Introduction
Hepatic fibrosis is a reversible wound healing
process elicited by various damaging factors, such as viruses,
parasites and alcohol, all of which lead to liver cell injury
accompanied by inflammatory responses. Upon stimulation by a
multitude of signals, hepatic stellate cells (HSCs) transform into
myofibroblast-like cells, with ensuing excessive production of
extracellular matrix (ECM), including type I and type III collagen.
Eventually, the disease may progress to liver cirrhosis and even
liver cancer (1,2). Therefore, it is crucial to arrest the
progression of hepatic fibrosis.
During the process of hepatic fibrosis, the
activation of HSCs is fundamental. Once activated, the HSCs
transform into myofibroblasts, which characteristically express
α-smooth muscle actin (SMA). Furthermore, pro-fibrotic factors are
generated, such as transforming growth factor (TGF)-β1 and tissue
inhibitors of metalloproteinases (TIMPs). It was previously
demonstrated that TGF-β signaling is key to the development of
hepatic fibrosis (3) and it
enhances the synthesis of hepatic fibrosis-related proteins, such
as type I collagen (COLI)α1, COLIα2 and TIMP1 (4–6).
Hence, molecular therapy targeting HSCs and inhibiting the TGF-β
signaling pathway may inhibit HSC activation and may block or even
reverse the pathological process of liver fibrosis. It is widely
accepted that TGF-β canonical signaling is essential for the
activation of HSCs; non-canonical signaling, which is associated
with multiple different pathways, such as mitogen-activated protein
kinase (MAPK), phosphoinositide 3 kinase (PI3K)/protein kinase B
(AKT) and Wnt pathways, may also contribute to the activation of
HSCs and liver fibrosis (7,8).
Recent studies have reported the role of Notch signaling in hepatic
fibrosis and the crosstalk between Notch and TGF-β signaling. There
is evidence that the expression of major components of the Notch
signaling pathway, including Notch3, Jagged1 and the downstream
transcription factor (TF) Hairy and Enhancer of Split 1 (HES1), is
induced by TGF-β canonical signaling via SMADs (9,10).
Another study reported that Notch signaling also contributes to
TGF-β-induced expression of a-SMA and COLI and, when inhibited,
a-SMA and COLI expression decreased (11). Furthermore, the present research
group has observed that overexpression of HES1 in the activated
HSCs can enhance the promoter activity of α-SMA and COLIα2
(12). Taken together, these
findings indicate that there may exist a positive feedback loop
between Notch and TGF-β canonical signaling. Other experiments have
already confirmed that HES1 can upregulate the expression of COLIα1
and COLIα2 on other cell types, such as fibroblast L929 cells and
MRC-5 cells (13). Therefore, this
positive feedback loop between Notch and TGF-β canonical signaling
may participate in HSC activation and hepatic fibrosis, with HES1
serving as an important TF in this crosstalk. HES1 belongs to the
family of bHLH TFs, which contain the bHLH motif; this motif
consists of ~60 amino acids, with a basic region and a helix
1-loop-helix 2, and the length of the loop differs between bHLH
proteins (14). The bHLH protein
family may be subdivided into three classes, according to their
structure and biochemical characteristics, and HES1 belongs to the
class C proteins, which bind to the class C DNA-binding domain
(CACGNG) (15,16). Furthermore, the authors of the
present study discovered that abundant Class C binding domains in
the promotor region of pro-fibrotic genes, include TGF-β, COLIα1,
COLIα2, TIMP1, α-SMA and Hes1 by the JASPAR database. The existence
of Class C binding domains indicate DNA-protein interaction between
the pro-fibrotic genes and Class C proteins may contribute to the
activation of HSC and hepatic fibrosis.
Decoy oligodeoxynucleotides (ODNs) are also known as
a TF ‘trap’. Short-chain DNA fragments containing the DNA-binding
site of specific TFs are artificially designed, synthesized and
transfected to competitively capture intracellular TFs, thereby
inhibiting downstream gene expression (17–19).
The present study hypothesized that competitively inhibiting Class
C proteins binding by Decoy ODN strategy may downregulate
expression of TGF-β, COLIα1, COLIα2, TIMP1, α-SMA and Hes1 and
consequently inhibit HSC activation and relieve liver fibrosis.
Materials and methods
Synthesis of ODNs and plasmid
construction
The decoy ODNs and scramble (Scr) decoy ODN
(Table I) were synthesized by
Sangon Biotech Co., Ltd. The eukaryotic expression plasmid
pGLuc-TRE-MiniTK was constructed when TGF-β-responsive element
(TRE) was cloned into the pGLuc-Mini-TK (New England Biolabs,
Inc.). The eukaryotic expression plasmids pGLuc-P-SMA,
pGLuc-P-COLIα1, pGLuc-P-COLIα2 and pGLuc-P-TIMP1 were constructed
when the promoters of α-SMA (P-SMA), COLIα1 (P-COLIα1), COLIα2
(P-COLIα2) and TIMP1 (P-TIMP1) were cloned into the pGLucBasic
vector (N8082S; New England Biolabs, Inc.) for luciferase assays
(Table II).
 | Table I.The sequences of each Decoy ODN. |
Table I.
The sequences of each Decoy ODN.
| Decoy ODN name | Sequence
(5′→3′)a |
|---|
| Class C1 Decoy
ODN | F: CGACACGTGATCACGTGGAC |
|
| R: GTCCACGTGATCACGTGTCG |
| Class C2 Decoy
ODN | F: CGACACGCGATCACGCGGAC |
|
| R:
GTCCGCGTGATCGCGTGTCG |
| Class C3 Decoy
ODN | F: CGACACGAGATCACGAGGAC |
|
| R:
GTCCTCGTGATCTCGTGTCG |
| Class C4 Decoy
ODN | F: CGACACGGGATCACGGGGAC |
|
| R:
GTCCCCGTGATCCCGTGTCG |
| Scramble | F:
CGAACGCTGATACGCTGGAC |
|
| R:
GTCCAGCGTATCAGCGTTCG |
 | Table II.Primers used for promoter cloning into
pGLucBasic vector. |
Table II.
Primers used for promoter cloning into
pGLucBasic vector.
| Gene | Primer sequence
(5′→3′) |
|---|
| P-Hairy and
Enhancer of Split 1 (HindIII) | F:
CGAAGCTTGAGCCTGAAGAGGTAGAGAGT |
|
| R:
ATGGATCCGCTTACGTCCCCTTTACTTGG |
| P-α-smooth muscle
actin (EcoR1) | F:
CCGGAATTCACGGTCCTTAAGCATGATATC |
|
| R:
CGGGATCCCTTACCCTGATGGCGACT |
| P-type I collagen
α1 (EcoR1) | F:
CCGGAATTCGCAGGTTCTCTACAGAGAGA |
|
| R:
CGGGATCCAGCCAATCAGAACT |
| P-tissue inhibitor
of metalloproteinase 1 (EcoR1) | F:
GCGGAATTCCAAACATCTTCACTGGTATG |
|
| R:
GCGGGATCCCTTTACTGGAAGCTATCAATG |
Cell culture
HSC-T6 cells, an immortalized rat HSC line provided
by the Huazhong University of Science and Technology, were cultured
in high-glucose DMEM (Invitrogen; Thermo fisher Scientific, Inc.)
supplemented with 10% newborn calf serum (Zhejiang Tianhang
Biotechnology Co., Ltd). HSC-T6 cells were seeded at a density of
6×105 cells/well in a 6-well-plate (Greiner GmbH) for
western blot assays or a 24-well-plate (Greiner GmbH) for
luciferase assays at 60% confluence per well and cultured in a
humidified atmosphere containing 5% CO2 for 24 h at
37°C.
Transfection and luciferase reporter
assays
The HSC-T6 cells were seeded at a density of
1×105 cells/well in a 24-well plate. After 24 h, the
cells had reached 70–80% confluence and were transfected with
different plasmids (1 µg per well) using the Tubofect Transfection
Reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The cells were then transfected with
different class C (C1/C2/C3/C4) decoy ODNs and Scr decoy ODN (at a
concentration of 20 nm/l; 2 µg per well for a 24-well plate), using
the Mirus Transfection Reagent (Mirus Bio LLC) after a further 24
h. The luciferase assays were performed using the
BioLux® Gaussia Luciferase Assay kit (New England
Biolabs, Inc.), according to the manufacturer's protocol. Briefly,
the supernatants were collected, the cells were lysed, and the
total intracellular protein concentration of the supernatant was
analyzed as described in the paragraph entitled ‘Western blotting’
of the Materials and methods section to estimate the cell number
per well. For normalization, the sampling size for each well was
adjusted according to the total intracellular protein levels to
detect the Gaussia Luciferase activity. The reactions were
examined using a fluorescence detector (Berthold Technologies).
Western blot analysis
The cells were collected for western blot assays
after decoy ODNs (at a concentration of 20 nm/l; 6 µg per well for
a 6-well plate) were transfected into HSC-T6 cells for 48 h, then
lysed in lysis buffer [25 mmol/l Tris-HCl (pH 7.5), 2.5 mmol/l
EDTA, 137 mmol/l NaCl, 2.7 mmol/l KCl, 1% sodium deoxycholic acid,
0.1% SDS, 1% Triton X-100, and 2 mmol/l PMSF] and protease
inhibitor cocktail for 30 min at 4°C. Cell lysates were cleared by
centrifugation at 7,200 × g for 10 min at 4°C and the supernatants
were collected. Protein concentration was measured using a BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.). An equal amount
of protein (40 µg loaded per lane) from each sample was separated
by 10% SDS-PAGE and transferred to a PVDF membrane. The membrane
was firstly incubated with blocker (5% defatted milk) for 2 h at
room temperature and subsequently incubated with the following
antibodies at 4°C overnight: Anti-TGF-β1 (1:1,000; cat. no. sc-146;
Santa Cruz Biotechnology, Inc.), anti-TIMP1(1:1,000; cat. no.
sc-6834; Santa Cruz Biotechnology, Inc.), anti-COLIα1 (1:1,000;
cat. no. sc-25974; Santa Cruz Biotechnology, Inc.), anti-COLIα2
(1:1,000; cat. no. sc-8788; Santa Cruz Biotechnology, Inc.),
anti-SMAD3 (1:3,000; cat. no. sc-133098; Santa Cruz Biotechnology,
Inc.) and anti-β-actin (1:3,000; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.). Following the primary antibody incubation,
membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies (1:3,000; cat nos. sc-2031 and sc-516721;
1:8,000; cat. no. sc-2354; Santa Cruz Biotechnology, Inc.) for 1 h
at room temperature. The membranes were treated using Immobilon
Western Detection Reagents (EMD Millipore). Chemiluminescence was
detected using the VersaDoc system (Bio-Rad Laboratories, Inc.).
Densitometric analyses of the band intensities were performed using
ImageJ software, version 1.38 (National Institutes of Health). All
the western blot analysis were repeated three times.
Bioinformatics analysis
The JASPAR 2020 database (http://jaspar.genereg.net) and UCSC Genome Browser
Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway) were used
for the bioinformatics analysis. Full-length Promoter sequences of
α-SMA, COLIα1, COLIα2 and TIMP1 were identified by UCSC Genome
Browser Gateway. Detailed information of Class C TFBS (Basic
helix-loop-helix factors) were identified by the JASPAR database.
The distribution of Class C TFBS on Promoters of α-SMA, COLIα1,
COLIα2 and TIMP1 were analyzed by the JASPAR database.
Statistical analysis
GraphPad Prism version 7.0 software (GraphPad
Software, Inc.) was used for the statistical analysis. Data are
presented as the mean ± SD and represent three independent
experimental repeats. Differences between three or more groups were
analyzed by one-way ANOVA and Tukey's post hoc test for multiple
comparisons. P<0.05 was considered to indicate a statistically
significant difference.
Results
The class C sequence is present in the
promoter region of TGF-β and its target genes
The JASPAR database is one of the most comprehensive
and reliable public databases of TFs and DNA-binding motifs, and
the data published there are rigorously screened from multiple
randomized experiments and integrated by computer-aided software.
This database was used in the present study to analyze the binding
potency between the promoters of TGF-β signaling pathway-related
genes and class C sequences. The present study found at least one
class C sequence that was present in the promoter region of TGF-β
and its downstream genes, namely COLIα1, TIMP1, HES1 and
α-SMA (Table III).
 | Table III.Analysis of the possible binding
sites on the promoters of TGF-β signal pathway-related genes for
four class C sequences by JASPAR database. |
Table III.
Analysis of the possible binding
sites on the promoters of TGF-β signal pathway-related genes for
four class C sequences by JASPAR database.
|
| Class |
|
|---|
|
|
|
|
|---|
| Gene promoter | C1 | C2 | C3 | C4 | Total |
|---|
| Transforming growth
factor-β | 1 | 6 | 1 | 3 | 11 |
| Type I collagen
α1 | 5 | 1 | 2 | 1 | 5 |
| Type I collagen
α2 | 0 | 0 | 0 | 0 | 0 |
| Tissue inhibitor of
metalloproteinase 1 | 0 | 0 | 2 | 3 | 5 |
| Hairy and Enhancer
of Split 1 | 4 | 1 | 3 | 2 | 5 |
| α-smooth muscle
actin | 1 | 0 | 3 | 1 | 5 |
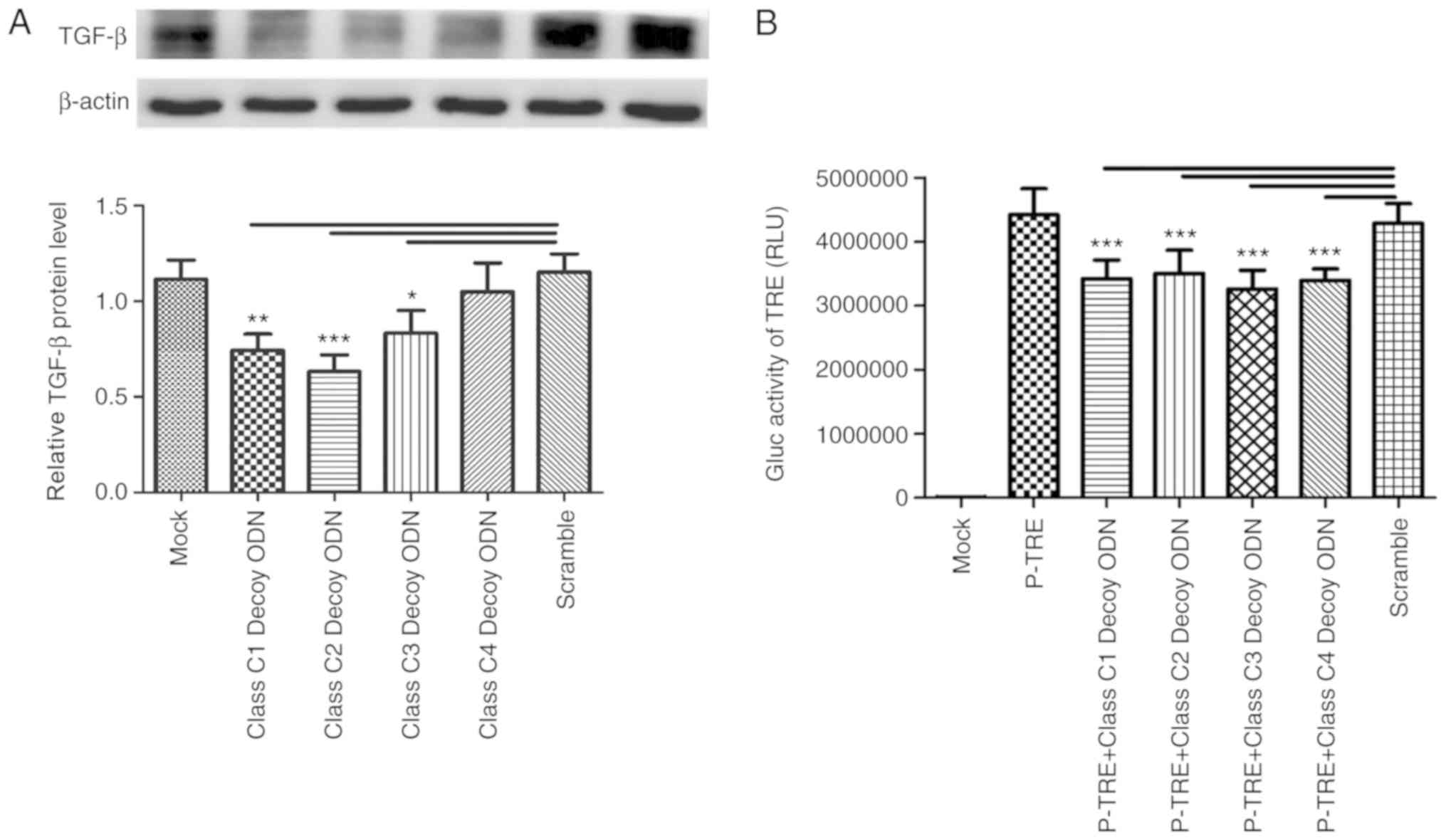
Class C decoy ODNs decrease TGF-β
synthesis in HSC-T6 cells
The bioinformatics analysis revealed that there was
at least one binding site in the promoter region of TGF-β
for each class C sequence. Class C decoy ODNs were transfected into
HSC-T6 cells for 48 h and the expression of TGF-β was tested
through western blot assays. Except for class C4 decoy ODNs, which
had no impact on TGF-β expression, the other three decoy
ODNs were able to significantly downregulate TGF-β
expression (P<0.05; Fig. 1A),
indicating that class C1/C2/C3 decoy ODNs can decrease TGF-β
synthesis in HSC-T6 cells. Furthermore, plasmid pTRE-Mini-TK-Gluc,
the Gaussia luciferase reporter gene for TRE was constructed
and transfected into HSC-T6 cells for 24 h; decoy ODNs were
transfected for another 24 h and the results revealed that the
luciferase activities of the pTRE-Mini-TK-Gluc had significantly
decreased in all class C decoy ODN groups compared with Scr
(P<0.001; Fig. 1B), suggesting
the four class C decoy ODNs can inhibit the transcriptional
activity of TRE.
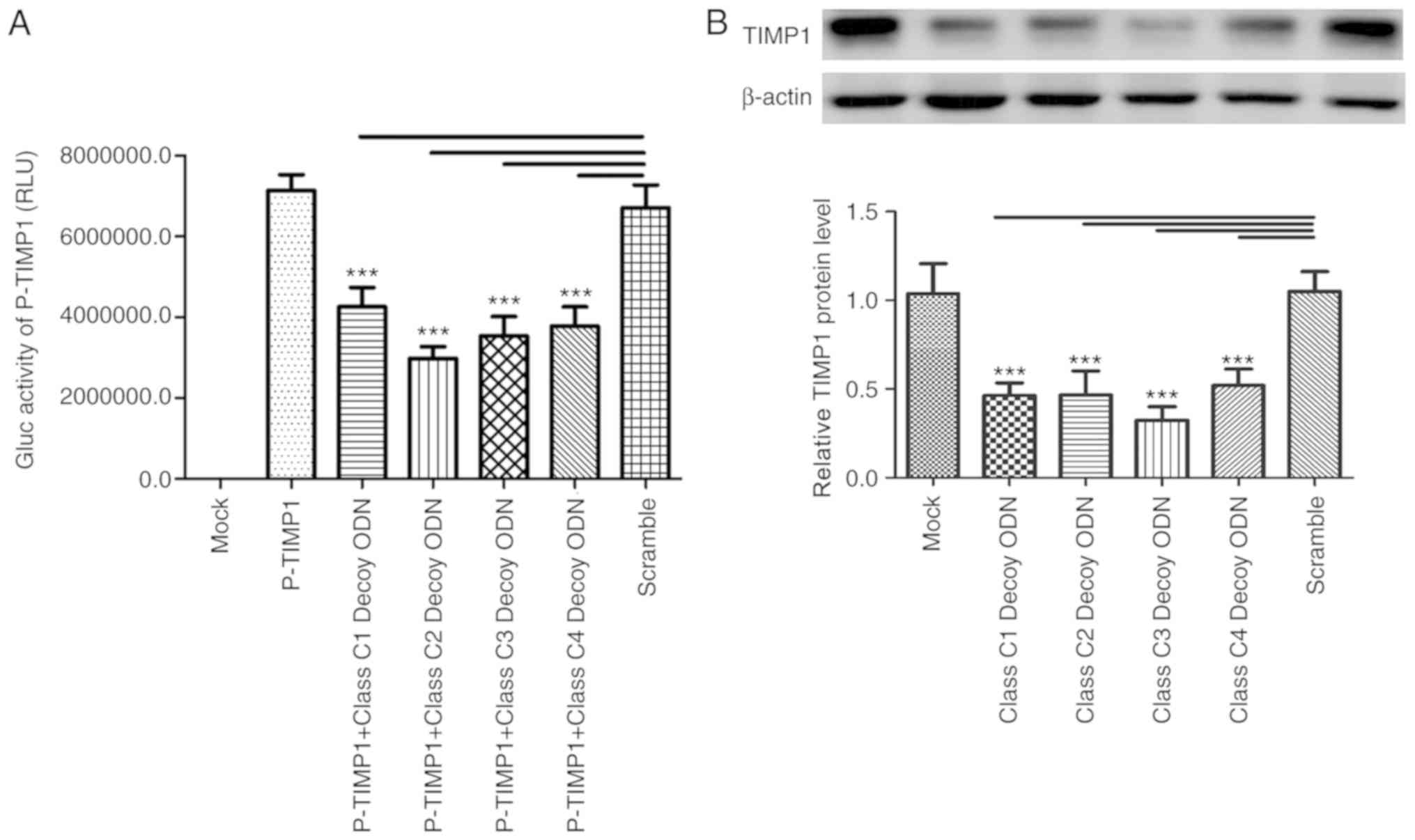
Class C decoy ODNs downregulate the
expression of TIMP1 in HSC-T6 cells
TIMP1 is a downstream gene of TGF-β (4). A total of five binding sites for
class C proteins in the promoter of TIMP1 were identified through
bioinformatics analysis. The Gaussia luciferase activities
of pTIMP1-GLuc-Basic significantly decreased in the four
experimental groups treated by Class C1-4 decoy ODNs compared with
Scr (P<0.001; Fig. 2A),
suggesting the four decoy ODNs can inhibit the activation of the
TIMP1 promoter. The expression of TIMP1 was also tested using
western blot assays. There were significant decreases in TIMP1
expression in the four class C decoy ODN groups compared with Scr
(Fig. 2B).
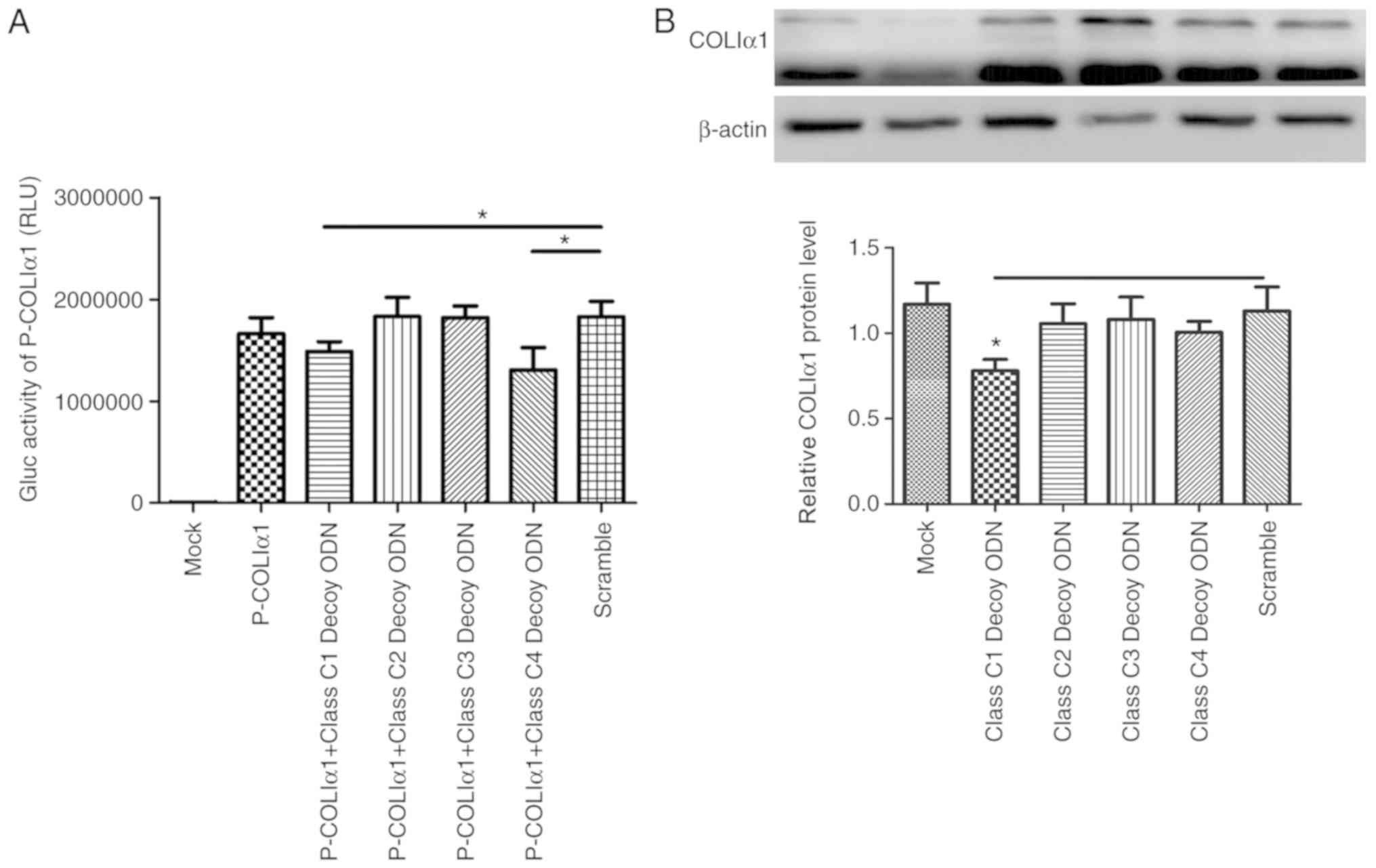
Class C decoy ODNs downregulate the
expression of COLIα1 in HSC-T6 cells
COLIα1 is one of the downstream genes of TGF-β and
it is positively correlated with TGF-β (4). Bioinformatics analysis demonstrated
that there are five binding sites for class C sequences in its
promoter. The results of the Gaussia luciferase assay
revealed that, in the class C1 and class C4 decoy ODN groups,
luciferase activity decreased compared with the Scr (Fig. 3A). The expression of COLIα1 was
also tested using western blot assays, but only class C1 decoy ODNs
were proven to downregulate the expression of COLIα1 (Fig. 3B).
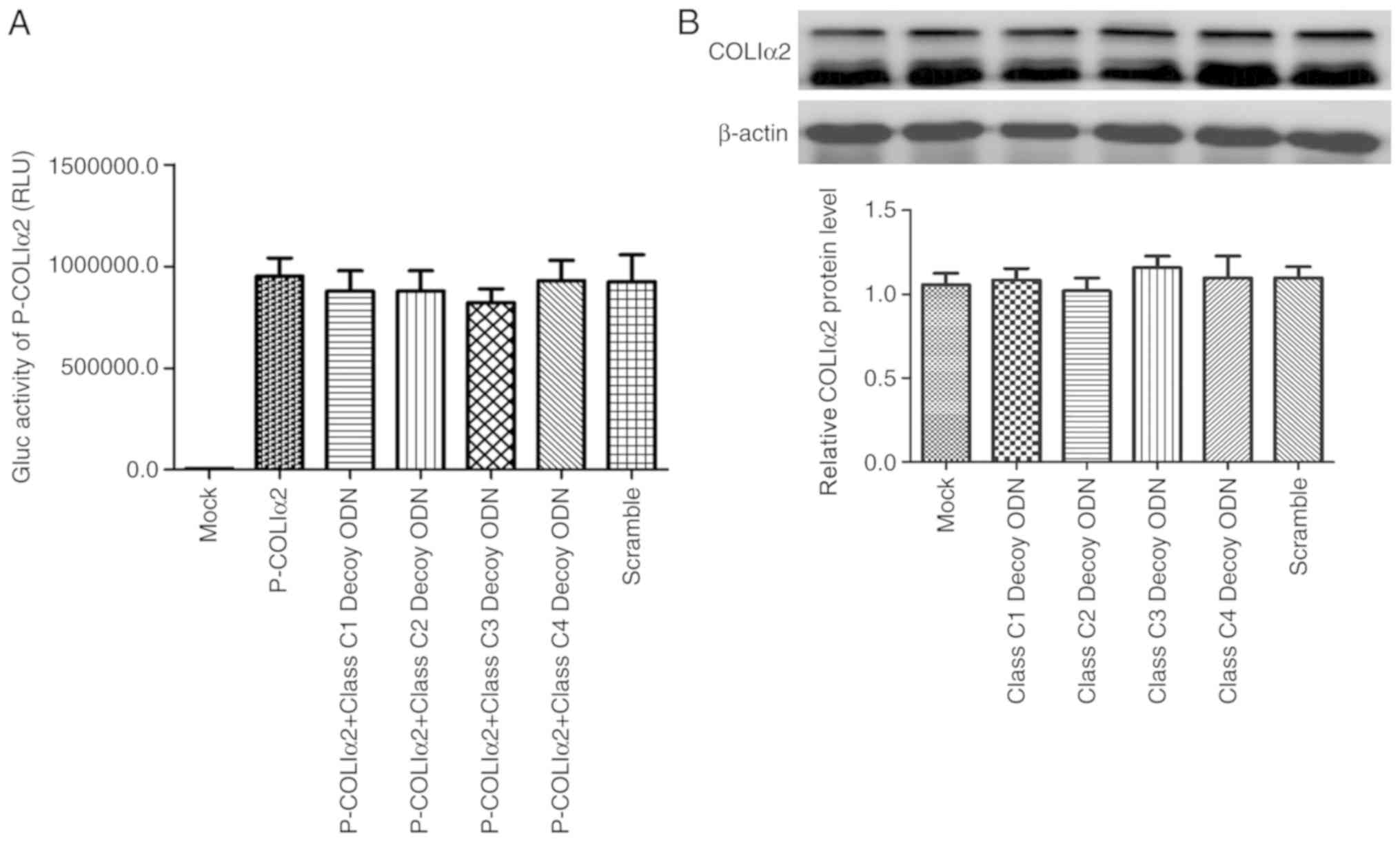
Class C decoy ODNs do not affect
COLIα2 expression in HSC-T6 cells
COLIα2 is also regulated by the TGF-β signaling
pathway (4). Using a
Gaussia luciferase assay, none of the four class C decoy
ODNs were found to exert any effect on COLIα2 promoter activity
(Fig. 4A). In addition, it was
also confirmed that the four class C decoy ODNs exerted no effect
on COLIα2 expression in HSC-T6 cells by western blotting (Fig. 4B).
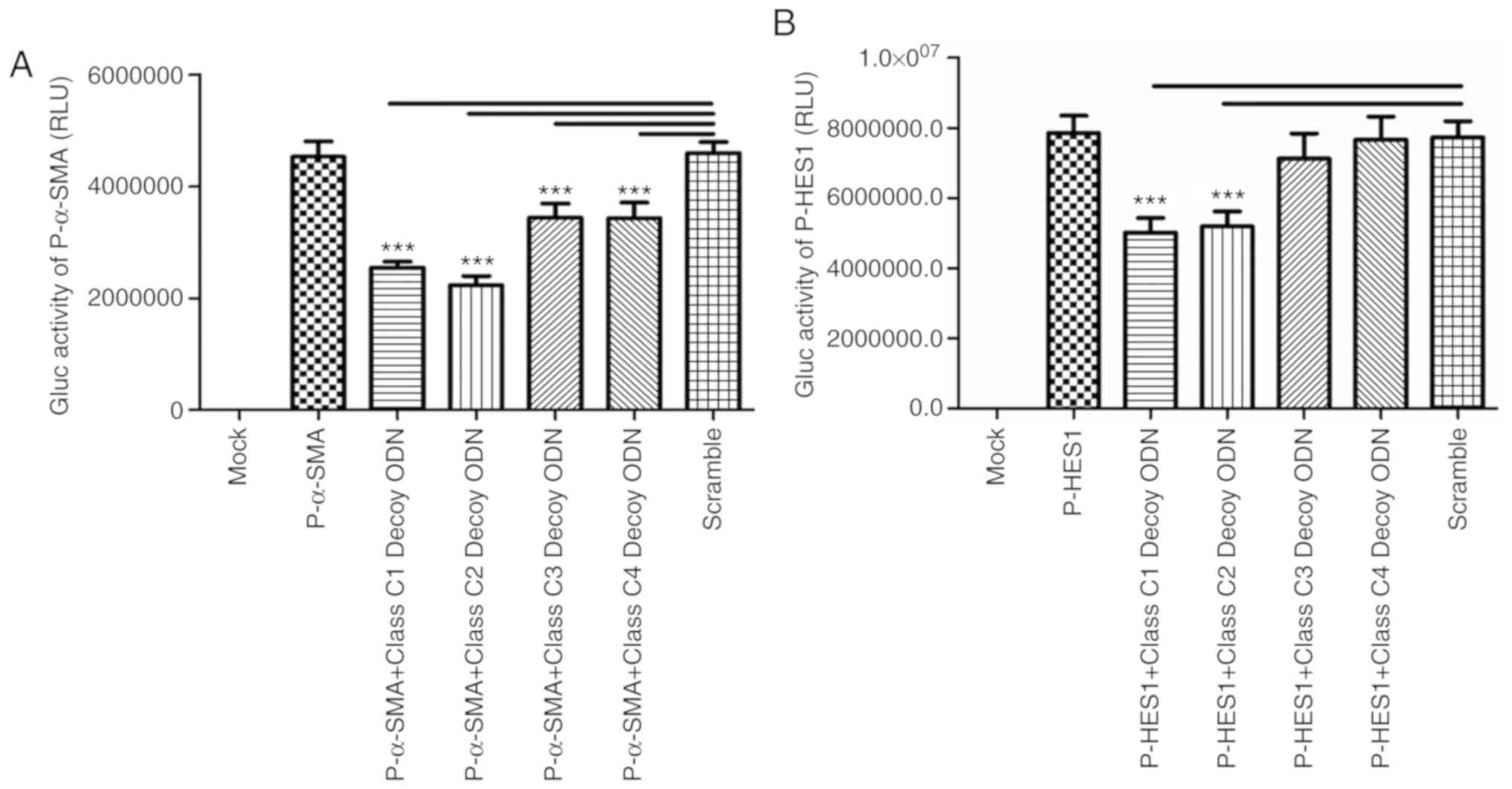
Class C decoy ODNs downregulate α-SMA
and HES1 promoter activity
Previous studies have demonstrated that TGF-β
regulates the expression of α-SMA and HES1 (20–23).
Bioinformatics analysis revealed that there are five and ten
binding sites for class C sequences in the promoters of α-SMA and
HES1, respectively. The Gaussia luciferase activities of
pSMA-GLuc-Basic were found to be significantly decreased in all
experimental groups compared with Scr (P<0.001; Fig. 5A), suggesting that the four class C
decoy ODNs can downregulate the activity of the α-SMA promoter,
whereas only class C1 and class C2 decoy ODNs affect the activity
of the HES1 promoter (Fig.
5B).
In conclusion, class C1 decoy ODNs exerted the most
prominent effect on TGF-β signaling pathway-related genes and it
downregulated the expression of TGF-β, TIMP1, HES1, α-SMA and
COL1α1.
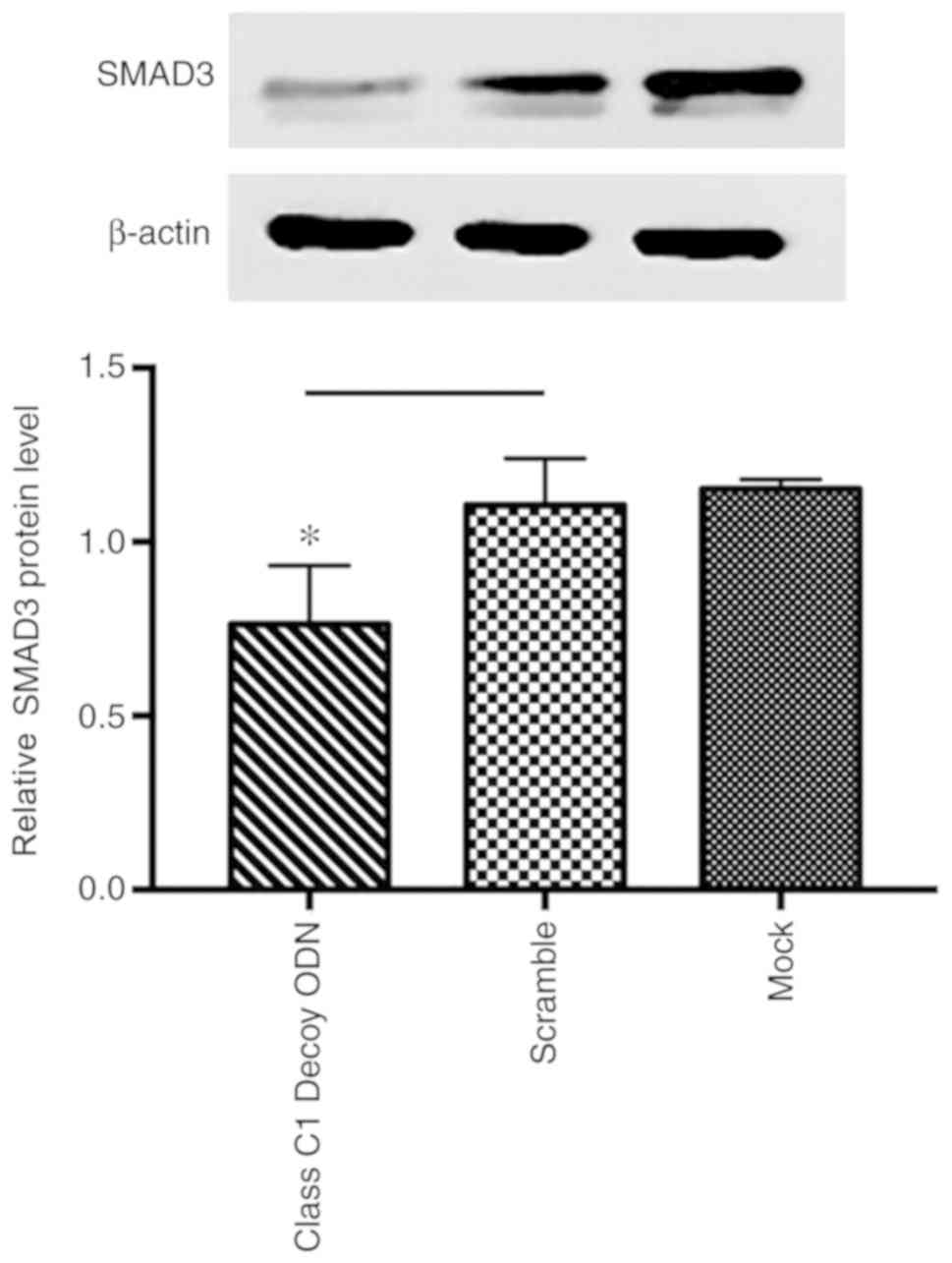
Class C1 decoy ODNs downregulate SMAD3
expression
Class C1 decoy ODNs were found to exert the broadest
and most prominent effect on TGF-β signaling pathway-related genes,
and it inhibited the promoter activity of TGF-β and its downstream
target genes, namely COLIα1, TIMP1 and α-SMA, and further
downregulated the protein expression of TGF-β, COLIα1 and TIMP1. To
investigate the mechanism through which class C1 decoy ODNs
downregulated TGF-β signaling pathway-related genes, the expression
of COLIα1 and SMAD3 was tested using western blot assays and proven
to be significantly downregulated by class C1 decoy ODNs
(P<0.05; Fig. 6).
Discussion
Liver fibrosis is an intermediate stage between
primary liver disease, liver cirrhosis, or even liver cancer. Thus,
reversing the process of liver fibrosis is key to preventing this
life-threatening progression. The major pathological characteristic
associated with liver fibrosis is disruption of the balance between
ECM synthesis and degradation (1,2).
Various cell-stimulating factors act on HSCs to promote their
activation and proliferation. Through proliferation, secretion of
ECM and contraction, activated HSCs are actively involved in the
occurrence of liver fibrosis and intrahepatic structural
remodeling, which is considered as the pathological basis of liver
fibrosis and portal hypertension. TGF-β1 is currently recognized as
the strongest pro-fibrosis factor by stimulating HSCs (1–3),
which mediate TGF-β1 signals from the cytoplasm to the nucleus,
ultimately inducing collagen (type I, II, III and others) synthesis
and secretion. Furthermore, it may also promote secretion of TIMPs
that can inhibit matrix metalloproteinases synthesis, resulting in
ineffective collagen degradation. It is broadly accepted that
TGF-β1 canonical signaling, also known as the TGF-β1/SMADs
signaling pathway, is crucial for the occurrence and progression of
hepatic fibrosis, whereas non-canonical signaling, which is
associated with multiple different pathways, such as MAPK, PI3K-AKT
and Wnt, also contributes to the activation of HSCs and liver
fibrosis (7,8). Several previous studies have revealed
the existence of crosstalk between Notch and TGF-β signaling in the
activation of HSCs, and the Notch downstream TF HES1 plays an
important role in this crosstalk (9–13).
Thus, blocking the signal transduction of TGF-β1 or regulating the
effect of SMADs on the expression of target genes in order to
decrease ECM synthesis and increase ECM degradation may be a
promising approach to reversing hepatic fibrosis.
HES1 belongs to the highly conserved bHLH family of
TFs, which are ~60 amino acids in length and named according to
their β helix-loop-helix structure. The C-type TF of the bHLH
family serves a role as a homologous or heterodimeric form that
binds to the class C sequence (CACGNG) (15). Bioinformatics analysis demonstrated
that the CACGNG sequence was present on the promoter region of the
TGF-β1, COLIα1, TIMP1, HES1 and α-SMA genes, indicating that the
C-type TF of the bHLH family may modulate the expression of those
pro-fibrotic genes. The results of the bioinformatics analysis were
consistent with the literature review (11–13).
Using the decoy ODN strategy, it was confirmed that class C decoy
ODNs have different capacities of inhibiting the expression of
pro-fibrotic genes, such as TGF-β, SMAD3, COLIα1 and TIMP1, and
downregulating the transcriptional activity of the HES1 and α-SMA
promoters, as well as TRE. Among the four decoy ODNs, class C1
decoy ODNs, which carry a class C TF trap sequence (CACGTG), are
the most efficient for downregulating those target genes, following
by Class C2, which indicate Class C1&2 DNA binding domains has
a greater affinity for Class C proteins than C3&4. It seemed to
be paradoxical that by bioinformatic analysis, the binding site of
Class C3&4 are outnumbered compared with the binding site of
Class C1&2. This is especially true in the promotor region of
Hes1, where Class1&2 has 1 binding site and Class C3&4 has
4 binding sites and in the promotor region of TIMP1, where
Class1&2 has 0 binding sites and Class C3&4 has 5 binding
sites. The present study assumed that the Class C proteins bind to
Class C3&4 binding sites in Hes1 and the TIMP1 promoter, and
after Class C1&2 decoy ODN which carried a binding domain with
better affinity was conducted into the cell, it captured Class C
proteins and competitively inhibited their binding with the Class
C3&4 binding site in the reporter plasmid, and then the
G-luciferase activity decreased. Similarly, Class C1&2 can
downregulate TIMP-1 promoter activity and its expression, which can
also be explained by the strong affinity to TFs of exogenous Class
C1&2 sequence.
By reducing COLIα1 synthesis and promoting ECM
degradation via downregulating TIMP1, as well as repressing HSC
transactivation via downregulating TGF-β and α-SMA, class C1 decoy
ODNs appear to be promising for preventing HSC activation and
hepatic fibrosis. The possible mechanism underlying the
anti-fibrotic effects of class C1 decoy ODNs is competitive binding
of class C TFs, including HES1, or indirect repression by
inhibiting the TGF-β/SMADs pathway, as the synthesis of TGF-β and
SMAD3 was downregulated and the transcriptional activity of TRE was
inhibited. However, the applicability of class C1 decoy ODNs in the
clinical setting requires further investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant. no. 81670555), The
Health Commission of Hubei Province Scientific Research Project
(grant. no. WJ2019H533) and The Hubei Provincial Department of
Education (grant. no. Q20181208).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JFW, LH and CBL conceived and designed the
experiments; CR, YRN, YMZ, YQZ and RTZ performed the experiments
and analyzed the data; CR and YRN wrote the manuscript. CBL was
responsible for the language editing of the manuscript. All authors
read and approved the final submitted version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Novo E, Cannito S, Paternostro C, Bocca C,
Miglietta A and Parola M: Cellular and molecular mechanisms in
liver fibrogenesis. Arch Biochem Biophys. 548:20–37. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yoshida K, Murata M, Yamaguchi T and
Matsuzaki K: TGF-β/Smad signaling during hepatic
fibro-carcinogenesis (review). Int J Oncol. 45:1363–1371. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sa Y, Li C, Li H and Guo H: TIMP-1 Induces
α-Smooth muscle actin in fibroblasts to promote urethral scar
formation. Cell Physiol Biochem. 35:2233–2243. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bi WR, Yang CQ and Shi Q: Transforming
growth factor-β1 induced epithelial-mesenchymal transition in
hepatic fibrosis. Hepatogastroenterology. 59:1960–1963.
2012.PubMed/NCBI
|
|
6
|
Okazaki I, Noro T, Tsutsui N, Yamanouchi
E, Kuroda H, Nakano M, Yokomori H and Inagaki Y: Fibrogenesis and
carcinogenesis in non-alcoholic steatohepatitis (NASH): Involvement
of matrix metalloproteinases (MMPs) and tissue inhibitors of
metalloproteinase (TIMPs). Cancers Basel). 6:1220–1255. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Poelstra K: Liver fibrosis in 2015:
Crucial steps towards an effective treatment. Nat Rev Gastroenterol
Hepatol. 13:67–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tacke F and Trautwein C: Mechanisms of
liver fibrosis resolution. J Hepatol. 63:1038–1039. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Shen RW, Han B, Li Z, Xiong L,
Zhang FY, Cong BB and Zhang B: Notch signaling mediated by
TGF-β/Smad pathway in concanavalin A-induced liver fibrosis in
rats. World J Gastroenterol. 23:2330–2336. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aimaiti Y, Jin X, Wang W, Chen Z and Li D:
TGF-β1 signaling regulates mouse hepatic stellate cell
differentiation via the Jagged1/Notch pathway. Life Sci.
192:221–230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen YX, Weng ZH, Qi D and Zhang SL:
Effect of Notch signaling on the activation of hepatic stellate
cells. Zhonghua Gan Zang Bing Za Zhi. 20:677–682. 2012.(In
Chinese). PubMed/NCBI
|
|
12
|
Zhang K, Zhang YQ, Ai WB, Hu QT, Zhang QJ,
Wan LY, Wang XL, Liu CB and Wu JF: Hes1, an important gene for
activation of hepatic stellate cells, is regulated by Notch1 and
TGF-β/BMP signaling. World J Gastroenterol. 21:878–887. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu M, Ou-Yang HF, Wu CG, Qu SY, Xu XT and
Wang P: Notch signaling regulates col1α1 and col1α2 expression in
airway fibroblasts. Exp Biol Med (Maywood. 239:1589–1596. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma PC, Rould MA, Weintraub H and Pabo CO:
Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on
DNA recognition and implications for transcriptional activation.
Cell. 77:451–459. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iso T, Kedes L and Hamamori Y: HES and
HERP families: Multiple effectors of the Notch signaling pathway. J
Cell Physiol. 194:237–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kobayashi T and Kageyama R: Expression
dynamics and functions of Hes factors in development and diseases.
Curr Top Dev Biol. 110:263–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tomita N, Ogihara T and Morishita R:
Transcription factors as molecular targets: Molecular mechanisms of
decoy ODN and their design. Curr Drug Targets. 4:603–608. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tomita N, Azuma H, Kaneda Y, Ogihara T and
Morishita R: Gene therapy with transcription factor decoy
oligonucleotides as a potential treatment for cardiovascular
diseases. Curr Drug Targets. 4:339–346. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jia D, Ni YR, Zhang YQ, Rao C, Hou J, Tang
HQ, Liu CB and Wu JF: SP1 and UTE1 Decoy ODNs inhibit activation
and proliferation of hepatic stellate cells by targeting tissue
inhibitors of metalloproteinase 1. Cell Biosci. 6:312016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krizhanovsky V, Yon M, Dickins RA, Hearn
S, Simon J, Miething C, Yee H, Zender L and Lowe SW: Senescence of
activated stellate cells limits liver fibrosis. Cell. 134:657–667.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li YH, Woo SH, Choi DH and Cho EH:
Succinate causes α-SMA production through GPR91 activation in
hepatic stellate cells. Biochem Biophys Res Commun. 463:853–858.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blokzijl A, Dahlqvist C, Reissmann E, Falk
A, Moliner A, Lendahl U and Ibáñez CF: Cross-talk between the Notch
and TGF-beta signaling pathways mediated by interaction of the
Notch intracellular domain with Smad3. J Cell Biol. 163:723–728.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu L, Gao C, Chen G, Li X, Li J, Wan Q
and Xu Y: Notch signaling molecules activate TGF-β in rat Mesangial
cells under high glucose conditions. J Diabetes Res.
2013:9797022013. View Article : Google Scholar : PubMed/NCBI
|