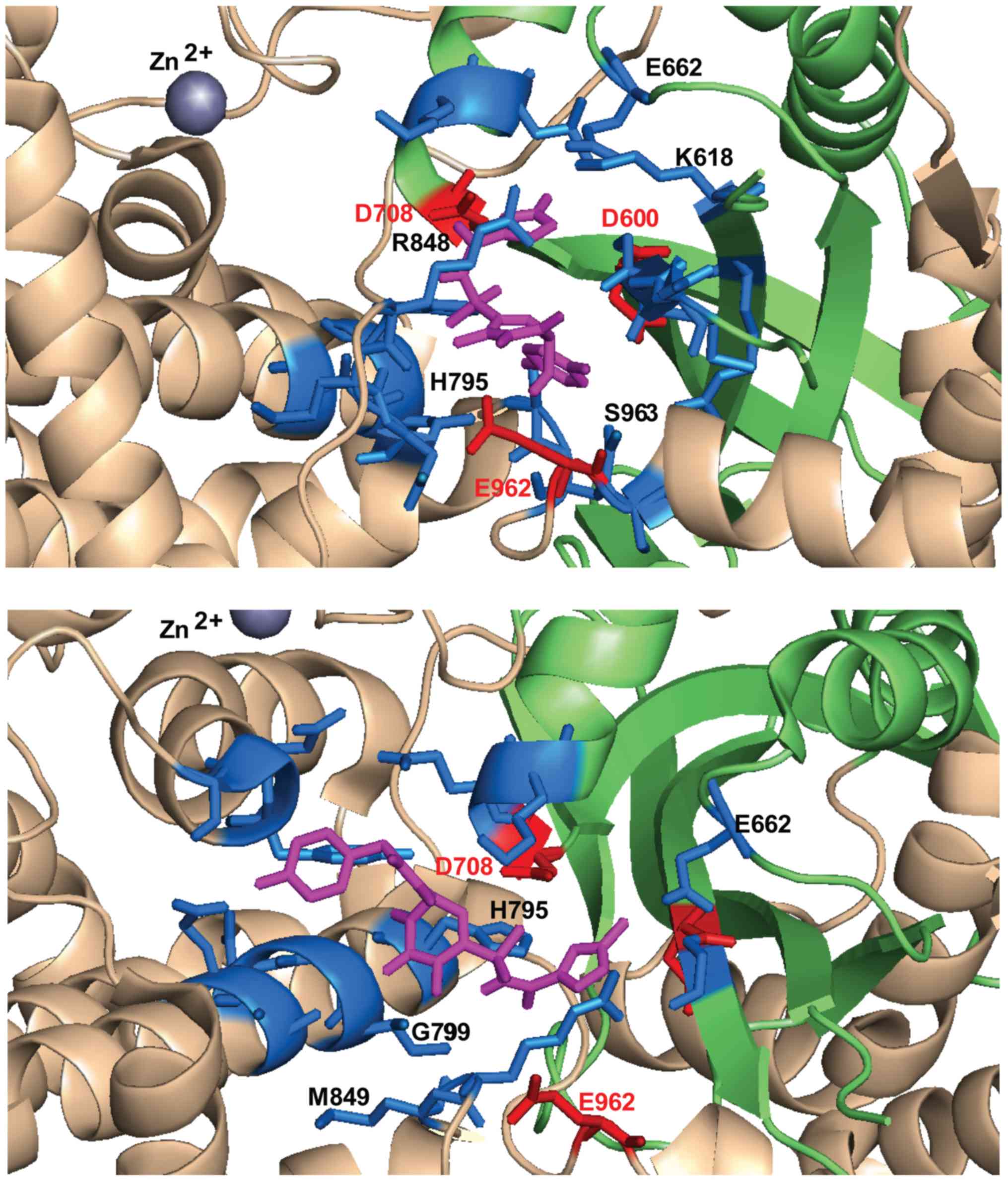
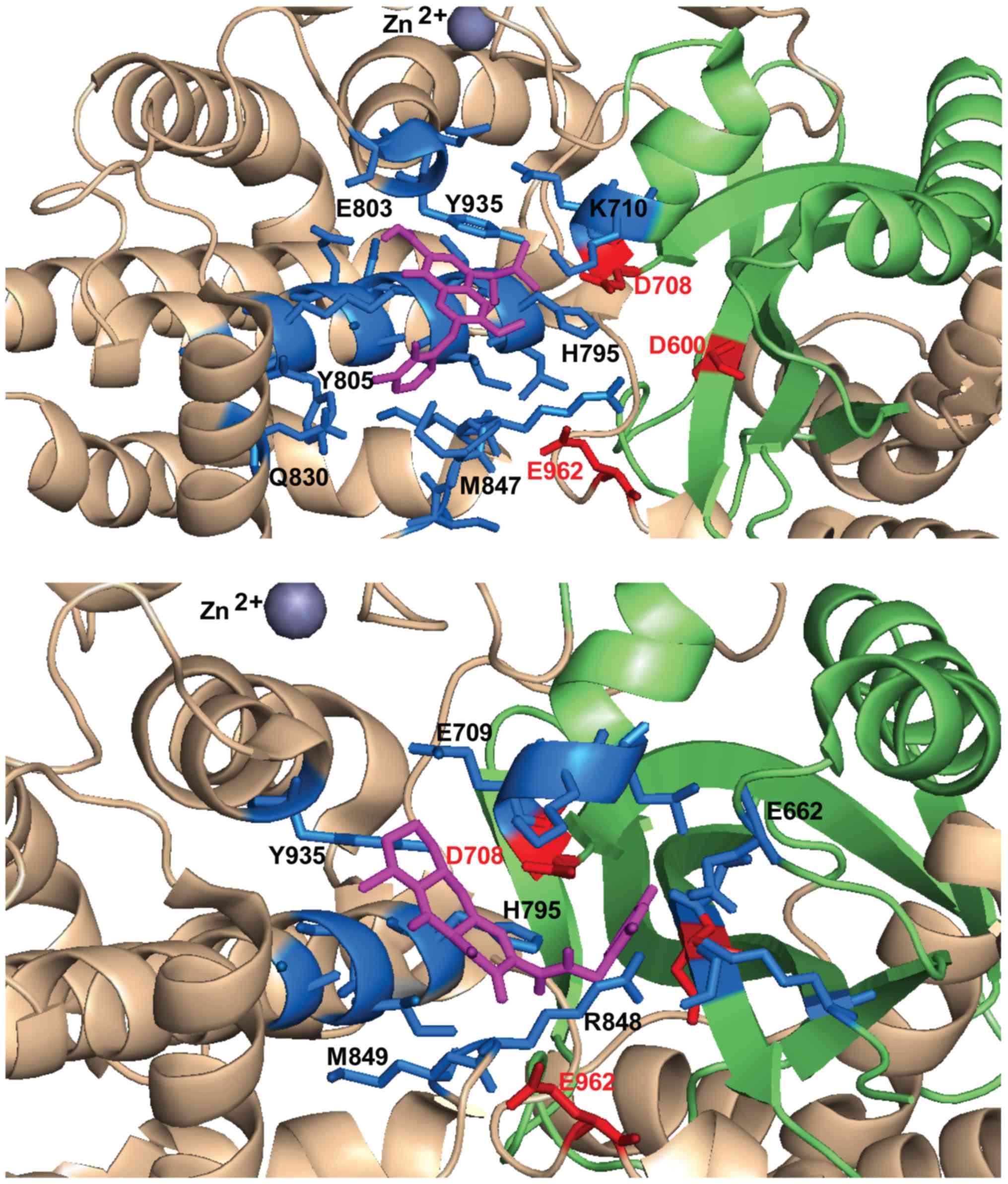
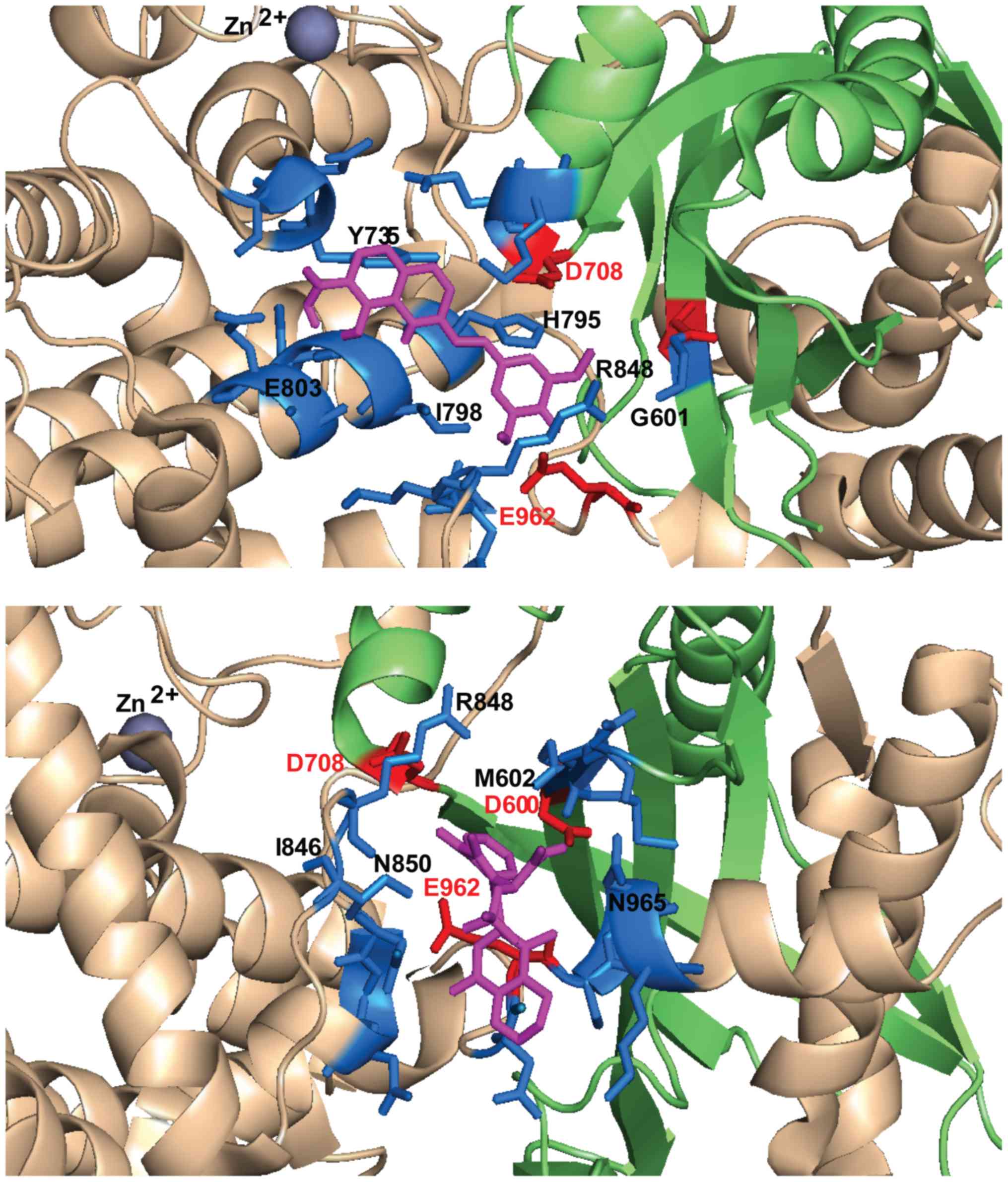
|
1
|
Pace JK II and Feschotte C: The
evolutionary history of human DNA transposons: Evidence for intense
activity in the primate lineage. Genome Res. 17:422–432. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jangam D, Feschotte C and Betrán E:
Transposable Element Domestication As an Adaptation to Evolutionary
Conflicts. Trends Genet. 33:817–831. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee SH, Oshige M, Durant ST, Rasila KK,
Williamson EA, Ramsey H, Kwan L, Nickoloff JA and Hromas R: The SET
domain protein Metnase mediates foreign DNA integration and links
integration to nonhomologous end-joining repair. Proc Natl Acad Sci
USA. 102:18075–18080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hickman AB and Dyda F: DNA Transposition
at Work. Chem Rev. 116:12758–12784. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McCLINTOCK B: The origin and behavior of
mutable loci in maize. Proc Natl Acad Sci USA. 36:344–355. 1950.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chandler M, de la Cruz F, Dyda F, Hickman
AB, Moncalian G and Ton-Hoang B: Breaking and joining
single-stranded DNA: The HUH endonuclease superfamily. Nat Rev
Microbiol. 11:525–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wicker T, Sabot F, Hua-Van A, Bennetzen
JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O,
et al: A unified classification system for eukaryotic transposable
elements. Nat Rev Genet. 8:973–982. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan YW and Wessler SR: The catalytic
domain of all eukaryotic cut-and-paste transposase superfamilies.
Proc Natl Acad Sci USA. 108:7884–7889. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lacroix C, Giovannini D, Combe A, Bargieri
DY, Späth S, Panchal D, Tawk L, Thiberge S, Carvalho TG, Barale JC,
et al: FLP/FRT-mediated conditional mutagenesis in pre-erythrocytic
stages of Plasmodium berghei. Nat Protoc. 6:1412–1428. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye J, Hong J and Ye F: Reprogramming rat
embryonic fibroblasts into induced pluripotent stem cells using
transposon vectors and their chondrogenic differentiation in
vitro. Mol Med Rep. 11:989–994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rice PA and Baker TA: Comparative
architecture of transposase and integrase complexes. Nat Struct
Biol. 8:302–307. 2001. View
Article : Google Scholar
|
|
12
|
Schatz DG and Ji Y: Recombination centres
and the orchestration of V(D)J recombination. Nat Rev Immunol.
11:251–263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai Y, Wong B, Yen Y-M, Oettinger MA, Kwon
J and Johnson RC: Determinants of HMGB proteins required to promote
RAG1/2-recombination signal sequence complex assembly and catalysis
during V(D)J recombination. Mol Cell Biol. 25:4413–4425. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nishana M, Nilavar NM, Kumari R, Pandey M
and Raghavan SC: HIV integrase inhibitor, Elvitegravir, impairs RAG
functions and inhibits V(D)J recombination. Cell Death Dis.
8:e28522017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Seegulam ME and Ratner L: Integrase
inhibitors effective against human T-cell leukemia virus type 1.
Antimicrob Agents Chemother. 55:2011–2017. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nadal M, Mas PJ, Blanco AG, Arnan C, Solà
M, Hart DJ and Coll M: Structure and inhibition of herpesvirus DNA
packaging terminase nuclease domain. Proc Natl Acad Sci USA.
107:16078–16083. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI
|
|
18
|
Kim MS, Lapkouski M, Yang W and Gellert M:
Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature.
518:507–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Avogadro, . Avogadro: an open-source
molecular builder and visualization tool. Version 1.0.3. http://AvogadroOpenmoleculesNet/2012
|
|
20
|
DeLano WL: The PyMOL Molecular Graphics
System, Version 1.8Schrödinger LLC; New York, NY: 2002
|
|
21
|
Kim MS, Chuenchor W, Chen X, Cui Y, Zhang
X, Zhou ZH, Gellert M and Yang W: Cracking the DNA Code for V(D)J
Recombination. Mol Cell. 70:358–370.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma Y, Pannicke U, Schwarz K and Lieber MR:
Hairpin opening and overhang processing by an Artemis/DNA-dependent
protein kinase complex in nonhomologous end joining and V(D)J
recombination. Cell. 108:781–794. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ru H, Chambers MG, Fu TM, Tong AB, Liao M
and Wu H: Molecular Mechanism of V(D)J Recombination from Synaptic
RAG1-RAG2 Complex Structures. Cell. 163:1138–1152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grazini U, Zanardi F, Citterio E, Casola
S, Goding CR and McBlane F: The RING domain of RAG1 ubiquitylates
histone H3: A novel activity in chromatin-mediated regulation of
V(D)J joining. Mol Cell. 37:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matthews AGW, Kuo AJ, Ramón-Maiques S, Han
S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone
DN, et al: RAG2 PHD finger couples histone H3 lysine 4
trimethylation with V(D)J recombination. Nature. 450:1106–1110.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang S, Tao X, Yuan S, Zhang Y, Li P,
Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, et al:
Discovery of an Active RAG Transposon Illuminates the Origins of
V(D)J Recombination. Cell. 166:102–114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Cheng TC, Huang G, Lu Q, Surleac
MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, et al:
Transposon molecular domestication and the evolution of the RAG
recombinase. Nature. 569:79–84. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kang YH, Son CY, Lee CH and Ryu CJ:
Aberrant V(D)J cleavages in T cell receptor β enhancer- and
p53-deficient lymphoma cells. Oncol Rep. 23:1463–1468.
2010.PubMed/NCBI
|
|
29
|
Lewis SM, Agard E, Suh S and Czyzyk L:
Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol.
17:3125–3136. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Papaemmanuil E, Rapado I, Li Y, Potter NE,
Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J,
et al: RAG-mediated recombination is the predominant driver of
oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia.
Nat Genet. 46:116–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Messier TL, O'Neill JP, Hou SM, Nicklas JA
and Finette BA: In vivo transposition mediated by V(D)J recombinase
in human T lymphocytes. EMBO J. 22:1381–1388. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reddy YVR, Perkins EJ and Ramsden DA:
Genomic instability due to V(D)J recombination-associated
transposition. Genes Dev. 20:1575–1582. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Z, Wu S, Wang J, Li W, Lin Y, Ji C, Xue
J and Chen J: Evaluation of the interactions of HIV-1 integrase
with small ubiquitin-like modifiers and their conjugation enzyme
Ubc9. Int J Mol Med. 30:1053–1060. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gupta K, Turkki V, Sherrill-Mix S, Hwang
Y, Eilers G, Taylor L, McDanal C, Wang P, Temelkoff D, Nolte RT, et
al: Structural Basis for Inhibitor-Induced Aggregation of HIV
Integrase. PLoS Biol. 14:e10025842016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lusic M and Siliciano RF: Nuclear
landscape of HIV-1 infection and integration. Nat Rev Microbiol.
15:69–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen JC-H, Krucinski J, Miercke LJW,
Finer-Moore JS, Tang AH, Leavitt AD and Stroud RM: Crystal
structure of the HIV-1 integrase catalytic core and C-terminal
domains: A model for viral DNA binding. Proc Natl Acad Sci USA.
97:8233–8238. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang W, Hendrickson WA, Crouch RJ and
Satow Y: Structure of ribonuclease H phased at 2 A resolution by
MAD analysis of the selenomethionyl protein. Science.
249:1398–1405. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Venanzi Rullo E, Ceccarelli M, Condorelli
F, Facciolà A, Visalli G, D'Aleo F, Paolucci I, Cacopardo B,
Pinzone MR, Di Rosa M, et al: Investigational drugs in HIV: Pros
and cons of entry and fusion inhibitors (Review). Mol Med Rep.
19:1987–1995. 2019.PubMed/NCBI
|
|
39
|
Wai JS, Egbertson MS, Payne LS, Fisher TE,
Embrey MW, Tran LO, Melamed JY, Langford HM, Guare JP Jr, Zhuang L,
et al: 4-Aryl-2,4-dioxobutanoic acid inhibitors of HIV-1 integrase
and viral replication in cells. J Med Chem. 43:4923–4926. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hazuda DJ, Felock P, Witmer M, Wolfe A,
Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau
C and Miller MD: Inhibitors of strand transfer that prevent
integration and inhibit HIV-1 replication in cells. Science.
287:646–650. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Summa V, Petrocchi A, Bonelli F, Crescenzi
B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda
DJ, et al: Discovery of raltegravir, a potent, selective orally
bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS
infection. J Med Chem. 51:5843–5855. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
U.S. Food & Drug Administration, . HIV
Timeline and History of Approvals. https://www.fda.gov/patients/hivaids/hiv-timeline-and-history-approvalsAugust
1–2018
|
|
43
|
Di Santo R: Inhibiting the HIV integration
process: Past, present, and the future. J Med Chem. 57:539–566.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sato M, Motomura T, Aramaki H, Matsuda T,
Yamashita M, Ito Y, Kawakami H, Matsuzaki Y, Watanabe W, Yamataka
K, et al: Novel HIV-1 integrase inhibitors derived from quinolone
antibiotics. J Med Chem. 49:1506–1508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee JSF, Calmy A, Andrieux-Meyer I and
Ford N: Review of the safety, efficacy, and pharmacokinetics of
elvitegravir with an emphasis on resource-limited settings. HIV
AIDS (Auckl). 4:5–15. 2012.PubMed/NCBI
|
|
46
|
Barnhart M and Shelton JD: ARVs: The next
generation. Going boldly together to new frontiers of HIV
treatment. Glob Health Sci Pract. 3:1–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Johns BA, Kawasuji T, Weatherhead JG,
Taishi T, Temelkoff DP, Yoshida H, Akiyama T, Taoda Y, Murai H,
Kiyama R, et al: Carbamoyl pyridone HIV-1 integrase inhibitors 3. A
diastereomeric approach to chiral nonracemic tricyclic ring systems
and the discovery of dolutegravir (S/GSK1349572) and
(S/GSK1265744). J Med Chem. 56:5901–5916. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hare S, Gupta SS, Valkov E, Engelman A and
Cherepanov P: Retroviral intasome assembly and inhibition of DNA
strand transfer. Nature. 464:232–236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hare S, Smith SJ, Métifiot M, Jaxa-Chamiec
A, Pommier Y, Hughes SH and Cherepanov P: Structural and functional
analyses of the second-generation integrase strand transfer
inhibitor dolutegravir (S/GSK1349572). Mol Pharmacol. 80:565–572.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hightower KE, Wang R, Deanda F, Johns BA,
Weaver K, Shen Y, Tomberlin GH, Carter HL III, Broderick T, Sigethy
S, et al: Dolutegravir (S/GSK1349572) exhibits significantly slower
dissociation than raltegravir and elvitegravir from wild-type and
integrase inhibitor-resistant HIV-1 integrase-DNA complexes.
Antimicrob Agents Chemother. 55:4552–4559. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yoshinaga T, Kobayashi M, Seki T, Miki S,
Wakasa-Morimoto C, Suyama-Kagitani A, Kawauchi-Miki S, Taishi T,
Kawasuji T, Johns BA, et al: Antiviral characteristics of
GSK1265744, an HIV integrase inhibitor dosed orally or by
long-acting injection. Antimicrob Agents Chemother. 59:397–406.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tsiang M, Jones GS, Goldsmith J, Mulato A,
Hansen D, Kan E, Tsai L, Bam RA, Stepan G, Stray KM, et al:
Antiviral activity of bictegravir (GS-9883), a novel potent HIV-1
integrase strand transfer inhibitor with an improved resistance
profile. Antimicrob Agents Chemother. 60:7086–7097. 2016.PubMed/NCBI
|
|
53
|
Mekouar K, Mouscadet JF, Desmaële D, Subra
F, Leh H, Savouré D, Auclair C and d'Angelo J: Styrylquinoline
derivatives: A new class of potent HIV-1 integrase inhibitors that
block HIV-1 replication in CEM cells. J Med Chem. 41:2846–2857.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Deprez E, Barbe S, Kolaski M, Leh H,
Zouhiri F, Auclair C, Brochon JC, Le Bret M and Mouscadet JF:
Mechanism of HIV-1 integrase inhibition by styrylquinoline
derivatives in vitro. Mol Pharmacol. 65:85–98. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Han Y-S, Xiao W-L, Quashie PK, Mesplède T,
Xu H, Deprez E, Delelis O, Pu JX, Sun HD and Wainberg MA:
Development of a fluorescence-based HIV-1 integrase DNA binding
assay for identification of novel HIV-1 integrase inhibitors.
Antiviral Res. 98:441–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bonnenfant S, Thomas CM, Vita C, Subra F,
Deprez E, Zouhiri F, Desmaële D, D'Angelo J, Mouscadet JF and Leh
H: Styrylquinolines, integrase inhibitors acting prior to
integration: A new mechanism of action for anti-integrase agents. J
Virol. 78:5728–5736. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Passos DO, Li M, Yang R, Rebensburg SV,
Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R and
Lyumkis D: Cryo-EM structures and atomic model of the HIV-1 strand
transfer complex intasome. Science. 355:89–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Quashie PK, Han YS, Hassounah S, Mesplède
T and Wainberg MA: Structural studies of the HIV-1 integrase
protein: Compound screening and characterization of a DNA-binding
inhibitor. PLoS One. 10:e01283102015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shkriabai N, Patil SS, Hess S, Budihas SR,
Craigie R, Burke TR Jr, Le Grice SF and Kvaratskhelia M:
Identification of an inhibitor-binding site to HIV-1 integrase with
affinity acetylation and mass spectrometry. Proc Natl Acad Sci USA.
101:6894–6899. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Du L, Zhao YX, Yang LM, Zheng YT, Tang Y,
Shen X and Jiang HL: Symmetrical 1-pyrrolidineacetamide showing
anti-HIV activity through a new binding site on HIV-1 integrase.
Acta Pharmacol Sin. 29:1261–1267. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ge H, Si Y and Roeder RG: Isolation of
cDNAs encoding novel transcription coactivators p52 and p75 reveals
an alternate regulatory mechanism of transcriptional activation.
EMBO J. 17:6723–6729. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Maertens G, Cherepanov P, Pluymers W,
Busschots K, De Clercq E, Debyser Z and Engelborghs Y: LEDGF/p75 is
essential for nuclear and chromosomal targeting of HIV-1 integrase
in human cells. J Biol Chem. 278:33528–33539. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Llano M, Delgado S, Vanegas M and Poeschla
EM: Lens epithelium-derived growth factor/p75 prevents proteasomal
degradation of HIV-1 integrase. J Biol Chem. 279:55570–55577. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
De Rijck J, Vandekerckhove L, Gijsbers R,
Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F and
Debyser Z: Overexpression of the lens epithelium-derived growth
factor/p75 integrase binding domain inhibits human immunodeficiency
virus replication. J Virol. 80:11498–11509. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Cherepanov P, Ambrosio ALB, Rahman S,
Ellenberger T and Engelman A: Structural basis for the recognition
between HIV-1 integrase and transcriptional coactivator p75. Proc
Natl Acad Sci USA. 102:17308–17313. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Du L, Zhao Y, Chen J, Yang L, Zheng Y,
Tang Y, Shen X and Jiang H: D77, one benzoic acid derivative,
functions as a novel anti-HIV-1 inhibitor targeting the interaction
between integrase and cellular LEDGF/p75. Biochem Biophys Res
Commun. 375:139–144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Christ F, Voet A, Marchand A, Nicolet S,
Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel
B, Strelkov SV, et al: Rational design of small-molecule inhibitors
of the LEDGF/p75-integrase interaction and HIV replication. Nat
Chem Biol. 6:442–448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Fader LD, Malenfant E, Parisien M, Carson
R, Bilodeau F, Landry S, Pesant M, Brochu C, Morin S, Chabot C, et
al: Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor
(NCINI) of HIV-1. ACS Med Chem Lett. 5:422–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tsiang M, Jones GS, Niedziela-Majka A, Kan
E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, et al:
New class of HIV-1 integrase (IN) inhibitors with a dual mode of
action. J Biol Chem. 287:21189–21203. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Melek M, Jones JM, O'Dea MH, Pais G, Burke
TR Jr, Pommier Y, Neamati N and Gellert M: Effect of HIV integrase
inhibitors on the RAG1/2 recombinase. Proc Natl Acad Sci USA.
99:134–137. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Goldgur Y, Craigie R, Cohen GH, Fujiwara
T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H and Davies
DR: Structure of the HIV-1 integrase catalytic domain complexed
with an inhibitor: A platform for antiviral drug design. Proc Natl
Acad Sci USA. 96:13040–13043. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
de Miguel R, Montejano R, Stella-Ascariz N
and Arribas JR: A safety evaluation of raltegravir for the
treatment of HIV. Expert Opin Drug Saf. 17:217–223. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Huhn GD, Badri S, Vibhakar S, Tverdek F,
Crank C, Lubelchek R, Max B, Simon D, Sha B, Adeyemi O, et al:
Early development of non-hodgkin lymphoma following initiation of
newer class antiretroviral therapy among HIV-infected patients -
implications for immune reconstitution. AIDS Res Ther. 7:442010.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Steigbigel RT, Cooper DA, Kumar PN, Eron
JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Gatell JM,
Rockstroh JK, et al BENCHMRK Study Teams, : Raltegravir with
optimized background therapy for resistant HIV-1 infection. N Engl
J Med. 359:339–354. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Barbaro G and Barbarini G: HIV infection
and cancer in the era of highly active antiretroviral therapy
(Review). Oncol Rep. 17:1121–1126. 2007.PubMed/NCBI
|
|
76
|
Gillead Sciences: sNDA 207561/S-014.
Genvoya (elvitegravir/cobicistat/emtricitabine/tenofovir
alafenamide). Clinical and Cross-Discipline Team Leader Review.
|
|
77
|
Huye LE, Purugganan MM, Jiang M-M and Roth
DB: Mutational analysis of all conserved basic amino acids in RAG-1
reveals catalytic, step arrest, and joining-deficient mutants in
the V(D)J recombinase. Mol Cell Biol. 22:3460–3473. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Davies DR, Goryshin IY, Reznikoff WS and
Rayment I: Three-dimensional structure of the Tn5 synaptic complex
transposition intermediate. Science. 289:77–85. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ason B, Knauss DJ, Balke AM, Merkel G,
Skalka AM and Reznikoff WS: Targeting Tn5 transposase identifies
human immunodeficiency virus type 1 inhibitors. Antimicrob Agents
Chemother. 49:2035–2043. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Czyz A, Stillmock KA, Hazuda DJ and
Reznikoff WS: Dissecting Tn5 transposition using HIV-1 integrase
diketoacid inhibitors. Biochemistry. 46:10776–10789. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Koh Y, Matreyek KA and Engelman A:
Differential sensitivities of retroviruses to integrase strand
transfer inhibitors. J Virol. 85:3677–3682. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Beck-Engeser GB, Eilat D, Harrer T, Jäck
HM and Wabl M: Early onset of autoimmune disease by the retroviral
integrase inhibitor raltegravir. Proc Natl Acad Sci USA.
106:20865–20870. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Stetson DB, Ko JS, Heidmann T and
Medzhitov R: Trex1 prevents cell-intrinsic initiation of
autoimmunity. Cell. 134:587–598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wolkowicz UM, Morris ER, Robson M,
Trubitsyna M and Richardson JM: Structural basis of Mos1
transposase inhibition by the anti-retroviral drug Raltegravir. ACS
Chem Biol. 9:743–751. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Shaheen M, Williamson E, Nickoloff J, Lee
SH and Hromas R: Metnase/SETMAR: A domesticated primate transposase
that enhances DNA repair, replication, and decatenation. Genetica.
138:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Williamson EA, Damiani L, Leitao A, Hu C,
Hathaway H, Oprea T, Sklar L, Shaheen M, Bauman J, Wang W, et al:
Targeting the transposase domain of the DNA repair component
Metnase to enhance chemotherapy. Cancer Res. 72:6200–6208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Marino-Merlo F, Mastino A, Grelli S,
Hermine O, Bazarbachi A and Macchi B: Future Perspectives on Drug
Targeting in Adult T Cell Leukemia-Lymphoma. Front Microbiol.
9:9252018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Rabaaoui S, Zouhiri F, Lançon A, Leh H,
d'Angelo J and Wattel E: Inhibitors of strand transfer that prevent
integration and inhibit human T-cell leukemia virus type 1 early
replication. Antimicrob Agents Chemother. 52:3532–3541. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Shimura K, Kodama E, Sakagami Y, Matsuzaki
Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, et
al: Broad antiretroviral activity and resistance profile of the
novel human immunodeficiency virus integrase inhibitor elvitegravir
(JTK-303/GS-9137). J Virol. 82:764–774. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Passos DO, Li M, Yang R, Rebensburg SV,
Ghirlando R, Jeon Y, Shkriabai N, Kvaratskhelia M, Craigie R and
Lyumkis D: Cryo-EM structures and atomic model of the HIV-1 strand
transfer complex intasome. Science. 355:89–92. 2017. View Article : Google Scholar : PubMed/NCBI
|