In the 1970s, circRNAs, a unique closed circular
form of non-coding RNA, were first discovered in plant viruses
(1) and were considered a
by-product of aberrant splicing reactions (2). In 2012, hundreds of circRNAs from
different cell types were detected by RNA sequencing technology
(3). With the development of
high-throughput sequencing technology, several circRNAs have been
detected in eukaryotes; however, they have been regarded as
splicing by-products without biological functions (4). Unlike linear RNAs, which possess
different 5′ and 3′ ends that indicate the start and stop sites of
transcription, circRNAs are the product of one or two exons being
spliced together via covalent bonding of the 3′ and 5′ ends,
resulting in establishment of a covalently closed continuous loop
(4–7). Therefore, circRNAs are unlikely to be
degraded by RNA exonuclease and are more stable than their linear
counterparts (7–9); circRNA functions may be linked to
their unique stability (9).
Notably, previous studies have indicated that circRNAs can sponge
microRNAs (miRNAs/miRs) or functional proteins to regulate specific
biological functions at the transcriptional or post-transcriptional
level (6,9–11).
Therefore, circRNAs may be considered promising diagnostic
biomarkers and therapeutic targets for numerous diseases.
Osteonecrosis of the femoral head (ONFH) is a
disabling clinical disease that is most common among young adults
(12). The mean age at diagnosis
is typically <50 years (13).
If no early intervention is provided, ~80% of patients progress to
femoral head collapse, hip dysfunction and permanent disability
(14), and ultimately hip
replacement becomes the only treatment option (13). However, postoperative infection,
pain, functional rehabilitation, prosthetic replacement and other
related complications (15–18)
result in vast economic burdens for patients and for society. The
mechanism by which ONFH develops remains unclear (19). The majority of experts agree that a
lack of blood supply to the femoral head and bone marrow causes
death of osteocytes, as explained by the microvascular injury
theory, osteoporosis theory, apoptosis theory, osteogenic
enhancement and adipogenic weakening theory of bone marrow
mesenchymal stem cells (20–24).
It has previously been reported that numerous circRNAs are
associated with ONFH, which suggested that they may have a critical
role in the development and progression of ONFH (25). This review aimed to describe the
evidence regarding the biogenesis and biological functions of
circRNAs, and to identify their potential mechanistic roles in
ONFH.
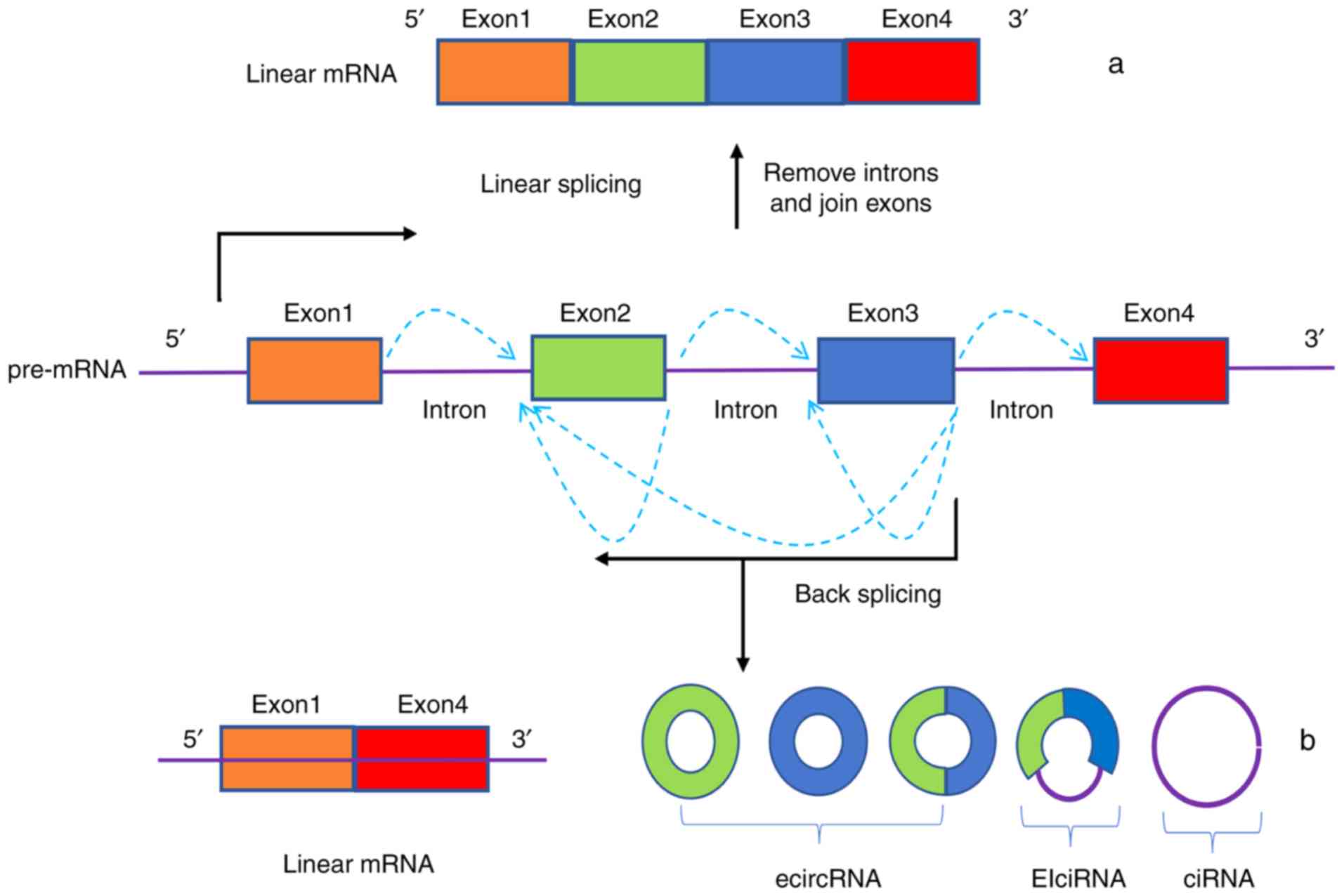
The manner by which circRNAs are generated is
different to how linear RNA transcripts are formed. Notably, the
methods for generating circRNAs are either direct splicing or
lasso-driven cyclization of precursor RNA, both with classical RNA
polymerase-mediated splicing out of introns and connecting of exons
via 5′ or 3′ polarities (6,7,9,10).
These two methods can form exon circRNAs (exon sequence only,
ecircRNAs), intron circRNAs (composed of intron sequences, ciRNAs)
and exon-intron circRNAs (exon and intron sequences, EIciRNAs)
(Fig. 1); ecircRNAs account for
>80% of total circRNAs (26).
The 3′ end of the upstream exon of the precursor mRNA is covalently
linked to the 5′ end of the downstream exon by a reverse splicing
process, after which the intron is cleaved to form an ecircRNA. If
introns are retained, then EIciRNAs are formed (27). CiRNAs contain an exclusive 2′-5′
linkage that distinguishes them from ecircRNAs, and their structure
relies on 7 nt GU-rich sequences near the 5′splice site and C-rich
sequences that are 11 nt in length (28). Another novel model of circRNA
biosynthesis has proposed that some RNA-binding proteins that have
intron-binding abilities interact with intron-binding sites to
promote the formation of circRNAs (29). Common RNA-binding proteins are
protein quaking (29) and
Drosophila muscle blind protein (30).
miRNAs are a class of non-coding RNA that contain
~20 nucleotides and bind to the 3′-untranslated region of mRNAs to
inhibit their translation (31).
Previous studies have offered evidence to suggest that some
circRNAs act as aggressive endogenous RNAs that compete for
miRNA-binding sites (10,11,32).
One study reported that circRNA E3 ubiquitin-protein ligase CHFR
serves as a sponge for miR-370, which typically targets forkhead
box protein (FOX)O1, and FOXO1 promotes the expression of cyclin D1
to drive the migration and proliferation of vascular smooth muscle
cells. This provides an example of a profound role that circRNAs
can serve in cardiovascular diseases (33). In addition, there are a number of
overlapping binding sites between circRNAs and miRNAs, and a single
circRNA can possibly interact with several miRNAs. For example, the
mouse sex determination region Y is a testis-specific circRNA that
has 16 binding sites and functions as a sponge for miR-138
(11), and circRNA
homeodomain-interacting protein kinase 3 (circHIPK3) acts as a
sponge for nine miRNAs (miR-654, miR-584, miR-379, etc.) and has 18
potential binding sites (34).
However, due to the numerous binding possibilities between circRNAs
and miRNAs, and the possibility of circRNAs interacting with
multiple miRNAs, the specific mechanisms underlying the
interactions between circRNAs and miRNAs requires further
research.
As they possess a partial translation initiation
codon and an open reading frame, the coding function of circRNAs
has been confirmed by various studies (35,36).
Therefore, a comprehensive database of annotated human circRNAs has
been constructed, which includes ~33,000 ecircRNAs (4). However, if an internal ribosome entry
point is inserted upstream of the start codon of circRNAs, some
proteins can be produced that are different from their linear
transcripts (37). Legnini et
al concluded that circRNA zinc finger protein 609 is associated
with the process of muscle development and acts as a linear coding
RNA that is translated to produce proteins, which provides a unique
example of protein-coding circRNAs in eukaryotes (38). The translation of circRNAs has been
confirmed and is closely related to the prognosis of certain
diseases (39). Furthermore, it
has been verified that circRNAs are modified by N6-methyladenosine
(m6A), which induces base modification in mRNA, and there is one
m6A site in circRNAs that is beneficial for promoting their
translation (40).
In addition to miRNAs, circRNAs also bind to
proteins to modulate their corresponding functions. ciRNAs and
EIciRNAs are mainly located in the nucleus and have unique tertiary
structures that act as binding sites for RNA-binding proteins;
therefore, these circRNAs can regulate gene transcription by
directly interacting with RNA-binding proteins (28,41).
For example, circRNA FOXO3 (circFOXO3) inhibited cell cycle
progression by binding to cyclin-dependent kinase 2 and
cyclin-dependent kinase inhibitor 1, leading to the formation of a
ternary complex (42). Notably, Du
et al (43) demonstrated
that overexpression of circFOXO3 reduced the binding between FOXO3
and murine double minute 2 (MDM2), and blunted the effect of MDM2
on modulating the polyubiquitination of FOXO3, which strengthened
FOXO3 activity and promoted cell apoptosis (43). These findings suggested that the
same circRNA may bind to different proteins in different tissues
and cells to perform various functions.
It has been reported that circRNAs regulate
alternative splicing, transcription, exosomal function and the
formation of circRNA pseudogenes, all of which affect the
occurrence and progression of disease (6,30,44,45).
Accumulating studies have demonstrated that circRNAs are highly
associated with various diseases, such as cancer, kidney disease,
diabetes, cardiovascular disease, Alzheimer's disease and
osteoarthritis (OA) (6,7,46).
However, the relationship between circRNAs and the initiation of
ONFH is still largely unclear; the present review provides a
summary of what is currently known.
Weakened osteogenic differentiation and increased
adipogenic differentiation of BMSCs are closely associated with the
formation of ONFH (47). miRNAs,
which can be negatively regulated by the competitive binding of
circRNAs, serve an extremely important role in regulating stem cell
differentiation (48).
Differential expression of 23 miRNAs has been identified in the
BMSCs of steroid-induced ONFH (SONFH) (49). Furthermore, various studies have
demonstrated that promoting osteogenic differentiation and
inhibiting adipogenic differentiation of BMSCs by regulating the
expression level of miRNAs can achieve the goal of effectively
treating ONFH (24,50,51).
A total of 231 preferentially expressed circRNAs
were detected in the BMSCs of SONFH compared with the control
group; 90 were upregulated and 141 were downregulated (25). CircRNAs were also differentially
expressed during the osteogenic differentiation of BMSCs, 95% of
which were protein-coding genes (52). In addition, silencing circRNA
immunoglobulin superfamily member 11 promotes osteoblast
differentiation and increases the expression of miR-199b-5p
(52), which exerts its role in
BMSC osteogenesis through the glycogen synthase kinase 3β/β-catenin
signaling pathway (53) and acts
as an important therapeutic targets during early-stage ONFH
(54). A recent report indicated
that circRNA FOXP1 acts as a sponge for several miRNAs and serves a
pivotal role in the regulation of MSC differentiation (55). Additionally, Kuang et al
(56) demonstrated that circRNA
ubiquitin-specific protease 45 can upregulate the expression of
phosphatase and tensin homolog through sponging miR-127-5p, which
inhibits the protein kinase B pathway and regulates bone mass in
rat SONFH. Therefore, circRNAs acting as miRNA sponges may be
associated with osteogenic differentiation of BMSCs and ONFH.
Although few studies have been performed on adipogenic
differentiation, the aforementioned observations provide a good
direction for studying circRNA, BMSCs and ONFH.
Osteoblasts are the primary cells that function in
bone formation, which are essential for mineralization of bone
matrix (57). Osteoclasts secrete
proteinases and hydrogen ions to degrade the organic bone matrix
and dissolve bone minerals, respectively (57). Maintaining a balance between
osteoblasts and osteoclasts is crucial for maintaining normal bone
mass (58). Bone cell metabolism
is irreversibly destroyed by local circulatory disorders, and leads
to the disappearance of osteoblasts, activation of osteoclasts, the
eventual destruction of trabecular bone and increased bone
fragility (59). This destructive
pathway was verified by the observation that osteoclast-related
activity is increased in the subchondral bone and necrotic areas of
ONFH tissues, whereas osteoblast activity is increased in the
sclerotic region (60).
circRNA 19142 (circ19142) and circRNA 5846
(circ5846) act as sponges for miR-7067-5p; this is a network that
is confirmed to function in osteoblast differentiation through a
circ19142/circ5846-targeted miRNA-mRNA axis (61). Dou et al (62) reported that 19 circRNAs were
upregulated and five circRNAs were downregulated during different
stages of mouse osteoclast formation. In addition, 260
differentially expressed circRNAs were detected between peripheral
blood lymphocytes from postmenopausal patients with osteoporosis
and controls (63). It has also
been demonstrated that the expression of circRNA 0001275 is
markedly increased in postmenopausal patients with osteoporosis;
therefore, it was concluded that it may be a potential diagnostic
biomarker (64). In summary, it
was hypothesized that circRNAs may have an important role in ONFH,
which is closely related to the activity of osteoblasts and
osteoclasts. Therefore, circRNAs may have a role as diagnostic
biomarkers. Identifying circRNAs that are preferentially expressed
in peripheral blood lymphocytes, during necrotic collapse or in
sclerotic bone tissue may provide a novel direction for studying
the relationship between circRNAs and ONFH.
Endothelial cell damage, intravascular coagulation
and disordered angiogenesis lead to ischemia, which is considered
to be one of the core pathological results of ONFH (20,22).
Endothelial cell tube formation is the first step in
neovascularization (65), and
blood vessels are critical in bone remodeling because they supply
nutrients (66). It has been
reported that SONFH may cause miRNA alterations in femoral head
bone microvascular endothelial cells that are not seen in controls
(67). The vascularity of the bone
in osteonecrosis is reduced by ~50% (68). Additionally, a reduction in bone
strength and vascularity of the bone precedes bone mass reduction
and microstructural deterioration (69). Previous studies have suggested that
ameliorating vascular endothelial cell proliferation, migration and
tube formation may positively promote bone vascularization in the
femoral head and prevent ONFH (70,71).
Notably, circRNAs are expressed in endothelial cells
and serve a biological role in angiogenesis (72). For example, circRNA 0003575 is
upregulated in oxidized low-density lipoprotein-induced human
umbilical vein endothelial cells (HUVECs), and promotes HUVEC
proliferation and angiogenesis (73). CircRNA 0010729 mediates vascular
endothelial cell apoptosis and proliferation by targeting the
miR-186/hypoxia inducible factor-1α axis (74). Furthermore, Shan et al
(75) reported that circHIPK3
expression is substantially upregulated in endothelial cells during
diabetic retinal vasculopathy, and in vitro endothelial cell
viability, proliferation, migration, and tube formation are altered
by silencing or overexpressing circHIPK3. Acting as an endogenous
miR-30a-3p sponge, in vivo silencing of circHIPK3 also
attenuates retinal vascular dysfunction (75). Therefore, it was hypothesized that
circRNAs may be involved in the endothelial cell damage and
disruption of angiogenesis that lead to the formation of ONFH;
circRNAs may be considered potential therapeutic targets.
It is generally believed that structural damage of
the subchondral bone in post-collapse cases of osteonecrosis
contributes to degeneration of articular cartilage (76). Notably, at the beginning of ONFH,
the subchondral bone receives reduced nutrition because the blood
supply to the area is impaired; that, coupled with degeneration of
the cartilage matrix, leads to degeneration of articular cartilage,
which can increase instability of the hip joint and accelerate the
development of ONFH (77,78). Therefore, prevention and early
treatment of hip cartilage damage are beneficial for ameliorating
the progression of ONFH (78).
Cartilage inflammation can be enhanced by
interleukin-21, which causes degradation of the cartilage in
patients with ONFH through the Janus kinase/signal transducers and
activators of transcription signaling pathway (79). However, studies on the role of
circRNAs in cartilage degeneration are currently more focused on OA
(80,81). In addition, it has been reported
that intermittent cyclic mechanical tension leads to differential
expression of miRNAs, which regulates stromal metabolism and
calcification of cartilage endplate chondrocytes via the
transforming growth factor-β signaling pathway (82). Furthermore, differentially
expressed circRNAs are detected in different regions of cartilage
in patients with OA; circRNA 100226 is associated with mechanical
tension, and it was demonstrated that circRNA 100226, acting as a
sponge for miR-875, promotes the degradation of cartilage matrix by
controlling the expression of tumor necrosis factor α (83). Therefore, it was concluded that
mechanical stress changes in the early stages and in the collapsed
areas of ONFH may lead to alterations in the expression of miRNAs
and circRNAs in the corresponding regional cartilage. The molecular
mechanism underlying the function of circRNAs in the degradation of
ONFH cartilage damage is essential for understanding the
pathogenesis of ONFH and exploring novel prospective therapeutic
targets.
CircRNAs were discovered decades ago. With advances
in research methods, increasing attention has been paid to
circRNAs. Notably, it has been reported that circRNAs are involved
in the development and progression of various diseases (6,7,46).
Although differential expression of circRNAs was observed in BMSCs
of ONFH and a possible connection was suggested (25), to the best of our knowledge, no
further functional or molecular mechanisms have been investigated.
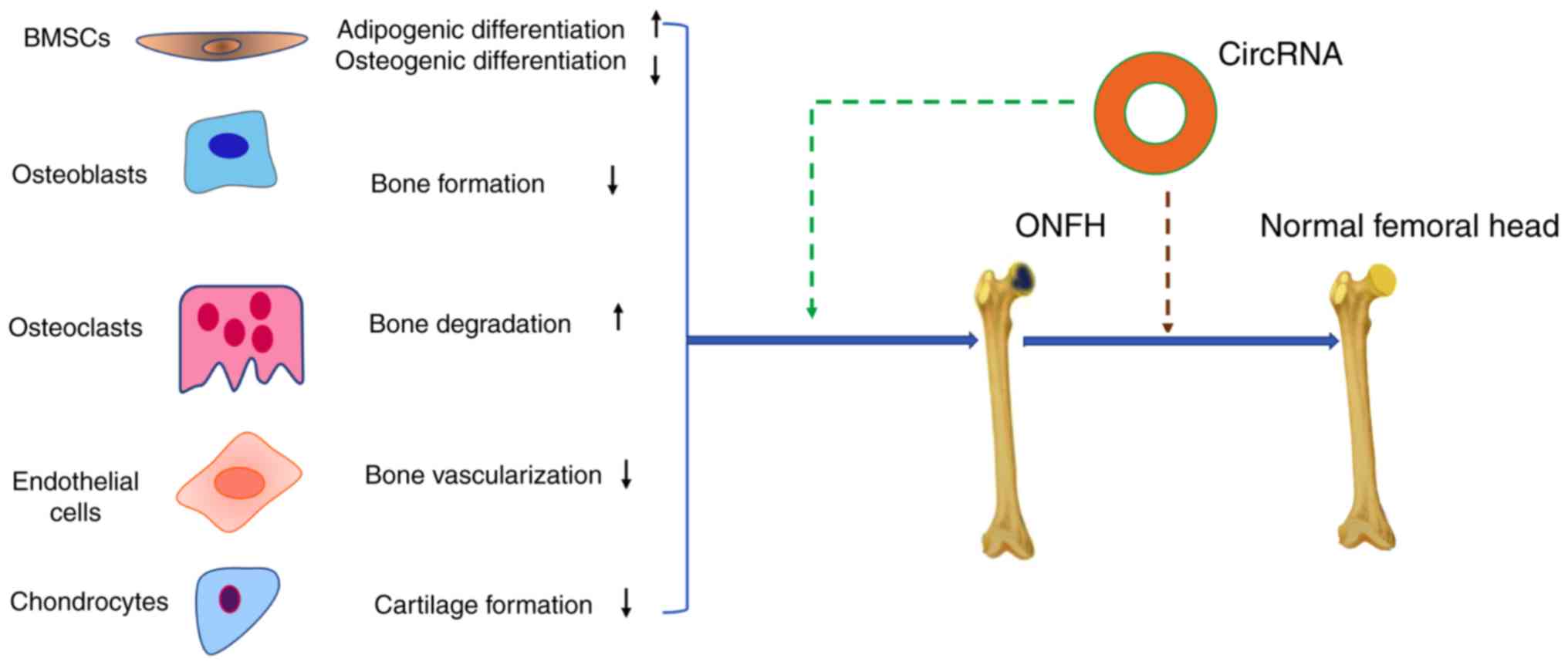
With respect to the potential pathological mechanisms of ONFH, this
review describes the potential relationship between BMSCs,
osteoblasts, osteoclasts, vascular endothelial cells, articular
cartilage and circRNAs (Fig. 2).
Addressing these problems may be beneficial to the prevention of
ONFH and the development of therapeutic targets for its
treatment.
CircRNAs can function as miRNA sponges, regulate
gene transcription and interact with RNA-binding proteins (10,11,28,29).
Most circRNA studies focus on their role as miRNA sponges, that is,
the related mechanisms and functions of the circRNA-miRNA-mRNA axis
in human diseases (84). However,
a single circRNA has numerous binding sites for one miRNA and can
interact with multiple miRNAs; therefore, more extensive research
is required to illuminate the functional interactions between
circRNAs and miRNAs. In addition, the role of circRNAs in
regulating protein translation and binding to RNA-binding proteins
in various diseases requires additional research.
In conclusion, circRNAs have a potential role in the
initiation mechanisms of ONFH, and they exhibit high stability
compared with linear RNAs. Therefore, it may be hypothesized that
circRNAs serve a unique role in the formation ONFH and could have a
role in ONFH therapy.
Not applicable.
No funding was received.
Not applicable.
JZ was responsible for reviewing the concept design,
and wrote and proofread the article. LM created the figures and
made important comments on the revision of the article. ZW, XF, XH,
XZ and XX participated in literature collection, analysis and
summary. XZ was responsible for project guidance. All authors read
and agree to the final text.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen X, Han P, Zhou T, Guo X, Song X and
Li Y: circRNADb: A comprehensive database for human circular RNAs
with protein-coding annotations. Sci Rep. 6:349852016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greene J, Baird AM, Brady L, Lim M, Gray
SG, McDermott R and Finn SP: Circular RNAs: Biogenesis, function
and role in human diseases. Front Mol Biosci. 4:382017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aufiero S, Reckman YJ, Pinto YM and
Creemers EE: Circular RNAs open a new chapter in cardiovascular
biology. Nat Rev Cardiol. 16:503–514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Enuka Y, Lauriola M, Feldman ME, Sas-Chen
A, Ulitsky I and Yarden Y: Circular RNAs are long-lived and display
only minimal early alterations in response to a growth factor.
Nucleic Acids Res. 44:1370–1383. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Müller S and Appel B: In vitro
circularization of RNA. RNA Biol. 14:1018–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui L, Zhuang Q, Lin J, Jin J, Zhang K,
Cao L, Lin J, Yan S, Guo W, He W, et al: Multicentric epidemiologic
study on six thousand three hundred and ninety five cases of
femoral head osteonecrosis in China. Int Orthop. 40:267–276. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lespasio MJ, Sodhi N and Mont MA:
Osteonecrosis of the Hip: A primer. Perm J. 23:18–100. 2019.
|
|
14
|
Kuroda Y, Matsuda S and Akiyama H:
Joint-preserving regenerative therapy for patients with early-stage
osteonecrosis of the femoral head. Inflamm Regen. 36:42016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mufarrih SH, Qureshi NQ, Sadruddin A,
Hashmi P, Mahmood SF, Zafar A and Noordin S: Relationship between
staphylococcus aureus carriage and surgical site infections
following total hip and knee arthroplasty in the South Asian
Population: Protocol for a prospective cohort study. JMIR Res
Protoc. 7:e102192018. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Robertson-Waters E, Berstock JR,
Whitehouse MR and Blom AW: Surgery for greater trochanteric pain
syndrome after total hip replacement confers a poor outcome. Int
Orthop. 42:77–85. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bandholm T, Wainwright TW and Kehlet H:
Rehabilitation strategies for optimisation of functional recovery
after major joint replacement. J Exp Orthop. 5:442018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hauer G, Vielgut I, Amerstorfer F,
Maurer-Ertl W, Leithner A and Sadoghi P: Survival rate of
Short-stem hip prostheses: A comparative analysis of clinical
studies and national arthroplasty registers. J Arthroplasty.
33:1800–1805. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arbab D and König DP: Atraumatic femoral
head necrosis in adults. Dtsch Arztebl Int. 113:31–38.
2016.PubMed/NCBI
|
|
20
|
Zhang Q, L VJ and Jin L: Role of
coagulopathy in glucocorticoid-induced osteonecrosis of the femoral
head. J Int Med Res. 46:2141–2148. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Petek D, Hannouche D and Suva D:
Osteonecrosis of the femoral head: Pathophysiology and current
concepts of treatment. EFORT Open Rev. 4:85–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kerachian MA, Séguin C and Harvey EJ:
Glucocorticoids in osteonecrosis of the femoral head: A new
understanding of the mechanisms of action. J Steroid Biochem Mol
Biol. 114:121–128. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zalavras C, Shah S, Birnbaum MJ and
Frenkel B: Role of apoptosis in glucocorticoid-induced osteoporosis
and osteonecrosis. Crit Rev Eukaryot Gene Expr. 13:221–235. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhun W, Donghai L, Zhouyuan Y, Haiyan Z
and Pengde K: Efficiency of cell therapy to GC-induced ONFH: BMSCs
with Dkk-1 interference is not superior to unmodified BMSCs. Stem
Cells Int. 2018:13402522018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiang Shuai: The study of changed
biological behavior and aberrantly expressed transcriptome in BMSCs
in seroid-induced osteonecrosis (D). Peking Union Medical College.
2018.
|
|
26
|
Wang W, Wang Y, Piao H, Li B, Huang M, Zhu
Z, Li D, Wang T, Xu R and Liu K: Circular RNAs as potential
biomarkers and therapeutics for cardiovascular disease. PeerJ.
7:e68312019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang L, Yang F, Zhao H, Wang M and Zhang
Y: Circular RNA circCHFR facilitates the proliferation and
migration of vascular smooth muscle via miR-370/FOXO1/Cyclin D1
pathway. Mol Ther Nucleic Acids. 16:434–441. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu J, Qi X, Liu L, Hu X, Liu J, Yang J,
Yang J, Lu L, Zhang Z, Ma S, et al: Emerging epigenetic regulation
of circular RNAs in human cancer. Mol Ther Nucleic Acids.
16:589–596. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing Glioma tumorigenesis. J Natl Cancer
Inst. 1102018.doi: 10.1093/jnci/djx166.
|
|
40
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and Characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL
and Yang L: CircRNA-derived pseudogenes. Cell Res. 26:747–750.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu CX and Sun S: An emerging role for
circular RNAs in osteoarthritis. Yonsei Med J. 59:349–355. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Houdek MT, Wyles CC, Packard BD, Terzic A,
Behfar A and Sierra RJ: Decreased osteogenic activity of
mesenchymal stem cells in patients with corticosteroid-induced
osteonecrosis of the femoral head. J Arthroplasty. 31:893–898.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ong SG, Lee WH, Kodo K and Wu JC:
MicroRNA-mediated regulation of differentiation and
trans-differentiation in stem cells. Adv Drug Deliv Rev. 88:3–15.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang B, Yu P, Li T, Bian Y and Weng X:
MicroRNA expression in bone marrow mesenchymal stem cells from mice
with steroid-induced osteonecrosis of the femoral head. Mol Med
Rep. 12:7447–7454. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li R, Lin QX, Liang XZ, Liu GB, Tang H,
Wang Y, Lu SB and Peng J: Stem cell therapy for treating
osteonecrosis of the femoral head: From clinical applications to
related basic research. Stem Cell Res Ther. 9:2912018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gu C, Xu Y, Zhang S, Guan H, Song S, Wang
X, Wang Y, Li Y and Zhao G: miR-27a attenuates adipogenesis and
promotes osteogenesis in steroid-induced rat BMSCs by targeting
PPARgamma and GREM1. Sci Rep. 6:384912016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang M, Jia L and Zheng Y: circRNA
expression profiles in human bone marrow stem cells undergoing
osteoblast differentiation. Stem Cell Rev. 15:126–138. 2019.
View Article : Google Scholar
|
|
53
|
Zhao R, Li Y, Lin Z, Wan J, Xu C, Zeng Y
and Zhu Y: miR-199b-5p modulates BMSC osteogenesis via suppressing
GSK-3β/β-catenin signaling pathway. Biochem Biophys Res Commun.
477:749–754. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Huang L, Wang Y, Jiang Y, Wu Y, Hu C and
Ouyang H: High levels of GSK-3β signalling reduce osteogenic
differentiation of stem cells in osteonecrosis of femoral head. J
Biochem. 163:243–251. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cherubini A, Barilani M, Rossi RL, Jalal
MMK, Rusconi F, Buono G, Ragni E, Cantarella G, Simpson H, Péault B
and Lazzari L: FOXP1 circular RNA sustains mesenchymal stem cell
identity via microRNA inhibition. Nucleic Acids Res. 47:5325–5340.
2019.PubMed/NCBI
|
|
56
|
Kuang MJ, Xing F, Wang D, Sun L, Ma JX and
Ma XL: CircUSP45 inhibited osteogenesis in glucocorticoid-induced
osteonecrosis of femoral head by sponging miR-127-5p through
PTEN/AKT signal pathway: Experimental studies. Biochem Biophys Res
Commun. 509:255–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mandelin J, Hukkanen M, Li TF, Korhonen M,
Liljeström M, Sillat T, Hanaemaijer R, Salo J, Santavirta S and
Konttinen YT: Human osteoblasts produce cathepsin K. Bone.
38:769–777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen X, Wang Z, Duan N, Zhu G, Schwarz EM
and Xie C: Osteoblast-osteoclast interactions. Connect Tissue Res.
59:99–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang C, Wang X, Xu XL, Yuan XL, Gou WL,
Wang AY, Guo QY, Peng J and Lu SB: Bone microstructure and regional
distribution of osteoblast and osteoclast activity in the
osteonecrotic femoral head. PLoS One. 9:e963612014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang C, Meng H, Wang Y, Zhao B, Zhao C,
Sun W, Zhu Y, Han B, Yuan X, Liu R, et al: Analysis of early stage
osteonecrosis of the human femoral head and the mechanism of
femoral head collapse. Int J Biol Sci. 14:156–164. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qian DY, Yan GB, Bai B, Chen Y, Zhang SJ,
Yao YC and Xia H: Differential circRNA expression profiles during
the BMP2-induced osteogenic differentiation of MC3T3-E1 cells.
Biomed Pharmacother. 90:492–499. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dou C, Cao Z, Yang B, Ding N, Hou T, Luo
F, Kang F, Li J, Yang X, Jiang H, et al: Changing expression
profiles of lncRNAs, mRNAs, circRNAs and miRNAs during
osteoclastogenesis. Sci Rep. 6:214992016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jin D, Wu X, Yu H, Jiang L, Zhou P, Yao X,
Meng J, Wang L, Zhang M and Zhang Y: Systematic analysis of
lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal
osteoporosis. Am J Transl Res. 10:1498–1510. 2018.PubMed/NCBI
|
|
64
|
Zhao K, Zhao Q, Guo Z, Chen Z, Hu Y, Su J,
Chen L, He Z, Cai X, Chen M, et al: Hsa_Circ_0001275: A potential
novel diagnostic biomarker for postmenopausal osteoporosis. Cell
Physiol Biochem. 46:2508–2516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Carulli C, Innocenti M and Brandi ML: Bone
vascularization in normal and disease conditions. Front Endocrinol
(Lausanne). 4:1062013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sivaraj KK and Adams RH: Blood vessel
formation and function in bone. Development. 143:2706–2715. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yue J, Wan F, Zhang Q, Wen P, Cheng L, Li
P and Guo W: Effect of glucocorticoids on miRNA expression spectrum
of rat femoral head microcirculation endothelial cells. Gene.
651:126–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lane NE: Glucocorticoid-induced
osteoporosis: New insights into the pathophysiology and treatments.
Curr Osteoporos Rep. 17:1–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Weinstein RS, Hogan EA, Borrelli MJ,
Liachenko S, O'Brien CA and Manolagas SC: The pathophysiological
sequence of glucocorticoid-induced osteonecrosis of the femoral
head in male mice. Endocrinology. 158:3817–3831. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu X, Li Q, Niu X, Hu B, Chen S, Song W,
Ding J, Zhang C and Wang Y: Exosomes secreted from human-induced
pluripotent stem cell-derived mesenchymal stem cells prevent
osteonecrosis of the femoral head by promoting angiogenesis. Int J
Biol Sci. 13:232–244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang Y, Yin J, Ding H, Zhang C and Gao
YS: Vitamin K2 ameliorates damage of blood vessels by
glucocorticoid: A potential mechanism for its protective effects in
glucocorticoid-induced osteonecrosis of the femoral head in a rat
model. Int J Biol Sci. 12:776–785. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Boeckel JN, Jaé N, Heumüller AW, Chen W,
Boon RA, Stellos K, Zeiher AM, John D, Uchida S and Dimmeler S:
Identification and characterization of hypoxia-regulated
endothelial circular RNA. Circ Res. 117:884–890. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li CY, Ma L and Yu B: Circular RNA
hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells
proliferation and angiogenesis. Biomed Pharmacother. 95:1514–1519.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dang RY, Liu FL and Li Y: Circular RNA
hsa_circ_0010729 regulates vascular endothelial cell proliferation
and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys
Res Commun. 490:104–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Shan K, Liu C, Liu BH, Chen X, Dong R, Liu
X, Zhang YY, Liu B, Zhang SJ, Wang JJ, et al: Circular noncoding
RNA HIPK3 mediates retinal vascular dysfunction in diabetes
mellitus. Circulation. 136:1629–1642. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Magnussen RA, Guilak F and Vail TP:
Cartilage degeneration in post-collapse cases of osteonecrosis of
the human femoral head: Altered mechanical properties in tension,
compression, and shear. J Orthop Res. 23:576–583. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen G, Zhong L, Wang Q, Li Z, Shang J,
Yang Q, Du Z, Wang J, Song Y and Zhang G: The expression of
chondrogenesis-related and arthritis-related genes in human ONFH
cartilage with different Ficat stages. PeerJ. 7:e63062019.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Xu R, Wei B, Li J, Huang C, Lin R, Tang C,
Xu Y, Yao Q and Wang L: Investigations of cartilage matrix
degeneration in patients with Early-stage femoral head necrosis.
Med Sci Monit. 23:5783–5792. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chen B, Liu Y and Cheng L: IL-21 enhances
the degradation of cartilage through the JAK-STAT signaling pathway
during osteonecrosis of femoral head cartilage. Inflammation.
41:595–605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhou Z, Du D, Chen A and Zhu L: Circular
RNA expression profile of articular chondrocytes in an
IL-1β-induced mouse model of osteoarthritis. Gene. 644:20–26. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Shen S, Wu Y, Chen J, Xie Z, Huang K, Wang
G, Yang Y, Ni W, Chen Z, Shi P, et al: CircSERPINE2 protects
against osteoarthritis by targeting miR-1271 and ETS-related gene.
Ann Rheum Dis. 78:826–836. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Feng C, Liu M, Fan X, Yang M, Liu H and
Zhou Y: Intermittent cyclic mechanical tension altered the microRNA
expression profile of human cartilage endplate chondrocytes. Mol
Med Rep. 17:5238–5246. 2018.PubMed/NCBI
|
|
83
|
Liu Q, Zhang X, Hu X, Yuan L, Cheng J,
Jiang Y and Ao Y: Emerging roles of circRNA related to the
mechanical stress in human cartilage degradation of osteoarthritis.
Mol Ther Nucleic Acids. 7:223–230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Rong D, Su H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|