Introduction
Non-alcoholic fatty liver disease (NAFLD) is one of
the most common chronic liver diseases worldwide and is an
important global public health issue (1). NAFLD is observed in metabolic
syndrome, and is typically accompanied by hepatic insulin
resistance, dyslipidemia and hyperglycemia (2); it consists of a wide spectrum of
liver pathologies ranging from simple steatosis to non-alcoholic
steatohepatitis (NASH) with fibrosis, which may progress to liver
cirrhosis and even hepatocellular carcinoma (3). Current non-pharmacological therapies
for NAFLD are mainly based on physical activity and dietary
modifications, such as the consumption of a Mediterranean diet,
which has been demonstrated to be effective (4–6).
Advanced glycation end-products (AGEs) are products
of non-enzymatic reactions of reducing sugars that have free amino
groups of nucleotides, lipids and peptides/proteins (7). AGEs may be formed endogenously as a
part of normal metabolism and aging, particularly under conditions
of high plasma glucose, or exogenously, entering the body through
the ingestion of certain foods, such as barbecued food, cheeses,
and foods high in fat and sugar. Dry-heat-cooked foods are typical
in the modern diet; these contain high levels of AGEs and their
consumption contributes to high levels of AGEs in the body
(8). Notably, the consumption of
modern diets containing excessive AGEs has increased in the past 20
years (9). The accumulation of
AGEs can exert detrimental effects and accelerate the progression
of AGE-related damage, such as diabetes, atherosclerosis and NAFLD
(10–12); however, the exact mechanisms
underlying the effects of dietary AGEs on NAFLD remain largely
unknown.
Microarray technology enables the determination of
mRNA profiles associated with human disease and provides a
comprehensive, unbiased approach to systematically analyze disease
processes, including NAFLD (13).
Additionally, some studies have revealed that the induction of a
high-fat or NASH diet may promote alterations at the genome level,
which are involved in NAFLD (13–15).
Accordingly, integrative analyses of genes and pathways associated
with modern diet-induced liver injury may provide an insight into
therapeutic targets and diagnostic biomarkers for NAFLD.
The present study aimed to analyze differentially
expressed genes (DEGs) in the liver tissues from mice with western
diet-induced liver disease and those that were administered a
regular diet using data downloaded from the Gene Expression Omnibus
(GEO). Hub genes were screened from a protein-protein interaction
(PPI) network and were verified using reverse
transcription-quantitative PCR (RT-qPCR) in a mouse model of NASH.
This integrative analysis identified candidate genes and pathways
in NAFLD, as well as DEGs and hub genes related to NAFLD
progression in silico and in vivo.
Materials and methods
Microarray data
The data were screened and analyzed by two
contributors using the following criteria for data analysis: i) The
mouse strain was C57BL/6; and ii) comparison was conducted between
NAFLD groups [high fat diet (HFD) or NASH diet, which mimics a
modern diet] and normal diet (ND) groups (negative control).
Datasets GSE57425 and GSE52748 were acquired from the GEO
(http://www.ncbi.nlm.nih.gov/geo; version
2.0) database for analysis (16,17).
In the GSE52748 dataset, C57BL/6 mice (male; age, 14 weeks) were
fed a ND or NASH-induced diet enriched with sucrose, cholesterol
and saturated fatty acids for 12 weeks. In the GSE57425 dataset,
C57BL/6 mice (male; age, 8 weeks) were fed a ND or HFD containing
60 kcal% of fat for 12 weeks. The probes were converted into the
corresponding gene symbols according to annotation information
provided by the platform.
Identification of DEGs
DEGs were analyzed using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r;
version 2.19.4), an online web tool that allows users to compare
two or more datasets in a GEO series (18). Probe sets without corresponding
gene symbols or genes with >1 probe set were averaged. Samples
with an absolute value of log fold-change >1 and P<0.05 were
considered DEGs.
Functional enrichment analysis
To investigate the biological characteristics and
functional enrichment of candidate DEGs, functional enrichment
analysis was performed using Database for Annotation, Visualization
and Integrated Discovery (https://david.ncifcrf.gov/; version 6.8). Results with
P<0.05 were considered significant. Additionally, Circos, a
visualization software (version 0.1.1) for comparative genomics
(19), was applied to identify
overlapping genes from the input gene lists and shared GO terms,
and a Venn diagram was plotted using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/;
version 1.0).
PPI network construction and module
analysis
A PPI network for DEGs was constructed using the
Search Tool for the Retrieval of Interacting Genes (STRING)
database (https://string-db.org/cgi/; version
11.0). Interactions with a combined score of >0.4 were
considered significant. The results were visualized using Cytoscape
software (version 3.7.1) (20).
MCODE, a Cytoscape plugin, was used to identify the most
significant module. The criteria for selection were as follows:
MCODE score ≥3, degree cutoff=2, node score cutoff=0.2 and max
depth=100.
Animal studies
The experimental protocol of this study was approved
by the Research Committee of the First Affiliated Hospital of
Nanchang University (Nanchang, China). In total, 20 C57BL/6J mice
(male; age, 12 weeks; weight, 25–28 g) were purchased from Hunan
SJA Laboratory Animal Co., Ltd., and maintained at 12 h light/dark
cycle with free access to food and water in a temperature- and
humidity-controlled environment of 20–24°C and 45–55% humidity.
Mice were divided into two groups, a ND group and high AGE diet
group, and fed normal chow and a baked diet, respectively, for 24
weeks. Dietary AGEs were produced by baking the food at 120°C for
15 min. The AGE content of the food was measured using ELISA
(21), which demonstrated that
baking the chow diet increased AGE levels by >2-fold from
3,194±330 to 6,639±750 ng/g. The NASH mouse model was established
using a baked diet that contains high levels of AGEs. The mice were
fed a baked diet for 24 weeks, and exhibited a NASH phenotype with
steatosis, liver injury and increased expression of inflammatory
and fibrogenic factors. These mice were sacrificed after the 24
weeks of feeding, and then the livers were harvested for
assessment.
Liver injury and histopathology
Blood samples were collected from the femoral artery
and were centrifuged at 2,500 × g for 15 min at 4°C to obtain
serum. Serum alanine aminotransferase (ALT) and aspartate
aminotransferase (AST) levels were evaluated to assess liver injury
using a Hitachi 7600 biochemical analyzer (Hitachi, Ltd.).
Hematoxylin & eosin (H&E) staining was performed in
paraffin-embedded liver tissue. The livers were fixed in 4%
paraformaldehyde solution for 24 h at room temperature and then
dehydrated. Sections were then embedded with paraffin, cut into
serial sections (thickness, 5 µm), dewaxed and rehydrated with
graded ethyl (100, 95, 80, 70, and 0%). For H&E staining, the
slides were first incubated with hematoxylin (cat. no. G1120;
Beijing Solarbio Science & Technology Co., Ltd.) for 6 min at
room temperature and then washed with 1% ethanol hydrochloride for
10 sec. After washing with water, the slides were stained with 1%
eosin for 3 min and dehydrated with graded ethyl concentrations.
Vacuoles were considered to have steatosis, as shown by H&E
staining (22). Oil red O staining
was performed on frozen tissue, as previously described (23). Liver tissues were cryosectioned
(thickness, 5 µm), fixed in 10% formalin solution at room
temperature for 10 min and dipped in 60% isopropanol for 3 min at
room temperature. The slides were then immersed in 1% ORO solution
for 3 min at room temperature and washed in 60% isopropanol
followed by distilled water. The slides were counterstained with
Mayer hematoxylin (cat. no. G1080, Beijing Solarbio Science &
Technology Co., Ltd.) for 30 sec at room temperature and were
mounted onto glycerin gelatin. The collagen content of the liver
was assessed by Sirius red staining, as described previously
(24). Sections were cut at 5 µm
and dewaxed in xylene, rehydrated in decreasing concentrations of
ethanol, and washed in 0.1 mmol/l PBS. The sections were stained
with picric acid-Sirius red (0.1% Sirius red in saturated aqueous
picric acid; Zhongshan Beijing Biotechnology Co., Ltd.) for 5 min
at 25°C. Slides were examined using a light microscope at
magnification ×200 and ×400 (Olympus Corporation). The lipid and
collagen staining areas were semi-quantified using ImageJ software
(version 1.8.0; National Institutes of Health).
Immunohistochemical (IHC)
staining
The levels of AGE, receptor for AGE (RAGE),
interleukin (IL)-1β, IL-6 and tumor necrosis factor (TNF)-α in the
liver tissues were measured by IHC staining in paraffin-embedded
sections (thickness, 5 µm), as previously described (25). The livers were fixed in 4%
paraformaldehyde solution for 24 h at room temperature and then
dehydrated. Liver sections were deparaffinized and hydrated by
sequential immersion in xylene and graded alcohol solution and
heated in a microwave for 3 min. Sections were then incubated with
methanol-3% H2O2 for 10 min at room
temperature and washed with PBS 3 times for 3 min. Then, 20% goat
serum (cat. no. SL2-10; Beijing Solarbio Science & Technology
Co., Ltd.) was used as blocking reagent for 30 min at 37°C.
Sections were incubated with the following primary antibodies:
Anti-AGE (1:10,000; Abcam cat. no. ab23722), anti-RAGE (1:100;
Abcam, ab3611), anti-IL-1β (15 µg/ml; R&D Systems, Inc.; cat.
no. AF-401-NA), anti-IL-6 (10 µg/ml; R&D Systems, Inc.; cat.
no. AF-406-NA) and anti-TNF-α (1:300; Novus Biologicals, LLC; cat.
no. NBP1-19532) overnight at 4°C, and then incubated with the Goat
Anti-Rabbit secondary antibody (1:5,000; ZB-2301; cat. no.
Zhongshan Beijing Biotechnology Co., Ltd.) at 37°C for 30 min. The
samples were washed with PBS 3 times for 3 min and stained with
0.03% DAB for 5 min at room temperature. The slides were washed,
dehydrated as above and observed using a light microscope at
magnification ×200 and ×400 (Olympus Corporation). The positive
staining area was semi-quantified using ImageJ software (version
1.8.0; National Institutes of Health) to analyze the mean optical
density.
RT-qPCR
Total RNA was extracted from the 200 mg liver
tissues using TRNzol reagent (cat. no. DP424; Tiangen Biotech Co.,
Ltd.), according to the manufacturer's protocol. Total RNA was
reverse transcribed into cDNA using the FastQuant RT kit (cat. no.
KR106; Tiangen Biotech Co., Ltd.), according to the manufacturer's
protocol. The thermocycling conditions used for qPCR were as
follows: Initial denaturation at 95°C for 15 min, followed by 40
cycles at 95°C for 10 sec, 60°C for 20 sec and 72°C for 20 sec.
qPCR was performed using the SuperReal PreMix Plus kit (cat. no.
FP205; Tiangen Biotech Co., Ltd.). The reactions were performed on
an iCycler (Bio-Rad Laboratories, Inc.). The primer pairs used for
qPCR are presented in Table SI.
Relative mRNA expression was quantified using the 2−ΔΔCq
method (26) and normalized to the
internal reference gene Gapdh.
Statistical analysis
Statistical analysis of all results was performed
using GraphPad Prism 7.0 software (GraphPad Software, Inc.). All
data are presented as the mean ± SD. Significant differences
between groups were determined using an unpaired Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of DEGs in NAFLD
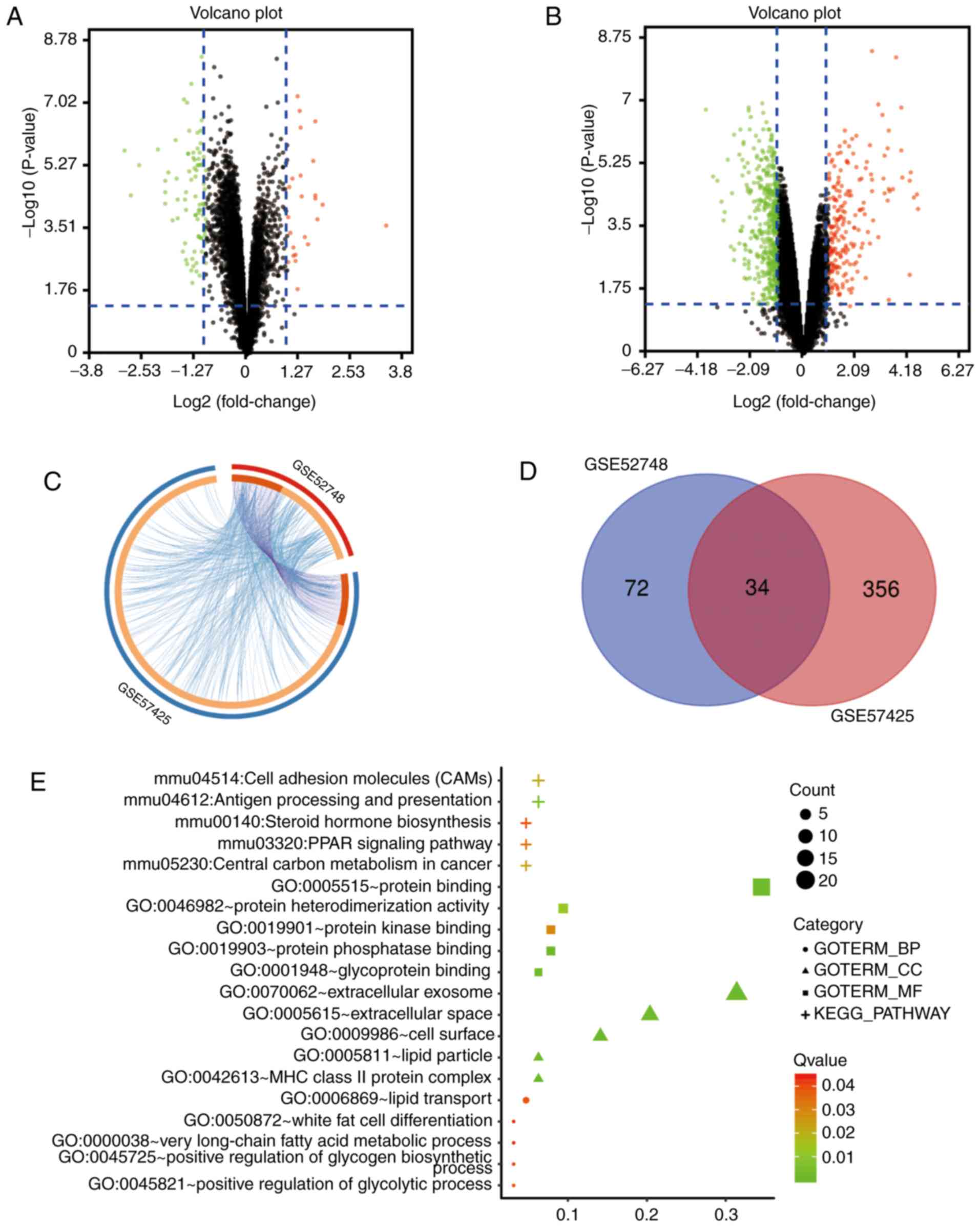
The microarray datasets GSE52748 and GSE57425 were
standardized and the results are presented in Fig. S1. A total of 106 DEGs were
identified from the GSE52748 dataset, including 84 upregulated
genes and 22 downregulated genes (Fig.
1A). Additionally, 390 DEGs were identified from the GSE57425
dataset, including 280 upregulated genes and 110 downregulated
genes (Fig. 1B). The overlap
between ontology terms associated with DEGs in GSE52748 and
GSE57425 was high (Fig. 1C); thus,
functional enrichment of these gene sets was analyzed together and
34 overlapping genes between the GSE52748 and GSE57425 datasets
were identified (Fig. 1D).
Functional enrichment analysis of
DEGs
Gene Ontology (GO) analysis identified that the DEGs
were significantly enriched in cellular components, including the
‘HC class II protein complex’, ‘extracellular exosomes’, ‘cell
surface’, ‘extracellular space’ and ‘lipid particle’ (Fig. 1E). In terms of molecular functions,
DEGs were mainly enriched in ‘protein phosphatase binding’,
‘protein binding’, ‘glycoprotein binding’, ‘protein
heterodimerization activity’ and ‘protein kinase binding’. In
addition, biological processes and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analyses demonstrated that the DEGs were
enriched in pathways involved in the ‘positive regulation of
glycolytic process’, ‘lipid transport’, ‘positive regulation of
glycogen biosynthetic process’, ‘white fat cell differentiation’,
‘very long-chain fatty acid metabolic process’, ‘antigen processing
and presentation’, ‘central carbon metabolism in cancer’ and ‘PPAR
signaling pathway’ (Fig. 1E).
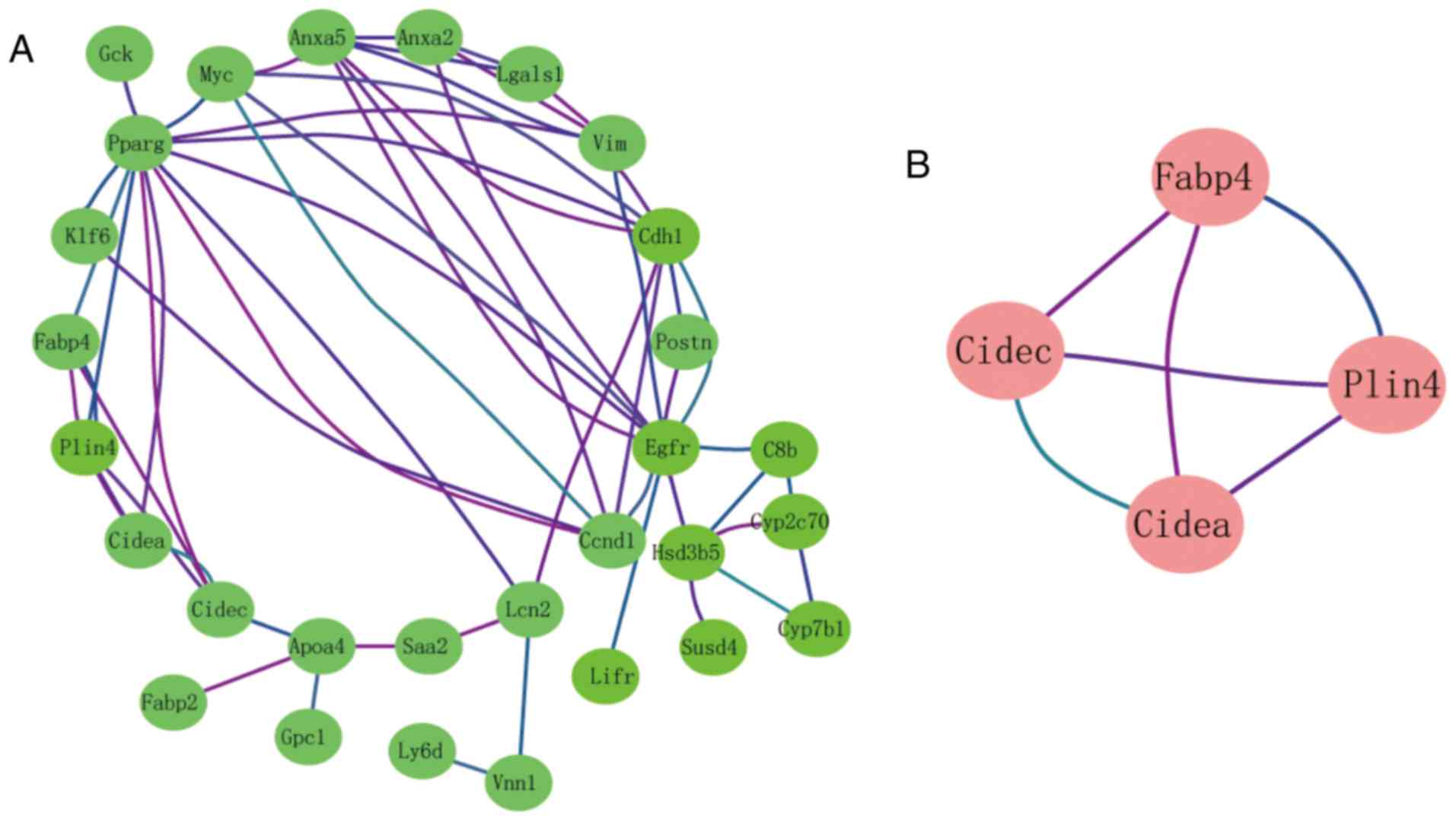
Module analysis from the PPI
network
The interactions of 34 DEGs were identified using
the STRING online database. A PPI network was generated with
Cytoscape, and the most significant modules were obtained using
MCODE (Fig. 2A). Cell
death-inducing DFFA-like effector a (Cidea), cell
death-inducing DFFA-like effector c (Cidec), perilipin 4
(Plin4) and fatty acid-binding protein 4 (Fabp4) were
identified as hub genes (Fig. 2B).
These genes were closely related to the term ‘regulation of
sequestering triglyceride’ and were enriched in the ‘PPAR signaling
pathway’ (Table I).
 | Table I.Functional analysis of the hub genes
identified from the protein-protein interaction network. |
Table I.
Functional analysis of the hub genes
identified from the protein-protein interaction network.
| Term | Description | Count | P-value |
|---|
| GO:0005811 | Lipid particle | 3 | 0.0000000157 |
| GO:0097194 | Execution phase of
apoptosis | 2 | 0.0000123000 |
| GO:0010890 | Positive regulation
of sequestering of triglyceride | 1 | 0.0007238700 |
| GO:0001816 | Cytokine
production | 2 | 0.0011631040 |
| GO:0010889 | Regulation of
sequestering of triglyceride | 1 | 0.0012406790 |
| mmu03320 | PPAR signaling
pathway | 2 | 0.0000299000 |
Liver injury and histopathology in
NASH model mice
Serum ALT and AST levels were significantly elevated
in the high AGE diet group compared with the ND group (Fig. 3B). H&E staining confirmed the
presence of steatosis (Fig. 3A);
the lipid content was significantly higher in the high AGE diet
group compared with the ND group (P<0.05; Fig. 3C and D). Collagen deposition in the
liver was also elevated in the high AGE diet group compared with
the ND group; however, the difference between groups was not
significant (P=0.1; Fig. 3E and
F). To determine whether inflammatory factors were altered in
high AGE diet-induced mice, the liver concentrations of IL-1β, IL-6
and TNF-α were measured. The results revealed that the levels of
these cytokines were all significantly increased in the high AGE
diet group compared with the ND group (Fig. 3G-L). These results were consistent
with the GSE57425 and GSE52748 datasets and suggested that mice
exhibited the NASH phenotype with increased expression of
inflammatory and fibrogenic factors, which is different from the
NAFLD phenotype that exhibits only benign simple steatosis
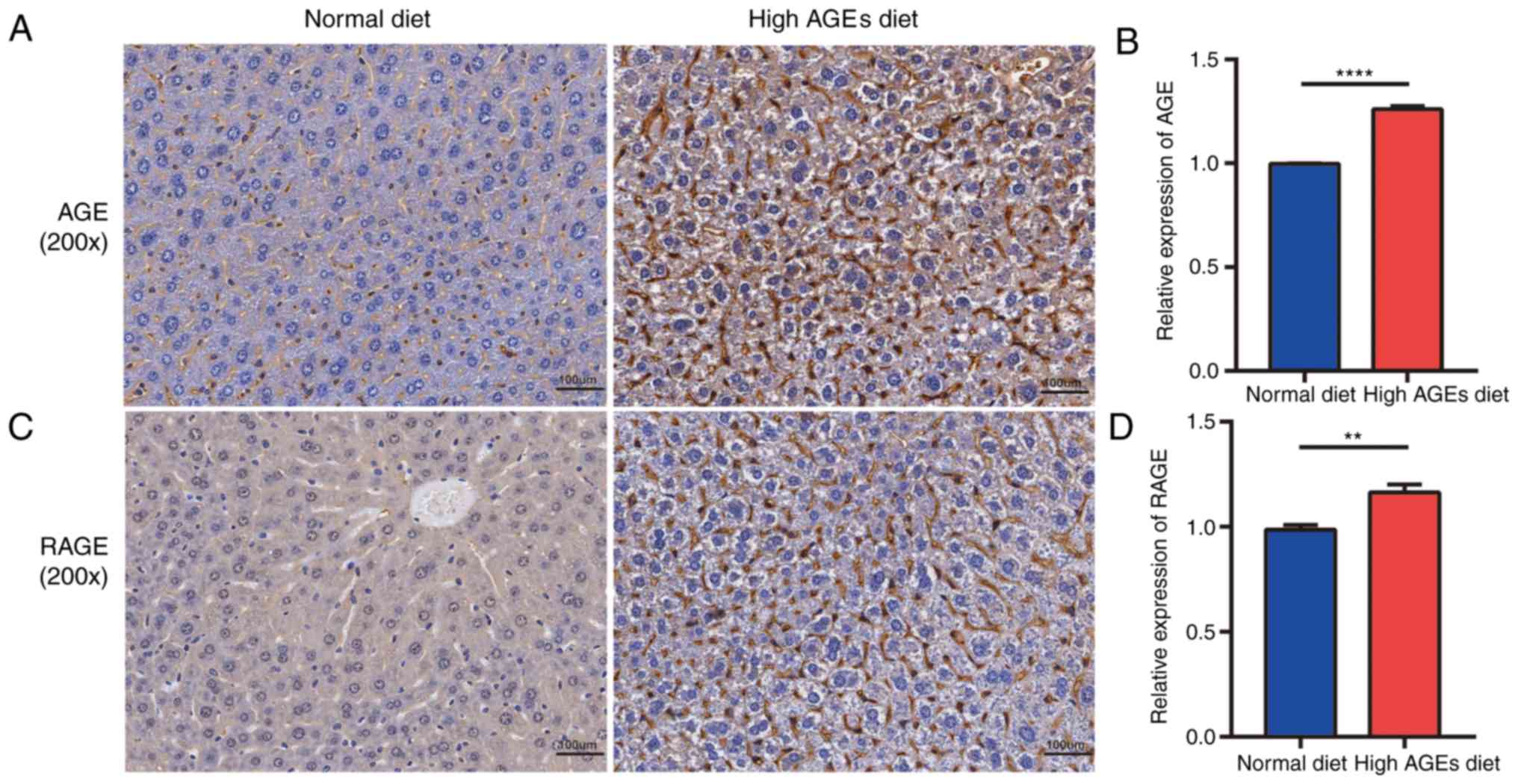
(27). In addition, the expression
levels of AGE and RAGE in the liver were significantly elevated in
the high AGE diet group compared with the ND group (Fig. 4), suggesting that the AGE/RAGE
signaling pathway is involved in the pathogenesis and progression
of NAFLD.
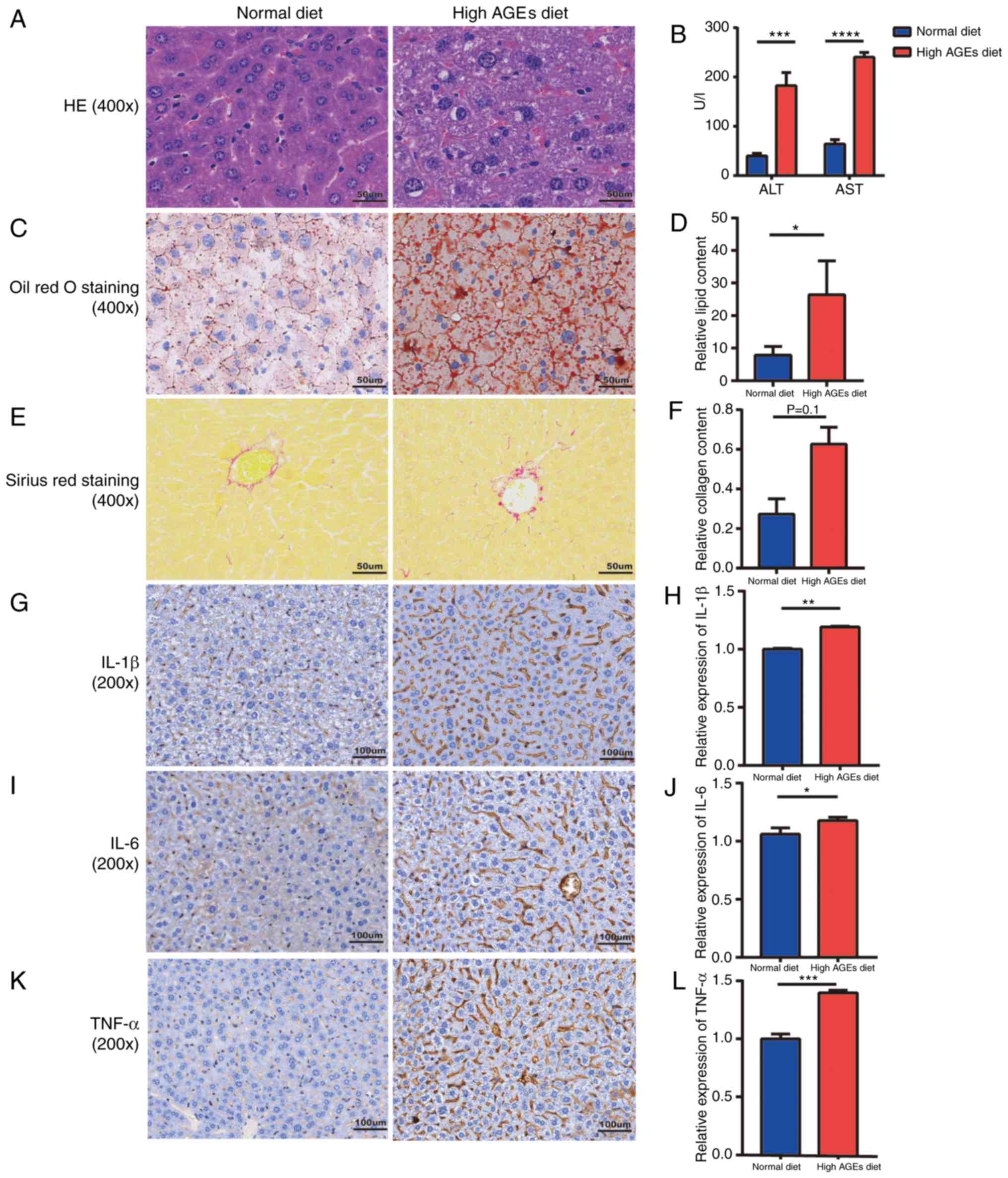 | Figure 3.Representative micrographs of liver
injury and histopathology in NASH model mice. (A) Hematoxylin &
eosin staining of mice livers (magnification, ×400). (B) Serum
levels of ALT and AST. (C) Oil red O staining of mice livers and
(D) semi-quantification of the lipid content. (E) Sirius red
staining of mice livers (magnification, ×400) and (F)
semi-quantification of the collagen deposition. (G-K)
Representative micrographs (magnification, ×200) and
semi-quantification of protein expression levels of (G and H)
IL-1β, (I and J) IL-6 and (K and L) TNF-α in liver tissues.
*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. AGE,
advanced glycation end-product; ALT, alanine aminotransferase; AST,
aspartate transaminase; HE, hematoxylin & eosin; IL,
interleukin; ND, normal diet; TNF, tumor necrosis factor. |
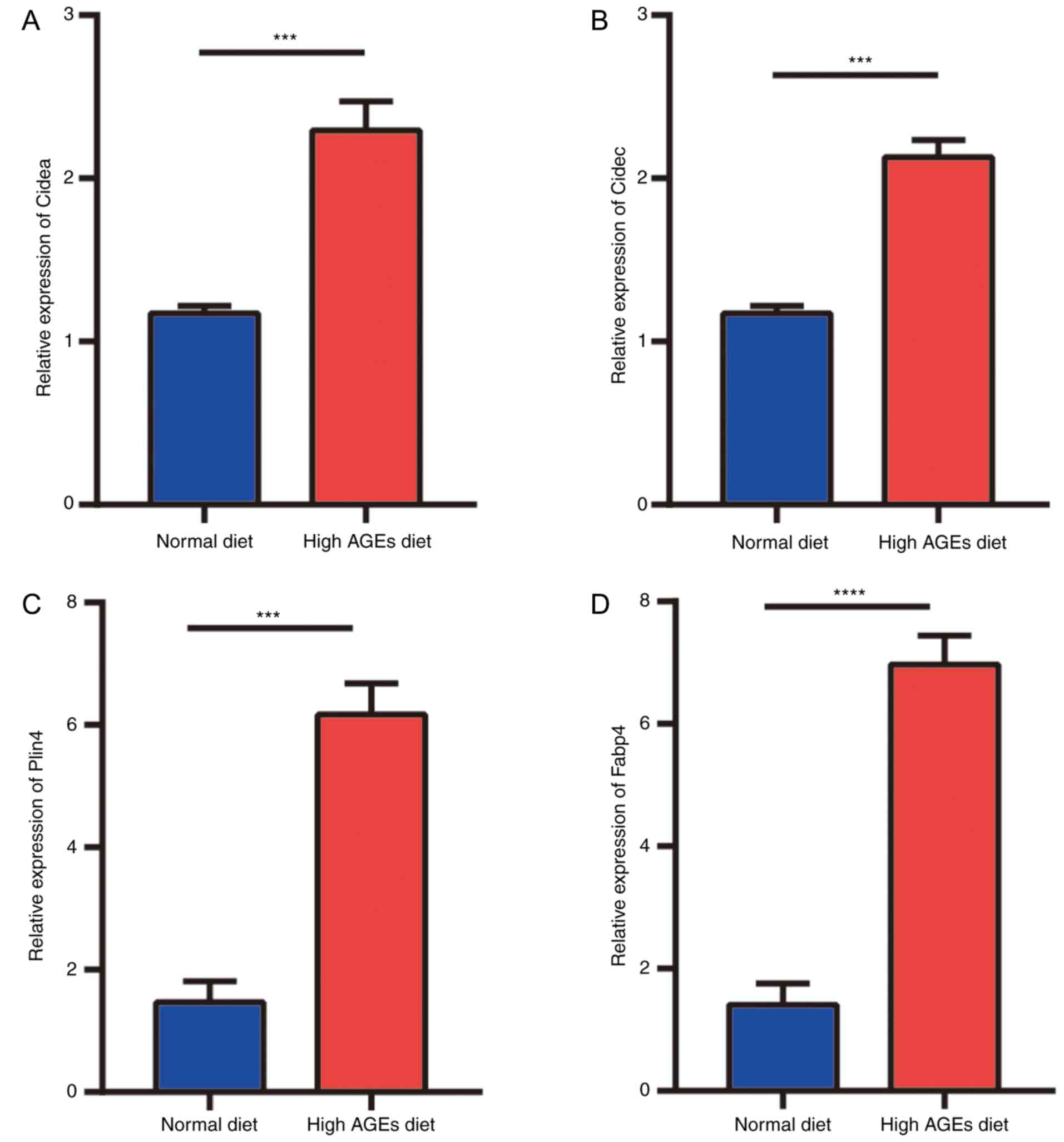
Verification of the hub genes in the
NASH model mice
To validate the hub genes identified by
bioinformatics analysis, RT-qPCR was performed in vivo. The
expression levels of Cidea, Cidec, Fabp4 and Plin4
were significantly elevated in the liver tissues of mice fed a high
AGE diet compared with those in the ND group (Fig. 5).
Discussion
The prevalence of NAFLD has increased dramatically
over the last decade, surpassing alcoholic liver disease and
ranking second amongst liver diseases following chronic hepatitis
(28). NAFLD is closely associated
with obesity and hepatic insulin resistance (29); however, the exact mechanisms of
NAFLD remain largely unclear and no specific drugs for NAFLD have
been approved. Thus, innovative treatment strategies are required
to prevent, treat and even reverse NAFLD. Transcriptional analysis
has deepened our understanding of the molecular mechanisms of human
disease, which are essential to identify genetic alterations and
establish potential therapeutic strategies. In the present study,
two transcriptional microarray datasets, including seven NAFLD and
seven normal samples were analyzed. In total, 34 overlapping DEGs
were identified in the two datasets. Functional enrichment analysis
revealed that the ‘PPAR signaling pathway’, ‘central carbon
metabolism in cancer’ and ‘cell adhesion molecules (CAMs)’ are
involved in NAFLD. Moreover, four hub genes (Cidea, Cidec,
Fabp4 and Plin4) were identified from the PPI network.
Upon experimentally verifying the four hub genes by RT-qPCR, the
expression levels of these genes in the NASH model mice were
consistent with the results from the bioinformatics analysis. NASH
is the second leading etiological factor of liver disease among
adults waiting for liver transplantations and it is mostly commonly
managed through lifestyle interventions combined with
pharmacological interventions (28). Thus, the NASH model is widely used
to explore pharmacological strategies for liver disease (28).
Previous studies have demonstrated that PPARs serve
an essential role in metabolic signaling networks and inflammation,
and are important regulators of the pathogenesis of NAFLD (30–32).
PPARs exist as three isotypes (PPARα, PPARβ/δ and PPARγ), which
have various tissue expression patterns and specificities. PPARα,
which is highly expressed in the liver, serves hepatoprotective
roles through mediating mitochondrial functions, and exhibiting
anti-inflammatory and antifibrotic effects (33,34).
PPARβ/δ, mainly expressed in the gastrointestinal tract, heart and
kidney, can improve hepatic steatosis through activating fatty acid
β-oxidation and reducing lipogenesis (35,36).
PPARγ, which is highly expressed in adipose tissues, has an
important role in transcription and glucose metabolism (37). Thus, PPAR modulators, including
dual or pan-PPAR agonists represent potential as therapeutic
targets in NAFLD.
Four hub genes (Cidea, Cidec, Fabp4 and
Plin4) were identified as having the highest scores in the
PPI network. Cidea and Cidec, which belong to a
family of cell death-inducing DNA fragmentation factor-a-like
effector proteins, serve important roles in hepatic lipid
metabolism (38). Additionally,
numerous studies have revealed that Cidea and Cidec protein
expression levels were highly elevated in the liver of mice fed HFD
(39,40). FABP4, an intracellular lipid
transporter, has a prominent role in lipid-mediated biological
processes and systemic metabolic homeostasis (41). PLIN4 is a known PPARγ target, which
is involved in the pathophysiology of NAFLD (42). Carino et al (43) reported that the expression levels
of Cidec, Cidea and Plin4 were increased in NASH
model mice, according to transcriptome analysis. In accordance with
previous studies, the results of the present study revealed that
the expression levels of Cidea, Cidec, Fabp4 and
Plin4 were significantly higher in the livers of NASH model
mice fed a high AGE diet compared with those in the livers of mice
in the ND group. CIDEA, CIDEC and PLIN4 localize to lipid droplets
to promote lipid droplet fusion and hepatic lipid storage under
high caloric intake, thus promoting liver steatosis (42,44).
FABP4 is highly upregulated by fatty acids and inflammatory
activation, and further promotes lipid infusion and inflammation in
hepatocytes (45). Thus, these
results suggested that the identified hub genes may be used as
therapeutic targets of NAFLD.
In addition, the present study demonstrated that the
NASH model mice were in a state of hepatic steatosis, inflammation
and fibrosis following the administration of a high AGE diet.
Collagen deposits were not significantly increased compared with
the ND group, which may be due to the inadequate AGE content in the
various food sources and preparation methods carried out in the
present study. Notably, RAGE expression levels were significantly
increased in high AGE diet mice, suggesting a role for the AGE/RAGE
axis in NAFLD. It is well known that AGEs promote liver injury,
inflammation and fibrosis through their interaction with RAGE,
which in turn activates oxidative and inflammatory events, creating
a positive feedback loop (46).
Thus, these results suggested that targeting the AGE/RAGE pathway
may be an effective therapeutic strategy for treating NASH. In
addition, previous studies have highlighted the effects of dietary
AGEs on the gut microbiota and their ability to cause metabolic
diseases, including NAFLD (47,48).
Owing to the increasing prevalence of western diets, alterations in
the microbiome by dietary AGEs are of particular interest (49); thus, this may represent a promising
therapeutic target.
In conclusion, the findings of the present study
revealed that high dietary AGEs can induce liver injury,
inflammation and even hepatic fibrosis. Additionally, four hub
genes involved in NAFLD progression were identified. These results
suggested that the restriction of dietary AGEs or pharmacological
strategies targeting the four hub genes may represent novel
approaches for treating and preventing NAFLD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
National Natural Science Funds of China (grant nos. 81760168 and
81560154) and The Innovation Fund Project in Jiangxi Province
(grant no. YC2016-S100).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW performed the animal study and wrote the
manuscript. HHL performed the histological examination of the
liver. GJX performed the data analysis. WC performed the RT-qPCR
analysis. JXX conceived and designed the study. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol of this study was approved
by the Research Committee of the First Affiliated Hospital of
Nanchang University (Nanchang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Estes C, Razavi H, Loomba R, Younossi Z
and Sanyal AJ: Modeling the epidemic of nonalcoholic fatty liver
disease demonstrates an exponential increase in burden of disease.
Hepatology. 67:123–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yki-Järvinen H: Non-alcoholic fatty liver
disease as a cause and a consequence of metabolic syndrome. Lancet
Diabetes Endocrinol. 2:901–910. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gluchowski NL, Becuwe M, Walther TC and
Farese RV Jr: Lipid droplets and liver disease: From basic biology
to clinical implications. Nat Rev Gastroenterol Hepatol.
14:343–355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Trovato FM, Castrogiovanni P, Malatino L
and Musumeci G: Nonalcoholic fatty liver disease (NAFLD)
prevention: Role of Mediterranean diet and physical activity.
Hepatobiliary Surg Nutr. 8:167–169. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Trovato FM, Martines GF, Brischetto D,
Catalano D, Musumeci G and Trovato GM: Fatty liver disease and
lifestyle in youngsters: Diet, food intake frequency, exercise,
sleep shortage and fashion. Liver Int. 36:427–433. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Trovato FM, Catalano D, Musumeci G and
Trovato GM: 4Ps medicine of the fatty liver: The research model of
predictive, preventive, personalized and participatory
medicine-recommendations for facing obesity, fatty liver and
fibrosis epidemics. EPMA J. 5:212014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh R, Barden A, Mori T and Beilin L:
Advanced glycation end-products: A review. Diabetologia.
44:129–146. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Uribarri J, Woodruff S, Goodman S, Cai W,
Chen X, Pyzik R, Yong A, Striker GE and Vlassara H: Advanced
glycation end products in foods and a practical guide to their
reduction in the diet. J Am Diet Assoc. 110:911–916.e12. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leung C, Herath CB, Jia Z, Goodwin M, Mak
KY, Watt MJ, Forbes JM and Angus PW: Dietary glycotoxins exacerbate
progression of experimental fatty liver disease. J Hepatol.
60:832–838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ravichandran G, Lakshmanan DK, Raju K,
Elangovan A, Nambirajan G, Devanesan AA and Thilagar S: Food
advanced glycation end products as potential endocrine disruptors:
An emerging threat to contemporary and future generation. Environ
Int. 123:486–500. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chaudhuri J, Bains Y, Guha S, Kahn A, Hall
D, Bose N, Gugliucci A and Kapahi P: The role of advanced glycation
end products in aging and metabolic diseases: Bridging association
and causality. Cell Metab. 28:337–352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Wang Y and Liu P: Omic studies
reveal the pathogenic lipid droplet proteins in non-alcoholic fatty
liver disease. Protein Cell. 8:4–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kahle M, Horsch M, Fridrich B, Seelig A,
Schultheiß J, Leonhardt J, Irmler M, Beckers J, Rathkolb B, Wolf E,
et al: Phenotypic comparison of common mouse strains developing
high-fat diet-induced hepatosteatosis. Mol Metab. 2:435–446. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou D, Hlady RA, Schafer MJ, White TA,
Liu C, Choi JH, Miller JD, Roberts LR, LeBrasseur NK and Robertson
KD: High fat diet and exercise lead to a disrupted and pathogenic
DNA methylome in mouse liver. Epigenetics. 12:55–69. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pruis MG, Lendvai A, Bloks VW, Zwier MV,
Baller JF, de Bruin A, Groen AK and Plösch T: Maternal western diet
primes non-alcoholic fatty liver disease in adult mouse offspring.
Acta Physiol (Oxf). 210:215–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dorn C, Engelmann JC, Saugspier M, Koch A,
Hartmann A, Müller M, Spang R, Bosserhoff A and Hellerbrand C:
Increased expression of c-Jun in nonalcoholic fatty liver disease.
Lab Invest. 94:394–408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu Y, Liu X, Jiao Y, Xiong X, Wang E, Wang
X, Zhang Z, Zhang H, Pan L, Guan Y, et al: Periostin promotes liver
steatosis and hypertriglyceridemia through downregulation of PPARα.
J Clin Invest. 124:3501–3513. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu Y, Ouyang Y and Yao W: shinyCircos: An
R/Shiny application for interactive creation of Circos plot.
Bioinformatics. 34:1229–1231. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sourris KC, Harcourt BE, Penfold SA, Yap
FY, Morley AL, Morgan PE, Davies MJ, Baker ST, Jerums G and Forbes
JM: Modulation of the cellular expression of circulating advanced
glycation end-product receptors in type 2 diabetic nephropathy. Exp
Diabetes Res. 2010:9746812010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kleiner DE, Brunt EM, Van Natta M, Behling
C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS,
Unalp-Arida A, et al: Design and validation of a histological
scoring system for nonalcoholic fatty liver disease. Hepatology.
41:1313–1321. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mehlem A, Hagberg CE, Muhl L, Eriksson U
and Falkevall A: Imaging of neutral lipids by oil red O for
analyzing the metabolic status in health and disease. Nat Protoc.
8:1149–1154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goodwin M, Herath C, Jia Z, Leung C,
Coughlan MT, Forbes J and Angus P: Advanced glycation end products
augment experimental hepatic fibrosis. J Gastroenterol Hepatol.
28:369–376. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trovato FM, Castrogiovanni P, Szychlinska
MA, Purrello F and Musumeci G: Early effects of high-fat diet,
extra-virgin olive oil and vitamin D in a sedentary rat model of
non-alcoholic fatty liver disease. Histol Histopathol.
33:1201–1213. 2018.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Farrell GC and Larter CZ: Nonalcoholic
fatty liver disease: From steatosis to cirrhosis. Hepatology. 43 (2
Suppl 1):S99–S112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong RJ, Aguilar M, Cheung R, Perumpail
RB, Harrison SA, Younossi ZM and Ahmed A: Nonalcoholic
steatohepatitis is the second leading etiology of liver disease
among adults awaiting liver transplantation in the United States.
Gastroenterology. 148:547–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Samuel VT and Shulman GI: Nonalcoholic
fatty liver disease as a nexus of metabolic and hepatic diseases.
Cell Metab. 27:22–41. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pawlak M, Lefebvre P and Staels B:
Molecular mechanism of PPARα action and its impact on lipid
metabolism, inflammation and fibrosis in non-alcoholic fatty liver
disease. J Hepatol. 62:720–733. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gross B, Pawlak M, Lefebvre P and Staels
B: PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat Rev
Endocrinol. 13:36–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liss KH and Finck BN: PPARs and
nonalcoholic fatty liver disease. Biochimie. 136:65–74. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pawlak M, Baugé E, Bourguet W, De Bosscher
K, Lalloyer F, Tailleux A, Lebherz C, Lefebvre P and Staels B: The
transrepressive activity of peroxisome proliferator-activated
receptor alpha is necessary and sufficient to prevent liver
fibrosis in mice. Hepatology. 60:1593–1606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koliaki C, Szendroedi J, Kaul K, Jelenik
T, Nowotny P, Jankowiak F, Herder C, Carstensen M, Krausch M,
Knoefel WT, et al: Adaptation of hepatic mitochondrial function in
humans with non-alcoholic fatty liver is lost in steatohepatitis.
Cell Metab. 21:739–746. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu HT, Chen CT, Cheng KC, Li YX, Yeh CH
and Cheng JT: Pharmacological activation of peroxisome
proliferator-activated receptor δ improves insulin resistance and
hepatic steatosis in high fat diet-induced diabetic mice. Horm
Metab Res. 43:631–635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bojic LA, Telford DE, Fullerton MD, Ford
RJ, Sutherland BG, Edwards JY, Sawyez CG, Gros R, Kemp BE,
Steinberg GR and Huff MW: PPARδ activation attenuates hepatic
steatosis in Ldlr-/- mice by enhanced fat oxidation, reduced
lipogenesis, and improved insulin sensitivity. J Lipid Res.
55:1254–1266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bensinger SJ and Tontonoz P: Integration
of metabolism and inflammation by lipid-activated nuclear
receptors. Nature. 454:470–477. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu L, Zhou L and Li P: CIDE proteins and
lipid metabolism. Arterioscler Thromb Vasc Biol. 32:1094–1098.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou L, Xu L, Ye J, Li D, Wang W, Li X, Wu
L, Wang H, Guan F and Li P: Cidea promotes hepatic steatosis by
sensing dietary fatty acids. Hepatology. 56:95–107. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Reynolds TH IV, Banerjee S, Sharma VM,
Donohue J, Couldwell S, Sosinsky A, Frulla A, Robinson A and Puri
V: Effects of a high fat diet and voluntary wheel running exercise
on cidea and cidec expression in liver and adipose tissue of mice.
PLoS One. 10:e01302592015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hotamisligil GS and Bernlohr DA: Metabolic
functions of FABPs-mechanisms and therapeutic implications. Nat Rev
Endocrinol. 11:592–605. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carr RM and Ahima RS: Pathophysiology of
lipid droplet proteins in liver diseases. Exp Cell Res.
340:187–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Carino A, Marchianò S, Biagioli M,
Fiorucci C, Zampella A, Monti MC, Morretta E, Bordoni M, Di Giorgio
C, Roselli R, et al: Transcriptome analysis of Dual FXR and GPBAR1
agonism in rodent model of NASH reveals modulation of lipid
droplets formation. Nutrients. 11(pii): E11322019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Slayton M, Gupta A, Balakrishnan B and
Puri V: CIDE proteins in human health and disease. Cells. 8(pii):
E2382019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Thompson KJ, Austin RG, Nazari SS, Gersin
KS, Iannitti DA and McKillop IH: Altered fatty acid-binding protein
4 (FABP4) expression and function in human and animal models of
hepatocellular carcinoma. Liver Int. 38:1074–1083. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Leung C, Herath CB, Jia Z, Andrikopoulos
S, Brown BE, Davies MJ, Rivera LR, Furness JB, Forbes JM and Angus
PW: Dietary advanced glycation end-products aggravate non-alcoholic
fatty liver disease. World J Gastroenterol. 22:8026–8040. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Delgado-Andrade C, Pastoriza de la Cueva
S, Peinado MJ, Rufián-Henares JÁ, Navarro MP and Rubio LA:
Modifications in bacterial groups and short chain fatty acid
production in the gut of healthy adult rats after long-term
consumption of dietary Maillard reaction products. Food Res Int.
100:134–142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Qu W, Yuan X, Zhao J, Zhang Y, Hu J, Wang
J and Li J: Dietary advanced glycation end products modify gut
microbial composition and partially increase colon permeability in
rats. Mol Nutr Food Res. 61:2017. View Article : Google Scholar
|
|
49
|
Snelson M and Coughlan MT: Dietary
advanced glycation end products: Digestion, metabolism and
modulation of gut microbial ecology. Nutrients. 11(pii): E2152019.
View Article : Google Scholar : PubMed/NCBI
|