteinatlas.org/ENSG00000144668-ITGA9/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000144668-ITGA9/tissue/gallbladder.
(G) PIK3R1, available from: https://www.proteinatlas.org/ENSG00000145675-PIK3R1/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000145675-PIK3R1/tissue/gallbladder.
(H) RAF1, available from: https://www.proteinatlas.org/ENSG00000132155-RAF1/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000132155-RAF1/tissue/gallbladder.
(I) TNN, available from: https://www.proteinatlas.org/ENSG00000120332-TNN/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000120332-TNN/tissue/gallbladder.
(J) SLC2A2, available from: https://www.proteinatlas.org/ENSG00000163581-SLC2A2/pathology/tissue/liver+cancer;
https://www.proteinatlas.org/ENSG00000163581-SLC2A2/tissue/gallbladder. |
 | Table IV.Binding sites for miR-132-3p of six
target genes. |
Table IV.
Binding sites for miR-132-3p of six
target genes.
| Target | miR-132-3p target
region (3′-untranslated region) | Seed match | Context score
percentile | Conserved branch
length | Pct |
|---|
| GHR | 234–240 | 7mer-m8 | 95 | 5.527 | 0.54 |
| HGF | 3,387-3,393 | 7mer-m8 | 92 | 1.625 | <0.1 |
| IGF1 | 6,684-6,690 | 7mer-1A | 44 | 0.775 | <0.1 |
| IRS1 | 1,875-1,881 | 7mer-m8 | 65 | 2.010 | <0.1 |
| ITGA9 | 292–298 | 7mer-1A | 44 | 5.855 | 0.29 |
| PIK3R1 | 1,083-1,089 | 7mer-m8 | 82 | 0.154 | <0.1 |
Discussion
To the best of our knowledge, the expression levels
and potential target genes of miR-132-3p in CCA have not been
investigated to date. The present study, by combining RT-qPCR,
miRNA-microarray and RNA-sequencing data, demonstrated that
miR-132-3p was significantly upregulated in 321 CCA tissues
compared with in 253 non-tumor controls. Furthermore, higher levels
of miR-132-3p were significantly associated with CCA initiation and
progression, which may be in part due to multiple target genes and
signaling pathways.
miR-132-3p has been reported to be differentially
expressed in numerous diseases. Notably, it is highly expressed in
sural nerve biopsies from patients with neuropathy exhibiting
neuropathic pain compared with those without pain (41), as well as in glioma tissues
(42) and pancreatic ductal
adenocarcinoma tissues (18).
Conversely, miR-132-3p downregulation has been reported in the gray
matter of Alzheimer's disease samples (43), mesothelioma (44) and gastric cancer tissues (19). Downregulated miR-132-3p may be a
promising novel diagnostic biomarker for malignant mesothelioma
with a sensitivity and specificity of 86 and 61%, respectively
(35). Therefore, miR-132-3p may
serve distinct roles in different diseases. To the best of our
knowledge, the present study is the first to detect overexpression
of miR-132-3p in CCA tissues compared with in non-cancerous
tissues, indicating that miR-132-3p may participate in the
tumorigenesis of CCA. This phenomenon is similar to what has been
documented in pancreatic ductal adenocarcinoma. Besides its
function in the carcinogenesis of various types of cancer,
miR-132-3p is associated with the development of tumors. A previous
study reported that the single nucleotide polymorphism rs1599795 in
CD80 3′-UTR contributed to the occurrence of gastric cancer through
disrupting the regulatory role of miR-132-3p, miR-212-3p and
miR-361-5p in CD80 expression (45). Notably, in the current study,
miR-132-3p upregulation was revealed to contribute to the
progression of CCA, as it was closely associated with clinical
stage and tumor differentiation. Therefore, miR-132-3p
overexpression may lead to the occurrence of CCA and may accelerate
disease progression.
The clinical role of a miRNA depends on its specific
targets. Regarding the target candidates of miR-132-3p, only a few
genes have been identified thus far. In breast cancer, miR-132-3p
contributes to the post-transcriptional regulation of BCRP/ABCG2
(8,17). In pancreatic ductal adenocarcinoma,
overexpressed miR-132-3p may regulate executioner caspase-7 and
contribute to malignant progression (18). In human osteosarcoma, miR-132-3p is
regulated by the long non-coding RNA (lncRNA) taurine up-regulated
1, and may promote proliferation and suppresses apoptosis in
osteosarcoma cell lines (46). In
colorectal cancer, miR-132-3p can be targeted by the lncRNA X
inactive specific transcript (47). To the best of our knowledge, no
target gene of miR-132-3p in CCA has been studied to date.
According to the outcome of bioinformatics analysis, the present
study revealed that the potential target genes of miR-132-3p in CCA
were principally enriched in ‘Fatty Acid β Oxidation’, ‘Focal
Adhesion-PI3K-Akt-mTOR-signaling pathway’ and ‘PDGFR-β pathway’,
based on WikiPathway cancer analysis. Of these, the current study
mainly focused on the ‘Focal Adhesion-PI3K-Akt-mTOR-signaling
pathway’, which has been reported to be associated with CCA. Focal
adhesion has been reported to be involved in CCA progression and
metastasis (20,21,26,27,48–51),
whereas the PI3K-Akt-mTOR signaling pathway serves an essential
role in regulating cell survival and proliferation in unresectable
and liver metastases of pancreatic cancer; notably, inhibition of
the PI3K/Akt/mTOR signaling pathway may serve as a promising
therapeutic strategy in the treatment of intrahepatic
cholangiocarcinoma (52).
According to previous studies, multiple molecules or drugs serve
vital roles in CCA via the mTOR signaling pathway, including Fyn
(53), compound C (54), sorafenib (55) and c-Myc (56). In terms of GO annotation, the
overlapped genes were principally enriched in ‘organic acid
metabolic process’, ‘mitochondrion’ and ‘coenzyme binding’, which
indicates that the targets of miR-132-3p may participate in
multiple steps of the metabolism of tumor cells.
According to previously published studies, the six
hub target genes identified in the current study are closely
associated with numerous types of cancer. GHR has been verified to
be involved in triple-negative breast cancer (57) and prostate cancer (58). HGF may participate in the
regulation of neuropilin-1, and may be involved in the growth and
metastasis of CCA cells (50). In
addition, activation of HGF-c-MET signaling is involved in cell
invasiveness and induces metastasis of biliary tract cancer
(59). IGF1, which is involved in
the regulation of Yes-associated protein, is associated with the
progression of CCA (60). IRS1 may
be a target of miR-664 and serves a role in suppressing cell
proliferation and invasion in breast cancer (61). Furthermore, IRS1 may also be
targeted by miR-497, thus inhibiting the tumor growth of colorectal
cancer (62). A previous study
reported that upregulation of ITGA9 in response to a decrease in
miR-125b levels in metastatic melanoma is responsible for melanoma
tumor cell migration and invasion (63). In addition, ITGA9 is associated
with other types of cancer, including nasopharyngeal carcinoma
(64), breast cancer (65) and colorectal cancer (66). PIK3R1 is a target of miR-29b and
miR-221, which may enhance chemosensitivity to gemcitabine in HuH28
human CCA cells (67). According
to the current findings, these hub genes may be the target genes of
miR-132-3p in CCA and may exert different functions; this requires
further verification in future studies.
The present study has several limitations. Firstly,
the sample size used for RT-qPCR testing was relatively small,
which may decrease the accuracy of the conclusions. Secondly, the
role of miR-132-3p in progression and survival requires further
study. Thirdly, the potential target genes of miRNA-132-3p were
only initially verified by their expression levels; therefore,
further in vitro or in vivo experiments are
necessary.
In conclusion, upregulation of miR-132-3p may serve
a pivotal role in the tumorigenesis and progression of CCA by
targeting different pathways. Further studies are required to
support the current findings.
Acknowledgements
Not applicable.
Funding
The study was supported by the Promoting Project of
Basic Capacity for Young and Middle-aged University Teachers in
Guangxi (grant no. 2017KY0111), Innovation Project of Guangxi
Graduate Education (grant no. YCBZ2017045) and the National Natural
Science Foundation of China (grant no. 31760319).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the TCGA (http://cancergenome.nih.gov/), the GEO (https://www.ncbi.nlm.nih.gov/geo/) and the SRA
(https://www.ncbi.nlm.nih.gov/sra/)
data portals. The RT-qPCR data from the present study can be
acquired from the correspondence author on reasonable request.
Authors' contributions
HYW performed RT-qPCR, analyzed and interpreted
data, and drafted the manuscript. SX, AGL, MDW, ZBC, YXL, YH, MJL,
QPH and SLP analyzed data from microarrays and miRNA
RNA-sequencing, and participated in all data processing, RT-qPCR
and paper draft writing. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
First Affiliated Hospital, Guangxi Medical University, China.
Informed written consent was obtained from all patients
participating in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chng KR, Chan SH, Ng AHQ, Li C, Jusakul A,
Bertrand D, Wilm A, Choo SP, Tan DMY, Lim KH, et al: Tissue
microbiome profiling identifies an enrichment of specific enteric
bacteria in opisthorchis viverrini associated cholangiocarcinoma.
EBioMedicine. 8:195–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Verathamjamras C, Weeraphan C,
Chokchaichamnankit D, Watcharatanyatip K, Subhasitanont P,
Diskul-Na-Ayudthaya P, Mingkwan K, Luevisadpaibul V,
Chutipongtanate S, Champattanachai V, et al: Secretomic profiling
of cells from hollow fiber bioreactor reveals PSMA3 as a potential
cholangiocarcinoma biomarker. Int J Oncol. 51:269–280. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rizvi S, Khan SA, Hallemeier CL, Kelley RK
and Gores GJ: Cholangiocarcinoma-evolving concepts and therapeutic
strategies. Nat Rev Clin Oncol. 15:95–111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou G, Yang Z, Wang X, Tao R and Zhou Y:
TRAIL enhances shikonin induced apoptosis through ROS/JNK signaling
in cholangiocarcinoma cells. Cell Physiol Biochem. 42:1073–1086.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mustafa MZ, Nguyen VH, Le Naour F, De
Martin E, Beleoken E, Guettier C, Johanet C, Samuel D,
Duclos-Vallee JC and Ballot E: Autoantibody signatures defined by
serological proteome analysis in sera from patients with
cholangiocarcinoma. J Transl Med. 14:172016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Su Z, Liu G, Fang T, Zhang K, Yang S,
Zhang H, Wang Y, Lv Z and Liu J: Expression and prognostic value of
glutamate dehydrogenase in extrahepatic cholangiocarcinoma
patients. Am J Transl Res. 9:2106–2118. 2017.PubMed/NCBI
|
|
7
|
Saha SK, Zhu AX, Fuchs CS and Brooks GA:
Forty-year trends in cholangiocarcinoma incidence in the U.S.:
Intrahepatic disease on the rise. Oncologist. 21:594–599. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ji W, Jiao J, Cheng C and Shao J:
MicroRNA-21 in the pathogenesis of traumatic brain injury.
Neurochem Res. 43:1863–1868. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang X, Hu S, Liu Q, Qian C, Liu Z and
Luo D: Exosomal microRNA remodels the tumor microenvironment.
PeerJ. 5:e41962017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun X, Wang M, Liu H and Wang J:
MicroRNA-423 enhances the invasiveness of hepatocellular carcinoma
via regulation of BRMS1. Am J Transl Res. 9:5576–5584.
2017.PubMed/NCBI
|
|
11
|
Wei Q, Liu H, Ai Z, Wu Y, Liu Y, Shi Z,
Ren X and Guo Z: SC1 promotes MiR124-3p expression to maintain the
self-renewal of mouse embryonic stem cells by inhibiting the
MEK/ERK pathway. Cell Physiol Biochem. 44:2057–2072. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Othman N and Nagoor NH: miR-608 regulates
apoptosis in human lung adenocarcinoma via regulation of AKT2. Int
J Oncol. 51:1757–1764. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun C, Zhu J, Wu B, Chen J, Zhu Z, Cai P,
Guo W, Gu Z, Wang J and Huang S: Diagnostic and prognostic value of
microRNAs in cholangiocarcinoma: A systematic review and
meta-analysis. Cancer Manag Res. 10:2125–2139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang Q, Liu L, Liu CH, You H, Shao F, Xie
F, Lin XS, Hu SY and Zhang CH: MicroRNA-21 regulates the invasion
and metastasis in cholangiocarcinoma and may be a potential
biomarker for cancer prognosis. Asian Pac J Cancer Prev.
14:829–834. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu YF, Li ZR, Cheng ZQ, Yin XM and Wu JS:
Decrease of miR-622 expression promoted the proliferation,
migration and invasion of cholangiocarcinoma cells by targeting
regulation of c-Myc. Biomed Pharmacother. 96:7–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Shen J, Chan MT and Wu WK: The role
of microRNAs in intrahepatic cholangiocarcinoma. J Cell Mol Med.
21:177–184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reustle A, Fisel P, Renner O, Büttner F,
Winter S, Rausch S, Kruck S, Nies AT, Hennenlotter J, Scharpf M, et
al: Characterization of the breast cancer resistance protein
(BCRP/ABCG2) in clear cell renal cell carcinoma. Int J Cancer.
143:3181–3193. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park JK, Doseff AI and Schmittgen TD:
MicroRNAs targeting caspase-3 and −7 in PANC-1 cells. Int J Mol
Sci. 19:E12062018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang T, Liu C, Huang S, Ma Y, Fang J and
Chen Y: A downmodulated microRNA profiling in patients with gastric
cancer. Gastroenterol Res Pract. 2017:15269812017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai Y, Wang W, Guo H, Li H, Xiao Y and
Zhang Y: miR-9-5p, miR-124-3p, and miR-132-3p regulate BCL2L11 in
tuberous sclerosis complex angiomyolipoma. Lab Invest. 98:856–870.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Luo D, Sun H, Qi Y, Xu W, Jin X,
Li C, Lin Z and Li G: MiR-132-3p regulates ADAMTS-5 expression and
promotes chondrogenic differentiation of rat mesenchymal stem
cells. J Cell Biochem. 119:2579–2587. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leinonen R, Sugawara H and Shumway M;
International Nucleotide Sequence Database Collaboration, : The
sequence read archive. Nucleic Acids Res. 39:D19–D21. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parkinson H, Kapushesky M, Shojatalab M,
Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N,
Lilja P, Lukk M, et al: ArrayExpress-a public database of
microarray experiments and gene expression profiles. Nucleic Acids
Res. 35:D747–D750. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gan BL, He RQ, Zhang Y, Wei DM, Hu XH and
Chen G: Downregulation of HOXA3 in lung adenocarcinoma and its
relevant molecular mechanism analysed by RT-qPCR, TCGA and in
silico analysis. Int J Oncol. 53:1557–1579. 2018.PubMed/NCBI
|
|
27
|
Deng Y, He R, Zhang R, Gan B, Zhang Y,
Chen G and Hu X: The expression of HOXA13 in lung adenocarcinoma
and its clinical significance: A study based on the cancer genome
atlas, oncomine and reverse transcription-quantitative polymerase
chain reaction. Oncol Lett. 15:8556–8572. 2018.PubMed/NCBI
|
|
28
|
Liang YY, Huang JC, Tang RX, Chen WJ, Chen
P, Cen WL, Shi K, Gao L, Gao X, Liu AG, et al: Clinical value of
miR-198-5p in lung squamous cell carcinoma assessed using
microarray and RT-qPCR. World J Surg Oncol. 16:222018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nikolayeva O and Robinson MD: edgeR for
differential RNA-seq and ChIP-seq analysis: An application to stem
cell biology. Methods Mol Biol. 1150:45–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gu YL, Rong XX, Wen LT, Zhu GX and Qian
MQ: miR-195 inhibits the proliferation and migration of
chondrocytes by targeting GIT1. Mol Med Rep. 15:194–200. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang N, Shen Q and Zhang P: miR-497
suppresses epithelial-mesenchymal transition and metastasis in
colorectal cancer cells by targeting fos-related antigen-1. Onco
Targets Ther. 9:6597–6604. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chi Y, Cui J, Wang Y, Du W, Chen F, Li Z,
Ma F, Song B, Xu F, Zhao Q, et al: Interferon-γ alters the microRNA
profile of umbilical cordderived mesenchymal stem cells. Mol Med
Rep. 14:4187–4197. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dweep H, Gretz N and Sticht C: miRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Vasaikar S, Shi Z, Greer M and
Zhang B: WebGestalt 2017: A more comprehensive, powerful, flexible
and interactive gene set enrichment analysis toolkit. Nucleic Acids
Res. 45:W130–W137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Z, Zhang G, Gao Z, Li S, Li Z, Bi J,
Liu X, Li Z and Kong C: Comprehensive analysis of differentially
expressed genes associated with PLK1 in bladder cancer. BMC Cancer.
17:8612017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zeng L, Yang K and Ge J: Uncovering the
pharmacological mechanism of astragalus salvia compound on
pregnancy-induced hypertension syndrome by a network pharmacology
approach. Sci Rep. 7:168492017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zeng L, Yang K, Liu H and Zhang G: A
network pharmacology approach to investigate the pharmacological
effects of Guizhi Fuling Wan on uterine fibroids. Exp Ther Med.
14:4697–4710. 2017.PubMed/NCBI
|
|
38
|
Wang A and Zhang G: Differential gene
expression analysis in glioblastoma cells and normal human brain
cells based on GEO database. Oncol Lett. 14:6040–6044.
2017.PubMed/NCBI
|
|
39
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:8.13.1–24. 2014. View Article : Google Scholar
|
|
40
|
Colwill K; Renewable Protein Binder
Working Group, ; Gräslund S: A roadmap to generate renewable
protein binders to the human proteome. Nat Methods. 8:551–558.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leinders M, Üçeyler N, Pritchard RA,
Sommer C and Sorkin LS: Increased miR-132-3p expression is
associated with chronic neuropathic pain. Exp Neurol. 283:276–286.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gu Y, Cai R, Zhang C, Xue Y, Pan Y, Wang J
and Zhang Z: miR-132-3p boosts caveolae-mediated transcellular
transport in glioma endothelial cells by targeting
PTEN/PI3K/PKB/Src/Cav-1 signaling pathway. FASEB J. 33:441–454.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pichler S, Gu W, Hartl D, Gasparoni G,
Leidinger P, Keller A, Meese E, Mayhaus M, Hampel H and
Riemenschneider M: The miRNome of Alzheimer's disease: Consistent
downregulation of the miR-132/212 cluster. Neurobiol Aging. 50:167
e1–e167 e10. 2017. View Article : Google Scholar
|
|
44
|
Weber DG, Gawrych K, Casjens S, Brik A,
Lehnert M, Taeger D, Pesch B, Kollmeier J, Bauer TT, Johnen G and
Brüning T: Circulating miR-132-3p as a candidate diagnostic
biomarker for malignant mesothelioma. Dis Markers.
2017:92801702017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu R, Li F, Zhu J, Tang R, Qi Q, Zhou X,
Li R, Wang W, Hua D and Chen W: A functional variant at miR-132-3p,
miR-212-3p, and miR-361-5p binding site in CD80 gene alters
susceptibility to gastric cancer in a Chinese Han population. Med
Oncol. 31:602014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li G, Liu K and Du X: Long non-coding RNA
TUG1 promotes proliferation and inhibits apoptosis of osteosarcoma
cells by sponging miR-132-3p and upregulating SOX4 expression.
Yonsei Med J. 59:226–235. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song H, He P, Shao T, Li Y, Li J and Zhang
Y: Long non-coding RNA XIST functions as an oncogene in human
colorectal cancer by targeting miR-132-3p. J BUON. 22:696–703.
2017.PubMed/NCBI
|
|
48
|
Phanthaphol N, Techasen A, Loilome W,
Thongchot S, Thanan R, Sungkhamanon S, Khuntikeo N, Yongvanit P and
Namwat N: Upregulation of TCTP is associated with
cholangiocarcinoma progression and metastasis. Oncol Lett.
14:5973–5979. 2017.PubMed/NCBI
|
|
49
|
Sae-Lao T, Luplertlop N, Janvilisri T,
Tohtong R, Bates DO and Wongprasert K: Sulfated galactans from the
red seaweed Gracilaria fisheri exerts anti-migration effect on
cholangiocarcinoma cells. Phytomedicine. 36:59–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhu H, Jiang X, Zhou X, Dong X, Xie K,
Yang C, Jiang H, Sun X and Lu J: Neuropilin-1 regulated by miR-320
contributes to the growth and metastasis of cholangiocarcinoma
cells. Liver Int. 38:125–135. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pak JH, Bashir Q, Kim IK, Hong SJ, Maeng
S, Bahk YY and Kim TS: Clonorchis sinensis excretory-secretory
products promote the migration and invasion of cholangiocarcinoma
cells by activating the integrin β4-FAK/Src signaling pathway. Mol
Biochem Parasitol. 214:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bian JL, Wang MM, Tong EJ, Sun J, Li M,
Miao ZB, Li YL, Zhu BH and Xu JJ: Benefit of everolimus in
treatment of an intrahepatic cholangiocarcinoma patient with a
PIK3CA mutation. World J Gastroenterol. 23:4311–4316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lyu SC, Han DD, Li XL, Ma J, Wu Q, Dong
HM, Bai C and He Q: Fyn knockdown inhibits migration and invasion
in cholangiocarcinoma through the activated AMPK/mTOR signaling
pathway. Oncol Lett. 15:2085–2090. 2018.PubMed/NCBI
|
|
54
|
Zhao X, Luo G, Cheng Y, Yu W, Chen R, Xiao
B, Xiang Y, Feng C, Fu W, Duan C, et al: Compound C induces
protective autophagy in human cholangiocarcinoma cells via
Akt/mTOR-independent pathway. J Cell Biochem. 119:5538–5550. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yokoi K, Kobayashi A, Motoyama H, Kitazawa
M, Shimizu A, Notake T, Yokoyama T, Matsumura T, Takeoka M and
Miyagawa SI: Survival pathway of cholangiocarcinoma via AKT/mTOR
signaling to escape RAF/MEK/ERK pathway inhibition by sorafenib.
Oncol Rep. 39:843–850. 2018.PubMed/NCBI
|
|
56
|
Luo G, Li B, Duan C, Cheng Y, Xiao B, Yao
F, Wei M, Tao Q, Feng C, Xia X, et al: cMyc promotes
cholangiocarcinoma cells to overcome contact inhibition via the
mTOR pathway. Oncol Rep. 38:2498–2506. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Girgert R, Emons G and Gründker C:
Inhibition of growth hormone receptor by Somavert reduces
expression of GPER and prevents growth stimulation of
triple-negative breast cancer by 17β-estradiol. Oncol Lett.
15:9559–9566. 2018.PubMed/NCBI
|
|
58
|
Recouvreux MV, Wu JB, Gao AC, Zonis S,
Chesnokova V, Bhowmick N, Chung LW and Melmed S: Androgen receptor
regulation of local growth hormone in prostate cancer cells.
Endocrinology. 158:2255–2268. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Heo MH, Kim HK, Lee H, Kim KM, Lee J, Park
SH, Park JO, Lim HY, Kang WK, Park YS and Kim ST: The clinical
impact of c-MET over-expression in advanced biliary tract cancer
(BTC). J Cancer. 8:1395–1399. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pei T, Li Y, Wang J, Wang H, Liang Y, Shi
H, Sun B, Yin D, Sun J, Song R, et al: YAP is a critical oncogene
in human cholangiocarcinoma. Oncotarget. 6:17206–17220. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu L, Li Y, Li J and Ma D: MicroRNA-664
targets insulin receptor substrate 1 to suppress cell proliferation
and invasion in breast cancer. Oncol Res. 27:459–467. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xu Y, Chen J, Gao C, Zhu D, Xu X, Wu C and
Jiang J: MicroRNA-497 inhibits tumor growth through targeting
insulin receptor substrate 1 in colorectal cancer. Oncol Lett.
14:6379–6386. 2017.PubMed/NCBI
|
|
63
|
Zhang J, Na S, Liu C, Pan S, Cai J and Qiu
J: MicroRNA-125b suppresses the epithelial-mesenchymal transition
and cell invasion by targeting ITGA9 in melanoma. Tumour Biol.
37:5941–5949. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Nawaz I, Hu LF, Du ZM, Moumad K, Ignatyev
I, Pavlova TV, Kashuba V, Almgren M, Zabarovsky ER and Ernberg I:
Integrin α9 gene promoter is hypermethylated and downregulated in
nasopharyngeal carcinoma. Oncotarget. 6:31493–31507. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mostovich LA, Prudnikova TY, Kondratov AG,
Loginova D, Vavilov PV, Rykova VI, Sidorov SV, Pavlova TV, Kashuba
VI, Zabarovsky ER and Grigorieva EV: Integrin alpha9 (ITGA9)
expression and epigenetic silencing in human breast tumors. Cell
Adh Migr. 5:395–401. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ou J, Li J, Pan F, Xie G, Zhou Q, Huang H
and Liang H: Endostatin suppresses colorectal tumor-induced
lymphangiogenesis by inhibiting expression of fibronectin extra
domain A and integrin α9. J Cell Biochem. 112:2106–2114. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Okamoto K, Miyoshi K and Murawaki Y:
miR-29b, miR-205 and miR-221 enhance chemosensitivity to
gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS One.
8:e776232013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sandhu V, Bowitz Lothe IM, Labori KJ,
Lingjærde OC, Buanes T, Dalsgaard AM, Skrede ML, Hamfjord J,
Haaland T, Eide TJ, et al: Molecular signatures of mRNAs and miRNAs
as prognostic biomarkers in pancreatobiliary and intestinal types
of periampullary adenocarcinomas. Mol Oncol. 9:758–771. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kojima M, Sudo H, Kawauchi J, Takizawa S,
Kondou S, Nobumasa H and Ochiai A: MicroRNA markers for the
diagnosis of pancreatic and biliary-tract cancers. PLoS One.
10:e01182202015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Plieskatt JL, Rinaldi G, Feng Y, Peng J,
Yonglitthipagon P, Easley S, Laha T, Pairojkul C, Bhudhisawasdi V,
Sripa B, et al: Distinct miRNA signatures associate with subtypes
of cholangiocarcinoma from infection with the tumourigenic liver
fluke opisthorchis viverrini. J Hepatol. 61:850–858. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Peng F, Jiang J, Yu Y, Tian R, Guo X, Li
X, Shen M, Xu M, Zhu F, Shi C, et al: Direct targeting of
SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma
tumourigenesis and metastasis. Br J Cancer. 109:3092–3104. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Oishi N, Kumar MR, Roessler S, Ji J,
Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, et al:
Transcriptomic profiling reveals hepatic stem-like gene signatures
and interplay of miR-200c and epithelial-mesenchymal transition in
intrahepatic cholangiocarcinoma. Hepatology. 56:1792–1803. 2012.
View Article : Google Scholar : PubMed/NCBI
|