Introduction
Renal cell carcinoma (RCC) is a common type of
cancer that is derived from renal tubular epithelial cells
(1). Clear cell RCC (CCRCC) has
been reported to be the most common histological subtype of RCC
(2,3). As for clinical treatment, RCC is
often resistant to radiotherapy, chemotherapy and hormonal
treatments (4). Although surgical
resection can effectively treat CCRCC, 20–40% of patients develop
local recurrence or distant metastasis following surgery (5). The observed survival rate of advanced
CCRCC is very low, which poses an obstacle in treating and managing
patients with CCRCC (6). As CCRCC
is a highly aggressive cancer with concomitant poor prognosis,
reliable biomarkers for predicting the susceptibility and survival
of patients with CCRCC are urgently required.
Circular RNAs (circRNAs) represent a series of
endogenous RNAs that modulate the expression of genes and do not
encode proteins (7). circRNAs are
commonly characterized by their stabilized structure and tissue
specificity, and are widely expressed in a variety of eukaryotic
cells (8,9). CircRNAs also have tissue specificity
and their expression is tissue specific in the eukaryotic
transcriptome (10). An
investigation into the regulation of competing endogenous RNAs
(ceRNAs) by Meng et al (9)
provided insight into the complex post-transcriptional interaction
network of various circRNAs and long non-coding RNAs; these
molecules function as microRNA (miRNA/miR) sponges, suppressing
their effects via miRNA response elements. Emerging evidence has
suggested that circRNAs may be considered robust biomarkers and
potential therapeutic targets in several diseases, including cancer
(11).
Numerous studies have confirmed the existence of the
regulatory role of ceRNAs in the circRNA-miRNA-mRNA network within
various diseases, including renal cancer (12,13).
For example, the novel circRNA circHIAT1 has been reported to be
downregulated in CCRCC tissues compared with normal tissues.
Analysis of androgen receptor-inhibited circHIAT1 revealed the
aberrant expression of miR-195-5p/29a-3p/29c-3p, which induced cell
division cycle 42 expression, promoting the migration and invasion
of CCRCC cells (12). Furthermore,
a previous study demonstrated that knockdown of circRNA_0001451
significantly enhanced tumor proliferation in vitro; the
levels of circRNA_0001451 were associated with the differentiation
grade of patients with CCRCC (13). In addition, circRNA ZNF609 has been
reported to serve as a ceRNA in modulating the expression of
Forkhead box P4 via sponging miR-138-5p in renal cancer; high
circ-ZNF609 expression was determined to enhance the growth and
invasive characteristics of renal cancer cells (14).
Despite improved understanding of the association
between circRNA expression and various types of human cancer, the
role of circRNAs in renal cancer remains unclear. The present study
identified several differentially expressed circRNAs, miRNAs and
genes by analyzing datasets of the Gene Expression Omnibus (GEO,
http://www.ncbi.nlm.nih.gov/gds/) and
The Cancer Genome Atlas (TCGA, http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
for CCRCC. Additionally, a circRNA-miRNA-mRNA regulatory network
was constructed using bioinformatics tools. The present findings
may improve understanding of the mechanisms underlying the
carcinogenesis of CCRCC.
Materials and methods
Microarray database
To identify datasets, ‘renal cellular cell
carcinoma’ and ‘circRNA’ were used as keywords to search the GEO;
datasets including cancer and normal groups was the main inclusion
criterion. The data were downloaded from the GEO of the National
Center for Biotechnology Information repository. The GSE100186
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100186)
circRNA expression microarray dataset of CCRCC was used, which
contained data from four CCRCC samples and four normal samples.
Arraystar circRNA microarray (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL21825)
analysis was used to examine the expression of circRNAs in CCRCC
and matched non-tumor tissues. mRNA expression and miRNA profiling
of TCGA CCRCC data was performed to identify differentially
expressed genes (DEGs) and differentially expressed miRNAs (DEMs)
between cancer and normal tissues. TCGA data were downloaded from
UCSC XENA (https://xena.ucsc.edu/).
Data processing
The limma package in R (version 3.6.0, http://www.r-project.org/) was used to analyze
differentially expressed circRNAs (DECs) between the groups. The
‘edge R’ package (version 3.26.8; http://www.bioconductor.org/packages/release/bioc/html/edgeR.html)
was employed to analyze the DEGs and DEMs between the groups.
P<0.05 and |log fold change|>2 were applied to determine
statistical significance.
circRNA-miRNA-mRNA regulatory
network
The Cancer-Specific circRNA Database (CSCD;
http://gb.whu.edu.cn/CSCD) can be used to predict
interactions between circRNAs and miRNAs (15). Using the CSCD, miRNAs that interact
with DECs were predicted. Subsequently, the DECs that interact with
the miRNAs were identified as CCRCC-specific miRNAs. TargetScan 7.2
(http://www.targetscan.org/vert_72/)
and miRDB 2.0 (http://www.mirdb.org/) were used to
predict the target genes of DEMs, which were matched to genes with
a mRNA expression profile that opposed the miRNA profile; the
expression of miRNAs is often inversely associated with that of the
target mRNA. Subsequently, the top five circRNAs with the highest
degrees of connectivity were selected as the hub circRNAs. The
circRNA-miRNA-mRNA regulatory network was constructed using
Cytoscape 3.7.0 (https://cytoscape.org/).
Gene Ontology (GO) and pathway
enrichment analyses
GO analysis is a useful bioinformatics strategy for
annotating genes or gene products, and comprises three categories:
Cellular component (CC), molecular function (MF) and biological
process (BP) (16). Kyoto
Encyclopedia of Genes and Genomes (KEGG) is a collection of
databases, which contain comprehensive information regarding
genomes, biological pathways, diseases and chemical substances
(17). In the present study, GO
and KEGG enrichment analyses were conducted using R package
clusterprofiler (version 3.12.0; http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html).
P<0.05 was considered to indicate a statistically significant
difference.
Prognostic value of circRNA-regulated
DEGs and DEMs
TCGA contains the survival information of patients
with various types of cancer. Using the survival 2.44 package
(https://cran.r-project.org/web/packages/survival/index.html)
in R, the prognostic value of circRNA-regulated DEGs and DEMs was
assessed. Additionally, a survival curve was plotted using
survminer 0.4.6 (https://cran.r-project.org/web/packages/survminer/index.html)
package in R.
Results
Identification of DECs
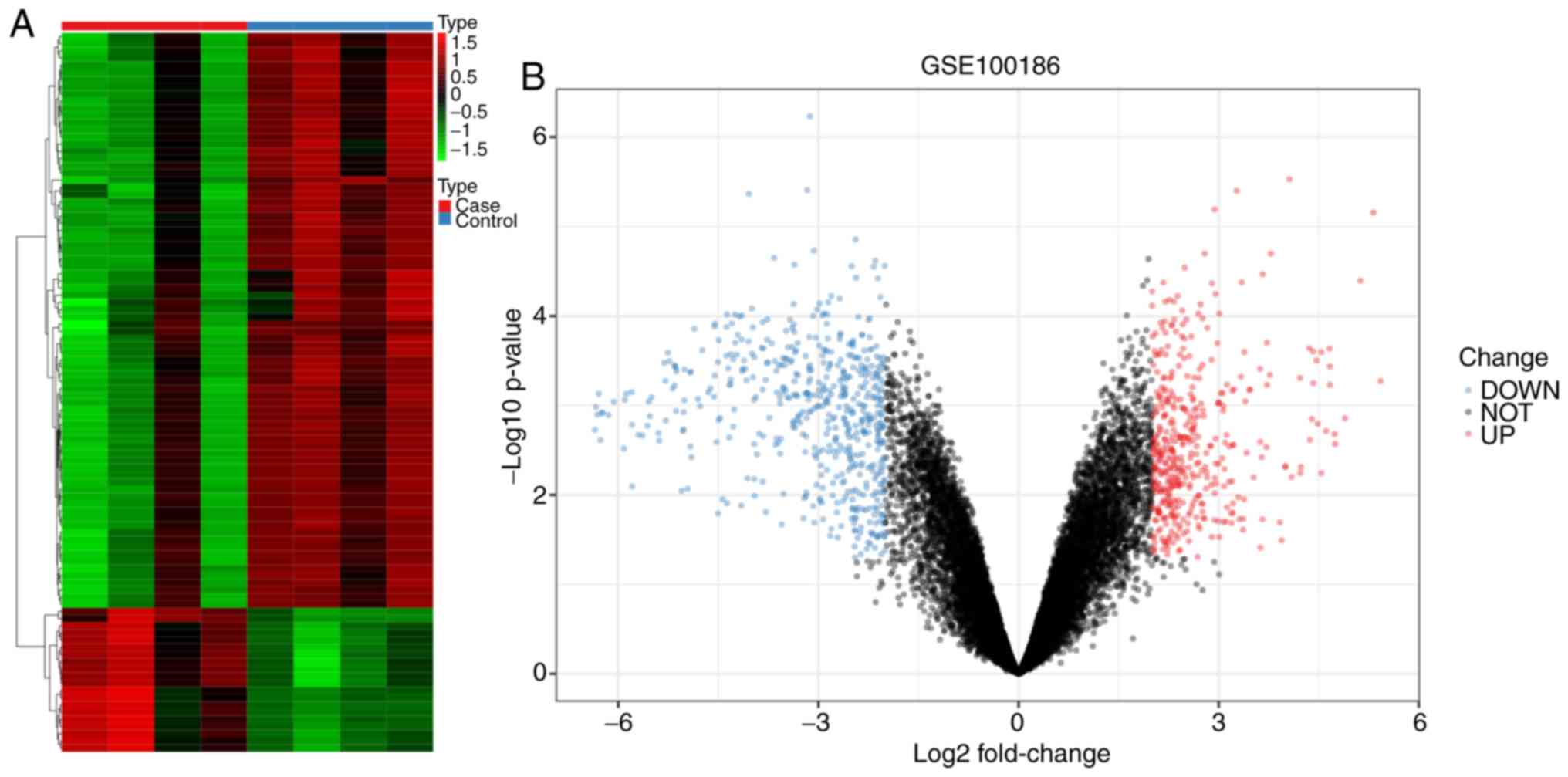
Following analysis of differential expression, a
total of 324 circRNAs in the GSE100186 dataset were downregulated
in the cancer group, whereas 218 circRNAs were upregulated in the
cancer group (Fig. 1 and Table SI). The top ten most significant
circRNAs according to P-value included hsa_circ_0031594,
hsa_circ_0001968, hsa_circ_0003596, hsa_circ_0058794,
hsa_circ_0001873, hsa_circ_0003748, hsa_circ_0003997,
hsa_circ_0000223, hsa_circ_0092367 and hsa_circ_0092360.
circRNA-miRNA-mRNA regulatory
network
According to the CSCD datasets, 2,363 miRNAs were
reported to interact with the identified DECs. After identifying
the DECs that interact with the 2,363 miRNAs, 42 miRNAs were
selected as CCRCC-specific miRNAs, including 32 upregulated miRNAs
and 10 downregulated miRNAs. Using TargetScan, miRDB and TCGA-DEGs,
a total of 244 downregulated genes and 85 upregulated genes were
selected as circRNA-regulated genes. Subsequently, the circRNAs
were sorted based on the degree of connectivity; the top five
circRNAs were selected as hub circRNAs in the regulatory network of
DEGs (Table I).
 | Table I.Information regarding the hub
circRNAs. |
Table I.
Information regarding the hub
circRNAs.
| Author, year | Alias | LogFC | Position | Strand | Genomic length
(bp) | Spliced length
(bp) | Gene symbol | (Refs.) |
|---|
| Salzman et
al, 2013 |
hsa_circ_0031594 | 5.320375 |
chr14:34398281-34400421 | − | 2,140 | 257 | EGLN3 | (35) |
| Salzman et
al, 2013; Jeck et al, 2013; Rybak et al,
2015 |
hsa_circ_0001968 | 4.893003 |
chr11:68359043-68367962 | + | 8,919 | 407 | PPP6R3 | (35–37) |
| Jeck et al,
2013 |
hsa_circ_0003596 | 4.73989 |
chr9:137716445-137717750 | + | 1,305 | 369 | COL5A1 | (36) |
| Rybak et al,
2015; Salzman et al, 2013 |
hsa_circ_0058794 | 4.668168 |
chr2:236626200-236659132 | + | 32,932 | 451 | AGAP1 | (35,37) |
| Memczak, 2013 |
hsa_circ_0001873 | 4.666399 |
chr9:93637042-93639999 | − | 2,957 | 2,957 | SYK | (38) |
| Jeck et al,
2013; Rybak et al, 2015; Salzman et al, 2013 |
hsa_circ_0003748 | −6.26453 |
chr3:48726970-48728915 | − | 1,945 | 352 | IP6K2 | (35–37) |
| Jeck et al,
2013; Rybak et al, 2015 |
hsa_circ_0003997 | −6.29312 |
chr11:122953792-122955421 | − | 1,629 | 493 | CLMP | (36,37) |
| Memczak et
al, 2013 |
hsa_circ_0000223 | −6.31392 |
chr10:17754818-17754937 | + | 119 | 119 | STAM | (38) |
| Zhang et al,
2013 |
hsa_circ_0092367 | −6.33682 |
chr15:25325262-25326442 | + | 1,180 | 1,180 | SNORD116-14 | (39) |
| Zhang et al,
2013 |
hsa_circ_0092360 | −6.34906 |
chr17:27047048-27047688 | + | 640 | 640 | RPL23A | (39) |
Construction of the
circRNA-miRNA-upregulated mRNA network
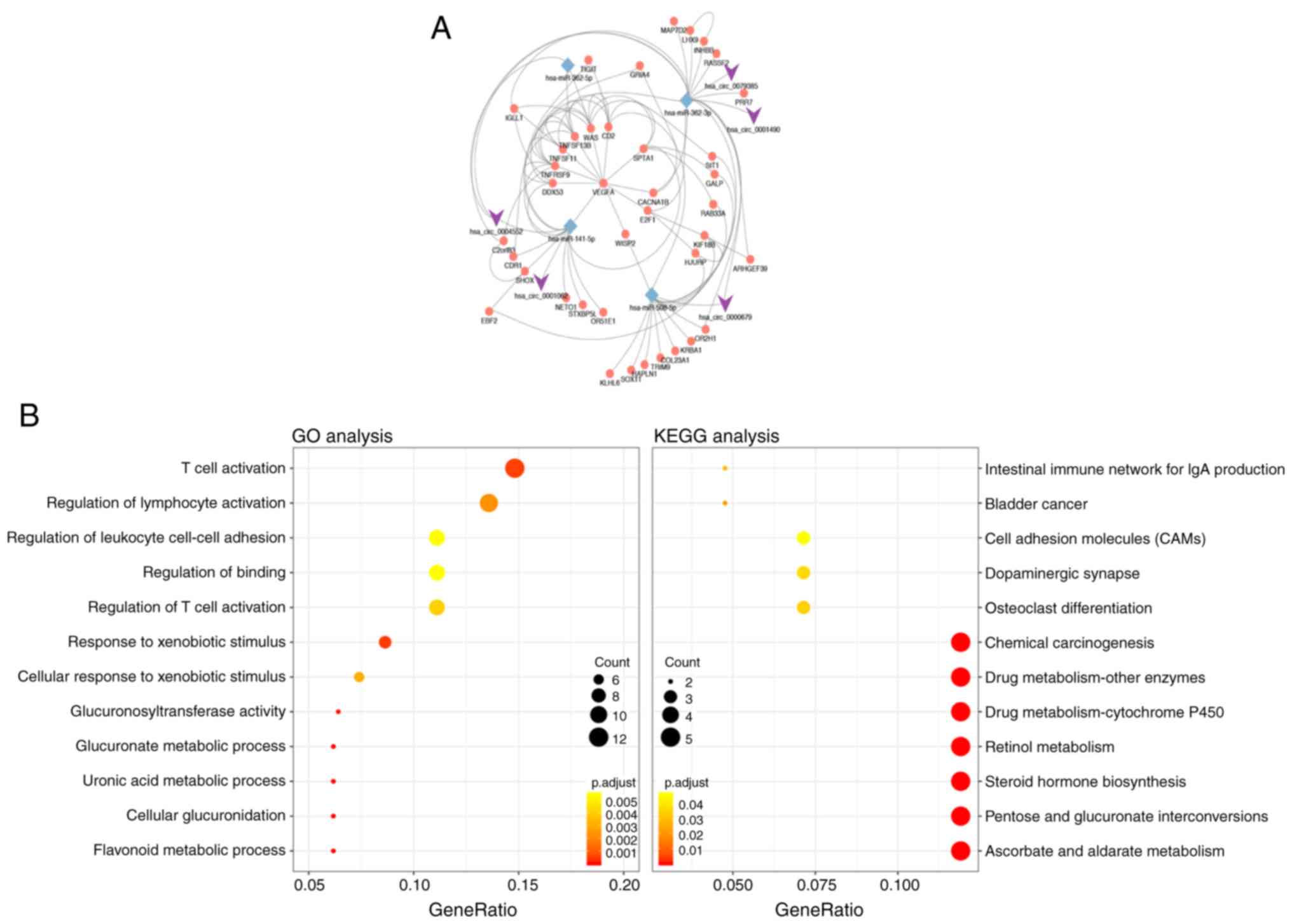
As presented in Fig.
2A, a hub circRNA-miRNA-upregulated mRNA network was built. The
results of enrichment analysis of the 85 upregulated genes are
presented in Fig. 2B and Table II. The upregulated genes were
mainly enriched in BP, including ‘flavonoid metabolic process’,
‘cellular glucuronidation’ and ‘T cell activation’. In addition,
these DEGs were enriched in MF, including ‘glucuronosyltransferase
activity’, ‘UDP-glycosyltransferase activity’ and ‘protein
heterodimerization activity’. KEGG pathway analysis suggested that
DEGs were associated with ‘ascorbate and aldarate metabolism’,
‘pentose and glucuronate interconversions’, ‘steroid hormone
biosynthesis’ and ‘retinol metabolism’.
 | Table II.Top five GO terms and KEGG pathways
enriched in the circRNA-miRNA-upregulated mRNA network. |
Table II.
Top five GO terms and KEGG pathways
enriched in the circRNA-miRNA-upregulated mRNA network.
| Term/pathway | ID | Description | Gene ratio | P-value | Count |
|---|
| BP | GO:0009812 | Flavonoid metabolic
process | 5/81 |
5.62×10−9 | 5 |
| BP | GO:0052695 | Cellular
glucuronidation | 5/81 |
1.15×10−8 | 5 |
| BP | GO:0006063 | Uronic acid
metabolic process | 5/81 |
4.80×10−8 | 5 |
| BP | GO:0019585 | Glucuronate
metabolic process | 5/81 |
4.80×10−8 | 5 |
| BP | GO:0009410 | Response to
xenobiotic stimulus | 7/81 |
1.20×10−6 | 7 |
| MF | GO:0015020 |
Glucuronosyltransferase activity | 5/78 |
4.05×10−7 | 5 |
| MF | GO:0046982 | Protein
heterodimerization activity | 10/78 |
7.20×10−5 | 10 |
| MF | GO:0001228 | DNA-binding
transcription activator activity, RNA polymerase II-specific | 9/78 |
9.24×10−5 | 9 |
| MF | GO:0008194 |
UDP-glycosyltransferase activity | 5/78 | 0.000492 | 5 |
| MF | GO:0005172 | Vascular
endothelial growth factor receptor binding | 2/78 | 0.001068 | 2 |
| KEGG | hsa00053 | Ascorbate and
aldarate metabolism | 5/42 |
3.24×10−7 | 5 |
| KEGG | hsa00040 | Pentose and
glucuronate interconversions | 5/42 |
1.09×10−6 | 5 |
| KEGG | hsa00860 | Porphyrin and
chlorophyll metabolism | 5/42 |
3.21×10−6 | 5 |
| KEGG | hsa00140 | Steroid hormone
biosynthesis | 5/42 |
1.91×10−5 | 5 |
| KEGG | hsa00830 | Retinol
metabolism | 5/42 |
3.29×10−5 | 5 |
Construction of the
circRNA-miRNA-downregulated mRNA interaction network
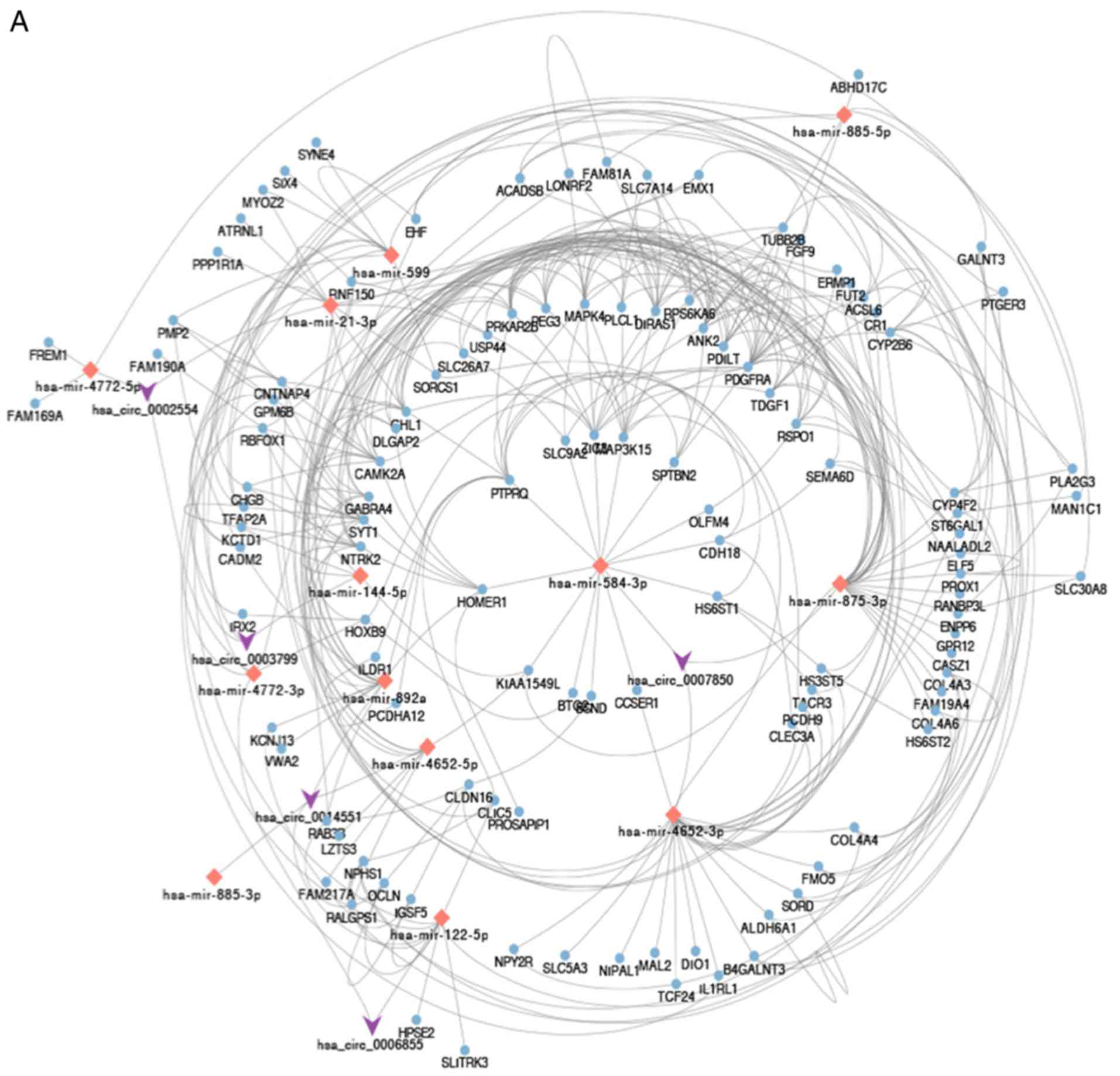
As shown in Fig.
3A, a hub circRNA-miRNA-downregulated mRNA network was built.
Enrichment analysis was performed on the 244 downregulated genes.
As presented in Fig. 3B and
Table III, DEGs were enriched in
BP, including ‘nephron development’, ‘kidney development’ and
‘renal system development’. CC analysis suggested that DEGs were
associated with ‘neuronal cell body’, ‘cell body’ and
‘extracellular matrix component’. In addition, these DEGs were
significantly enriched in MF, including ‘heparan sulfate
sulfotransferase activity’. KEGG enrichment pathway analysis
revealed DEGs to be involved in ‘ECM-receptor interaction’,
‘glycosaminoglycan biosynthesis-heparan sulfate/heparin’ and ‘cell
adhesion molecules (CAMs)’ (Fig.
3C and Table III).
 | Table III.Top five GO terms and KEGG pathways
enriched in the circRNA-miRNA-downregulated mRNA network. |
Table III.
Top five GO terms and KEGG pathways
enriched in the circRNA-miRNA-downregulated mRNA network.
| Term/pathway | ID | Description | Gene ratio | P-value | Count |
|---|
| BP | GO:0072006 | Nephron
development | 14/230 |
4.20×10−9 | 14 |
| BP | GO:0001822 | Kidney
development | 18/230 |
1.50×10−8 | 18 |
| BP | GO:0072001 | Renal system
development | 18/230 |
3.72×10−8 | 18 |
| BP | GO:0072073 | Kidney epithelium
development | 13/230 |
4.06×10−8 | 13 |
| BP | GO:0072009 | Nephron epithelium
development | 11/230 |
2.11×10−7 | 11 |
| CC | GO:0043025 | Neuronal cell
body | 20/240 |
1.12×10−6 | 20 |
| CC | GO:0044297 | Cell body | 20/240 |
8.82×10−6 | 20 |
| CC | GO:0044420 | Extracellular
matrix component | 9/240 |
2.67×10−5 | 9 |
| CC | GO:0005604 | Basement
membrane | 8/240 |
2.80×10−5 | 8 |
| CC | GO:0098644 | Complex of collagen
trimers | 4/240 | 0.000113 | 4 |
| MF | GO:0034483 | Heparan sulfate
sulfotransferase activity | 4/228 |
3.54×10−5 | 4 |
| KEGG | hsa00534 | Glycosaminoglycan
biosynthesis-heparan sulfate/heparin | 4/113 | 0.000418 | 4 |
| KEGG | hsa04514 | Cell adhesion
molecules (CAMs) | 8/113 | 0.001462 | 8 |
| KEGG | hsa04974 | Protein digestion
and absorption | 6/113 | 0.00233 | 6 |
| KEGG | hsa04512 | ECM-receptor
interaction | 5/113 | 0.007853 | 5 |
| KEGG | hsa00590 | Arachidonic acid
metabolism | 4/113 | 0.014923 | 4 |
Prognostic value of miRNAs and mRNAs
regulated by circRNAs
To identify potential prognostic indicators of
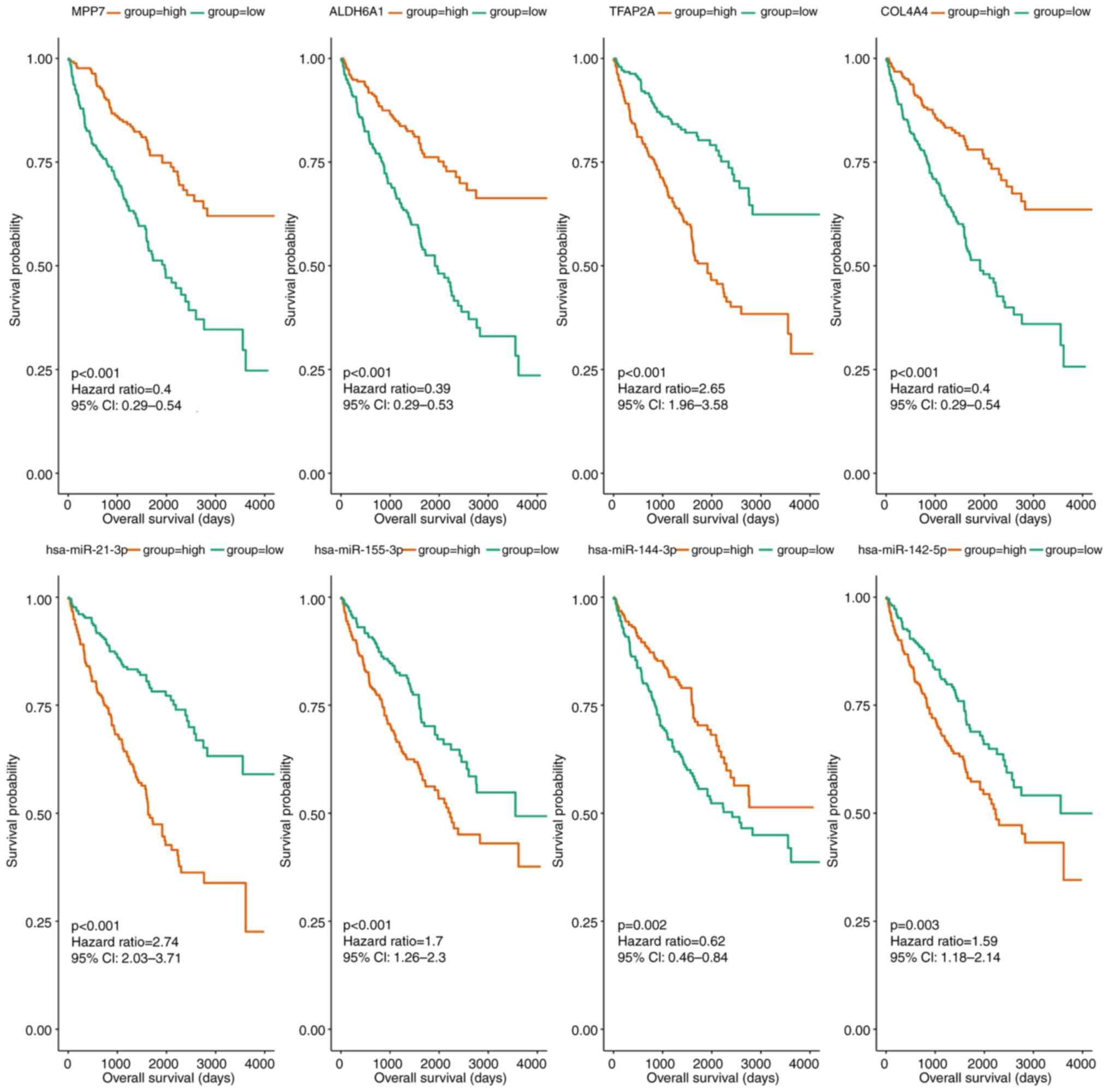
CCRCC, the miRNAs and mRNAs regulated by circRNAs were analyzed.
Following prognostic analysis, 125 genes and 10 miRNAs were
associated with the prognosis of CCRCC. Subsequently, as shown in
Fig. 4 and Table IV, the genes and miRNAs were
sorted based on P-value, after which the top 10 genes and miRNAs
were selected as hub genes [membrane palmitoylated protein 7
(MPP7), aldehyde dehydrogenase 6 family member A1 (ALDH6A1),
transcription factor AP-2α (TFAP2A), collagen type IV α 4 chain
(COL4A4), nuclear receptor subfamily 3 group C member 2 (NR3C2),
plasminogen (PLG), Holliday junction recognition protein (HJURP),
claudin 10 (CLDN10), kinesin family member 18B (KIF18B) and thyroid
hormone receptor β (THRB)] and hub miRNAs (hsa-miR-21-3p,
hsa-miR-155-3p, hsa-miR-144-3p, hsa-miR-142-5p, hsa-miR-875-3p,
hsa-miR-885-3p, hsa-miR-3941, hsa-miR-224-3p, hsa-miR-584-3p and
hsa-miR-138-1-3p).
 | Table IV.Prognostic analysis of hub genes and
miRNAs. |
Table IV.
Prognostic analysis of hub genes and
miRNAs.
| Gene | HR | 95% CI | P-value |
|---|
| MPP7 | 0.397 | 0.293–0.538 |
1.42×10−9 |
| ALDH6A1 | 0.388 | 0.287–0.525 |
1.70×10−9 |
| TFAP2A | 2.649 | 1.961–3.578 |
3.63×10−9 |
| COL4A4 | 0.398 | 0.295–0.538 |
7.88×10−9 |
| NR3C2 | 0.396 | 0.293–0.536 |
1.79×10−8 |
| PLG | 0.411 | 0.304–0.556 |
3.15×10−8 |
| HJURP | 2.455 | 1.817–3.319 |
5.84×10−8 |
| CLDN10 | 0.432 | 0.319–0.584 |
8.22×10−8 |
| KIF18B | 2.415 | 1.787–3.265 |
9.02×10−8 |
| THRB | 0.441 | 0.326–0.596 |
9.42×10−8 |
| hsa-mir-21-3p | 2.745 | 2.029–3.712 |
7.12×10−11 |
| hsa-mir-155-3p | 1.702 | 1.262–2.295 |
5.72×10−4 |
| hsa-mir-144-3p | 0.625 | 0.463–0.843 | 0.002 |
| hsa-mir-142-5p | 1.587 | 1.176–2.14 | 0.003 |
| hsa-mir-875-3p | 0.626 | 0.462–0.849 | 0.004 |
| hsa-mir-885-3p | 0.649 | 0.481–0.876 | 0.005 |
| hsa-mir-3941 | 0.71 | 0.526–0.958 | 0.025 |
| hsa-mir-224-3p | 1.371 | 1.017–1.849 | 0.039 |
| hsa-mir-584-3p | 0.734 | 0.544–0.989 | 0.045 |
|
hsa-mir-138-1-3p | 1.35 | 1.001–1.822 | 0.048 |
Discussion
ceRNAs are involved in a complex regulatory network
associated with the transcriptome; the present findings may improve
understanding of the regulatory mechanism underlying gene
expression. The present findings, along with other studies
(9,11,18),
suggest the importance of ceRNAs in the carcinogenesis of various
types of cancer. The present study analyzed GEO and TCGA datasets
to identify DECs, DEMs and DEGs in CCRCC. In addition, ceRNA
regulatory networks in CCRCC were constructed using these circRNAs,
miRNAs and genes. Additionally, the prognostic value of miRNAs and
genes regulated by circRNAs was determined to identify indicators
that may predict the prognosis of CCRCC.
By analyzing the data from a circRNA expression
microarray of CCRCC (GSE100186), which contained four CCRCC samples
and four normal samples, 324 downregulated and 218 upregulated
circRNAs were reported in the cancer group. The mRNA and miRNA
expression profiles of TCGA CCRCC dataset were used to identify
DEGs and DEMs between cancer and normal samples. Using the CSCD,
TargetScan and miRDB, a circRNA-miRNA-mRNA regulatory network was
constructed in CCRCC based on the ceRNA theory.
The results of enrichment analysis of the 85
upregulated genes in the circRNA-miRNA-upregulated mRNA network
suggested associated BP terms, including ‘flavonoid metabolic
process’, ‘cellular glucuronidation’, ‘uronic acid metabolic
process’, ‘glucuronate metabolic process’ and ‘response to
xenobiotic stimulus’. According to the results, CCRCC was
associated with various metabolic processes, including flavonoid,
uronic acid and glucuronate metabolism. The exact role of the
various metabolic pathways in the initiation, progression and
treatment of CCRCC requires further investigation. KEGG enrichment
analysis indicated the importance of ‘ascorbate and aldarate
metabolism’, ‘pentose and glucuronate interconversions’, ‘steroid
hormone biosynthesis’ and ‘retinol metabolism’. Steroid hormones
serve a critical role in the regulation of metabolism,
inflammation, immune functions, salt and water balance, the
development of sexual characteristics, and the ability to withstand
illness and injury (19). It has
previously been reported that glucocorticoids inhibit the
development of renal cancer by increasing the levels of Na and the
expression of K-ATPase β-1 subunit, which suggests the possible
benefits of glucocorticoids as a supplementary treatment in RCC
management (20). In addition,
aldosterone mediates the metastatic spread of renal cancer via its
G protein-coupled estrogen receptor (GPER); therefore, GPER
inhibitors may be considered promising therapeutic agents for
inhibiting metastatic spread (21). Further study into the specific
molecular mechanisms underlying the effects of steroid hormones on
CCRCC development is required.
In the present study, enrichment analysis was
performed on the 244 downregulated genes associated with the
circRNA-miRNA-downregulated mRNA network. The results indicated
critical BP terms, including ‘nephron development’, ‘kidney
development’ and ‘renal system development’. In addition, MF
analysis revealed ‘heparan sulfate sulfotransferase activity’ was
enriched in this network, whereas KEGG pathway enrichment analysis
suggested the importance of ‘ECM-receptor interaction’,
‘glycosaminoglycan biosynthesis-heparan sulfate/heparin’ and ‘cell
adhesion molecules (CAMs)’, ‘ECM-receptor interaction’ and
‘arachidonic acid metabolism’. CAMs are a subset of proteins that
maintain cellular polarity and inhibit tumor growth (22). Cell adhesion molecule 4, which is
one of the immunoglobulin-superfamily CAM proteins, has been
proposed to be involved in suppressing tumor invasion and formation
in CCRCC and nude mice (23). In
addition, dysregulated methylation and suppression of the tumor
inhibitor cell adhesion molecule 2 (CADM2) have been linked to
human renal cell carcinogenesis; therefore, CADM2 could be a
possible therapeutic target (24).
These findings suggested that analysis of biological terms may
provide novel insight into the complex mechanisms underlying the
development and progression of CCRCC.
In order to identify prognostic indicators of CCRCC,
the miRNAs and genes regulated by circRNAs were investigated.
According to the analysis, the top 10 genes and miRNAs were
selected as hub genes (MPP7, ALDH6A1, TFAP2A, COL4A4, NR3C2, PLG,
HJURP, CLDN10, KIF18B and THRB) and hub miRNAs (miR-21-3p,
miR-155-3p, miR-144-3p, miR-142-5p, miR-875-3p, miR-885-3p,
miR-3941, miR-224-3p, miR-584-3p and miR-138-1-3p). The tumor
suppressor gene TFAP2A has been reported to be hypermethylated and
markedly downregulated in RCC. Therefore, analysis of TFAP2A
methylation in cells obtained from urine or blood samples may be
valuable in early diagnosis (25).
The suppressive role of NR3C2 has been reported in various types of
cancer; low NR3C2 expression levels are correlated with aggressive
characteristics and poorer survival in non-distant metastatic CCRCC
(26). In a study investigating
the role of gene copy number variation in relation to the clinical
parameters of metastatic CCRCC, the loss of PLG was associated with
advanced tumor stage and Fuhrman grade (27). As for the hub miRNAs, miR-21 has
been widely studied in renal cancer for its regulatory roles in
cellular proliferation and metastasis (28–30).
In addition, the hub miRNA, miR-155, was determined to regulate the
growth and invasion of CCRCC cells by interacting with E2F
transcription factor 2 (31).
miR-155 has also been suggested to modulate the proliferation,
invasion and apoptosis of renal carcinoma cells by altering the
glycogen synthase kinase-3β/β-catenin pathway (32). Furthermore, miR-144-3p could
promote cell proliferation and migration in CCRCC by downregulating
AT-rich interactive domain-containing protein 1A (33), which was also regarded as a
possible novel plasma biomarker for the diagnosis of CCRCC
(34). This study identified
numerous ceRNAs that may serve a critical role in the development
of CCRCC; however, the specific mechanism as to how these ceRNAs
function requires further investigation.
In the present study, a series of circRNAs, miRNAs
and genes, which may be implicated in CCRCC, were identified. In
addition, the ceRNA regulatory network of circRNAs-miRNAs-genes was
constructed, and could serve as a wide-scale profile of the complex
regulation underlying the development of CCRCC. These findings may
not only provide insight into the etiology of CCRCC, but could aid
developments into the treatment of this disease.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The dataset generated and/or analyzed during the
current study are available in TCGA (https://cancergenome.nih.gov/)and GEO (https://www.ncbi.nlm.nih.gov/gds/).
Authors' contributions
CM, JQ and XLW analysed the data, CM, JPZ and DJW
wrote the manuscript, and XNL designed the study and wrote and
revised the article manuscript. JPZ and DJW identified the
databases and reviewed the data analysis. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schmidt LS and Linehan WM: Genetic
predisposition to kidney cancer. Semin Oncol. 43:566–574. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morris MR and Latif F: The epigenetic
landscape of renal cancer. Nat Rev Nephrol. 13:47–60. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moch H, Srigley J, Delahunt B, Montironi
R, Egevad L and Tan PH: Biomarkers in renal cancer. Virchows Arch.
464:359–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dabestani S, Marconi L and Bex A:
Metastasis therapies for renal cancer. Curr Opin Urol. 26:566–572.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu CH, Ou YC, Huang LH, Weng WC, Chang YK,
Chen HL, Hsu CY and Tung MC: Early dutasteride monotherapy in
patients with elevated serum prostate-specific antigen levels
following robot-assisted radical prostatectomy. Front Oncol.
9:6912019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Corgna E, Betti M, Gatta G, Roila F and De
Mulder PH: Renal cancer. Crit Rev Oncol Hematol. 64:247–262. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.PubMed/NCBI
|
|
10
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang K, Sun Y, Tao W, Fei X and Chang C:
Androgen receptor (AR) promotes clear cell renal cell carcinoma
(ccRCC) migration and invasion via altering the
circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett.
394:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang G, Xue W, Jian W, Liu P, Wang Z, Wang
C, Li H, Yu Y, Zhang D and Zhang C: The effect of Hsa_circ_0001451
in clear cell renal cell carcinoma cells and its relationship with
clinicopathological features. J Cancer. 9:3269–3277. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiong Y, Zhang J and Song C: CircRNA
ZNF609 functions as a competitive endogenous RNA to regulate FOXP4
expression by sponging miR-138-5p in renal carcinoma. J Cell
Physiol. 234:10646–10654. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai
F, Jin Y, Gao Y, Xia L, Chang H, et al: CSCD: A database for
cancer-specific circular RNAs. Nucleic Acids Res. 46:D925–D929.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
The Gene Ontology (GO) project in 2006.
Nucleic Acids Res. 34:D322–D326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sanchez-Mejias A and Tay Y: Competing
endogenous RNA networks: Tying the essential knots for cancer
biology and therapeutics. J Hematol Oncol. 8:302015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
D'Uva G and Lauriola M: Towards the
emerging crosstalk: ERBB family and steroid hormones. Semin Cell
Dev Biol. 50:143–152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huynh TP, Barwe SP, Lee SJ, McSpadden R,
Franco OE, Hayward SW, Damoiseaux R, Grubbs SS, Petrelli NJ and
Rajasekaran AK: Glucocorticoids suppress renal cell carcinoma
progression by enhancing Na,K-ATPase beta-1 subunit expression.
PLoS One. 10:e01224422015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feldman RD, Ding Q, Hussain Y, Limbird LE,
Pickering JG and Gros R: Aldosterone mediates metastatic spread of
renal cancer via the G protein-coupled estrogen receptor (GPER).
FASEB J. 30:2086–2096. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong X, Drgonova J, Li CY and Uhl GR:
Human cell adhesion molecules: Annotated functional subtypes and
overrepresentation of addiction-associated genes. Ann N Y Acad Sci.
1349:83–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nagata M, Sakurai-Yageta M, Yamada D, Goto
A, Ito A, Fukuhara H, Kume H, Morikawa T, Fukayama M, Homma Y and
Murakami Y: Aberrations of a cell adhesion molecule CADM4 in renal
clear cell carcinoma. Int J Cancer. 130:1329–1337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He W, Li X, Xu S, Ai J, Gong Y, Gregg JL,
Guan R, Qiu W, Xin D, Gingrich JR, et al: Aberrant methylation and
loss of CADM2 tumor suppressor expression is associated with human
renal cell carcinoma tumor progression. Biochem Biophys Res Commun.
435:526–532. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dalgin GS, Drever M, Williams T, King T,
DeLisi C and Liou LS: Identification of novel epigenetic markers
for clear cell renal cell carcinoma. J Urol. 180:1126–1130. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Z, Zhang M, Duan X, Deng T, Qiu H and
Zeng G: Low NR3C2 levels correlate with aggressive features and
poor prognosis in non-distant metastatic clear-cell renal cell
carcinoma. J Cell Physiol. 233:6825–6838. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nouhaud FX, Blanchard F, Sesboue R, Flaman
JM, Sabourin JC, Pfister C and Di Fiore F: Clinical relevance of
gene copy number variation in metastatic clear cell renal cell
carcinoma. Clin Genitourin Cancer. 16:e795–e805. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
An F, Liu Y and Hu Y: miR-21 inhibition of
LATS1 promotes proliferation and metastasis of renal cancer cells
and tumor stem cell phenotype. Oncol Lett. 14:4684–4688. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bera A, Ghosh-Choudhury N, Dey N, Das F,
Kasinath BS, Abboud HE and Choudhury GG: NFκB-mediated cyclin D1
expression by microRNA-21 influences renal cancer cell
proliferation. Cell Signal. 25:2575–2586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuan H, Xin S, Huang Y, Bao Y, Jiang H,
Zhou L, Ren X, Li L, Wang Q and Zhang J: Downregulation of PDCD4 by
miR-21 suppresses tumor transformation and proliferation in a nude
mouse renal cancer model. Oncol Lett. 14:3371–3378. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao Y, Ma X, Yao Y, Li H, Fan Y, Zhang Y,
Zhao C, Wang L, Ma M, Lei Z and Zhang X: miR-155 regulates the
proliferation and invasion of clear cell renal cell carcinoma cells
by targeting E2F2. Oncotarget. 7:20324–20337. 2016.PubMed/NCBI
|
|
32
|
Wei RJ, Zhang CH and Yang WZ: MiR-155
affects renal carcinoma cell proliferation, invasion and apoptosis
through regulating GSK-3β/β-catenin signaling pathway. Eur Rev Med
Pharmacol Sci. 21:5034–5041. 2017.PubMed/NCBI
|
|
33
|
Xiao W, Lou N, Ruan H, Bao L, Xiong Z,
Yuan C, Tong J, Xu G, Zhou Y, Qu Y, et al: Mir-144-3p promotes cell
proliferation, metastasis, sunitinib resistance in clear cell renal
cell carcinoma by downregulating ARID1A. Cell Physiol Biochem.
43:2420–2433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lou N, Ruan AM, Qiu B, Bao L, Xu YC, Zhao
Y, Sun RL, Zhang ST, Xu GH, Ruan HL, et al: miR-144-3p as a novel
plasma diagnostic biomarker for clear cell renal cell carcinoma.
Urol Oncol. 35:36.e7–36.e14. 2017. View Article : Google Scholar
|
|
35
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|