Introduction
Klebsiella pneumoniae (K. pneumoniae)
isolates with carbapenemases are often resistant to the majority of
antibiotics available. Infections caused by K. pneumoniae
with carbapenemases often increase mortality rates, which range
between 23 and 75% (1), and are
partially attributed to the lack of efficacious antimicrobial
agents (2). Therefore, timely
identification of the drug-resistance profile of K.
pneumoniae isolates and selection of appropriate antimicrobial
agents are essential for anti-infective therapy. Usually,
antibiotic susceptibility testing is performed using minimal
inhibition concentration (MIC) or agar diffusion methods in
clinical laboratories. However, the determination of KPC-producing
strains remains intractable, as it has been reported that 7–87% of
KPC-producing K. pneumoniae clinical isolates are
susceptible to imipenem or meropenem (3). Therefore, the identification of
carbapenem-resistance phenotypes with automatic systems is not
always reliable. The majority of influencing factors remain unclear
at present, although the majority of mechanisms correlate with the
production of carbapenemase (4,5).
Numerous factors, including deletions of the upstream genetic
environment (6),
blaKPC copy numbers (7), outer membrane and porins (8–10),
biofilm formation (11), the
AcrAB-TolC efflux system (12) and
transcriptional start site (13),
have been identified to affect carbapenem MICs to varying
degrees.
In our previous study, a series of
blaKPC-2-harboring plasmids that possessed a
backbone region, in which the KlcAHS gene
coexisted with the blaKPC-2 gene, were identified
(14). Of note, the subscript HS
is used in this context to distinguish the KlcA sequence
investigated in the present study from other KlcA sequences.
KlcAHS deletion and complementation experiments
were performed to investigate the association between carbapenem
MICs and the KlcAHS gene, revealing that the
KlcAHS gene can increase carbapenem MICs by
upregulating the gene expression of blaKPC-2.
Materials and methods
Strains, plasmids and antibiotics
The pHS10842 and pHS092839 plasmids were extracted
from Escherichia coli (E. coli) and K.
pneumoiae clinical isolates, respectively, in the clinical
laboratory at Huashan Hospital (Shanghai, China). The pIJ790
plasmid was used as a helper plasmid to generate λ-RED (gam, bet,
exo) proteins to knock out the KlcAHS gene in
pHS092839. The pET24a expression plasmid was used to generate the
recombinant plasmid pET24a-KlcA. The E. coli BL21 (DE3)
strain was selected as a host strain to produce
ΔKlcAHSpHS10842, and the E. coli BW25113
strain was selected as a host strain to produce
ΔKlcAHSpHS092839. The E. coli strain
NM1049 was selected as a host strain to harbor wild-type and mutant
pHS092839. This strain, which carries a type I A restriction and
modification system (15), was
provided by Professor Dryden (University of Edinburgh, UK).
Ampicillin (AMP, 100 µg/ml), chloramphenicol (Cm, 25
µg/ml), kanamycin (Kan, 25 µg/ml) and L-arabinose were all
purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany), and
added to the culture medium as indicated. Mueller-Hinton (MH) agar
was obtained from BD Biosciences (Franklin Lakes, NJ, USA).
L-arabinose (final concentration, 0.2%) was added into
Luria-Bertani (LB) medium (Beijing Lablead Biotech Co., Ltd.) to
induce the expression of genes under the control of the pBAD
promoter (16). All enzymes used
in the present study were purchased from New England BioLabs,
Inc.
Construction of the recombinant
plasmid pET24a-KlcAHS
The KlcAHS gene was amplified from
the pHS092839 plasmid by polymerase chain reaction (PCR) using the
primers P1/P2, in which the NdeI and EcoRI
recognition sites were pre-located and underlined (Table I). PCR was carried out in a 25 µl
reaction mixture, which contained 20 ng template DNA, 0.15 U Taq
DNA polymerase, 2.0 mM dNTPs, 1X Taq DNA polymerase buffer (Takara
Biotechnology Co., Ltd.) and 10 µM primer. DNA amplification was
done under the following conditions: 95°C for 5 min, followed by 30
cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec,
with a final extension at 72°C for 5 min. The PCR products
obtained, together with the pET24a vector, were digested with
NdeI and EcoRI, purified and ligated with T4 DNA
ligase, prior to being transformed into CaCl2-treated
competent BL21 (DE3) cells (Sangon Biotech Co., Ltd.). The correct
clones were screened using colony PCR. Briefly, a masterbatch was
prepared for PCR amplification and placed in the PCR tube. A
sterile toothpick for was sued for each colony to remove a small
number of bacterial colonies directly from the plate to be tested.
The toothpick was placed in the PCR tube with the masterbatch and
the bacteria scraped into the PCR mixture. PCR was performed: 94°C
for 5 min, 30 cycles: 94°C for 30 sec, 55°C for 30 sec, 72°C for 1
min and a final extension of 72°C for 5 min. To ensure no errors in
the sequence had been introduced during the PCR amplification step,
the 426 bp PCR product of KlcAHS amplification was
sequenced using primers P3/P4 (Table
I). Each sequencing reaction was performed using 0.5 µl of
BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and 1.6 µl of each primer (1 µM) in
10 µl final volume per reaction. Dye-labelled products were
sequenced using an ABI 3500 sequencer (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Sequencing chromatograms were edited
manually using Sequencher 4.7 software (Gene Codes
Corporation).
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| No. | Primer name | Sequence
(5′-3′) | Product (bp) |
|---|
| P1 |
KlcAHS-F | ACGGTGTCATATGATGCAAACAGAACTTAA |
|
| P2 |
KlcAHS-R | GCTAGAATTCCTAGTCTATTGCGGCCAAG | 426 |
| P3 | T7 primer F |
TAATACGACTCACTATAGG |
|
| P4 | T7 terminator
R |
GCTAGTTATTGCTCAGCGG |
|
| P5 | FP |
GTCAATACCGGAGAACTCCGCAACAGAAAGCCCCGGTGATG |
|
| P6 | RP |
CATCACCGGGGCTTTCTGTTGCGGAGTTCTCCGGTATTGAC |
|
| P7 | CS-F |
CCGCTTGACACAATAGGC |
|
| P8 | CS-R |
ATCGCCGTTCCTCACTAC |
|
| P9 | Upstream F |
GAGAACTCCGATGATCCAC |
|
| P10 | Upstream R |
CCTACATACCTCGCTCTGTTGCATCATCGGAGTTCTC | 407 |
| P11 | Kan-F |
GAGAACTCCGATGATGCAACAGAGCGAGGTATGTAGG |
|
| P12 | Kan-R |
TGTTGCTAGTCTATTGCGGCAAATATGTATCCGCTCATG | 463 |
| P13 | Downstream F |
CATGAGCGGATACATATTTGCCGCAATAGACTAGCAACA |
|
| P14 | Downstream R |
CCACTGAGAGGCTTACTAAG | 1,184 |
| P15 | KPC-2F |
TTGCTGGACTTTGTTGAG |
|
| P16 | KPC-2R |
CTATCTCTTCGTTGCCATC | 106 |
| P17 | 16S-F |
CGTATTCACCGTGGCATTCT |
|
| P18 | 16S-R |
GAGCAAGCGGACCTCATAA | 110 |
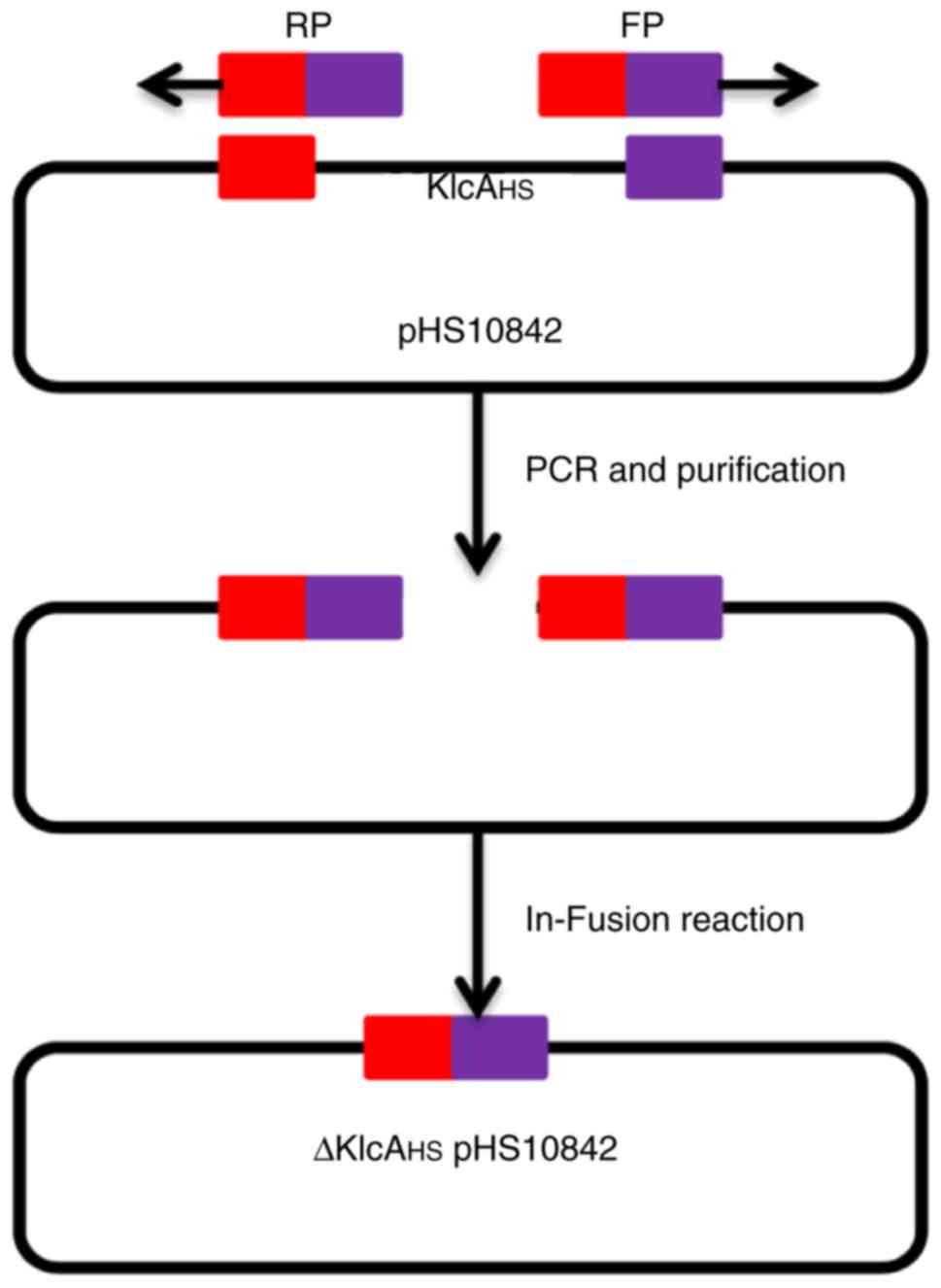
Knockout of the KlcAHS gene
in pHS10842
The KlcAHS gene in pHS10842
(accession no. KP125892) was knocked out using the modified
protocol described by Zhang et al (17). The primers P5/P6 were designed
based on the adjacent sequences of the two outer extremes of
KlcAHS with Primer Premier 6.22 software (Premier
Biosoft International, Palo Alto, CA, USA), and were used to
amplify the entire sequence of pHS10842 with the exception of
KlcAHS (Fig. 1).
A High-Fidelity PCR kit was used to amplify the target DNA
fragments (Takara Biotechnology Co., Ltd., Dalian, China). The PCR
product was subjected to electrophoresis, and the target band was
recovered and purified using the E.Z.N.A® Gel Extraction
kit (Omega Bio-Tek, Inc., Norcross, GA, USA). The product had a
41-bp homology region at each terminus. The linear DNA was cyclized
into ΔKlcAHSpHS10842 using the HieffClone One Step
Cloning kit (Shanghai Yeasen Biotech Co., Ltd., Shanghai, China)
according to the manufacturer's instructions, and transformed into
BL21 (DE3) competent cells. The correct clones were screened by
primer P7/P8 (Table I) and colony
PCR protocol in the above steps. inoculated into 3 ml fresh LB
supplemented with AMP and incubated overnight at 37°C. The mutant
pHS10842 was obtained.
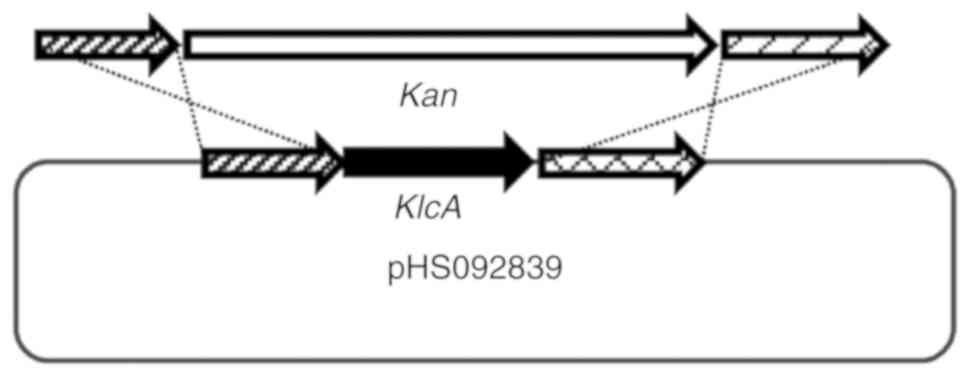
Knockout of the KlcAHS gene in
pHS092839. The KlcAHS gene in pHS092839
(accession no. KF724506) was knocked out according to the protocol
described by Derbise et al (18). Briefly, a four-step PCR procedure
was used to generate a Kan cassette with homologous regions
flanking the KlcAHS sequence in pHS092839. The
specific four-step PCR program was: First, using the plasmid
pHS092839 as a template, the 407- and 463-bp regions flanking the
KlcAHS gene were amplified with the primer pairs
P9/P10 and P13/P14 (Table I),
respectively, with the following amplification cycles: 94°C for 5
min, 30 cycles: 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min
and a final extension of 72°C for 5 min. The 20-bp homologous
region at each end of the Kan cassette was pre-designed at the 5′
end of the primers P10 and P13. Second, the Kan cassette was
amplified with the pET24a plasmid as a template using primers
P11/P12 (Table I), the PCR
reaction procedure for this step was the same as the previous step.
The PCR products had a homologous region of 20 bp at their 5′ end,
with the upstream and downstream DNA fragments, respectively. In
this manner, a 40-bp homologous region was formed at each extreme
of the Kan cassette with the upstream and downstream DNA fragments,
respectively. Third, three separate DNA fragments were integrated
into the entire Kan cassette using a series of PCRs. The entire Kan
cassette produced in the last step had two homologous regions
flanking the KlcAHS sequence in pHS092839.
Subsequently, the target fragments were recovered and purified with
the E.Z.N.A.® Gel Extraction kit. In total, 200 ng
purified linear PCR fragments were transformed into
electro-competent E. coli BW25113 cells carrying pIJ790
(CmR) and the desired pHS092839 (AMPR). The
shocked cells were incubated in 0.95 ml LB at 37°C for 1 h and
plated onto LB plates (3×106/plate) containing AMP and
Kan. The transformants were streaked onto LB plates containing Cm
to confirm the loss of temperature-sensitive pIJ790
(CmR) during incubation at 37°C. The transformants
(CmS AMPR KanR) were selected,
inoculated into 3 ml LB supplemented with AMP and Kan, and
incubated overnight. The mutant ΔKlcAHSpHS092839
was extracted and confirmed by DNA sequencing with primers P9/P14
(Fig. 2).
Antimicrobial susceptibility
tests
The plasmids pHS10842, mutant
ΔKlcAHSpHS10842, pET24a and recombinant
pET24a-KlcAHS were transformed into BL21 (DE3) competent
cells. All the transformants were selected to detect the imipenem
or Kan MICs using microdilution in cation-adjusted MH broth with
inoculation of 5×105 CFU/ml according to the CLSI M07-A9
guidelines (19). Serial dilutions
from this stock solution were added to freshly prepared MH (BD
Biosciences) to achieve a final concentration in the plates of
1–128 µg/ml for imipenem and 32–2,048 µg/ml for Kan. The MIC values
were assessed following incubation at 37°C for 18 h. The
experiments were performed in triplicate for each antibiotic and
each strain. The lowest concentrations of imipenem or Kan showing
no bacterial growth were considered as the MIC values. The assay
was repeated in triplicate.
In addition, another
blaKPC-2-harboring plasmid, pHS092839, and
ΔKlcAHSpHS092839 were transformed into the NM1049
E. coli strain. The imipenem and meropenem MICs of the two
transformants were detected using the fully automated microbial
identification system VITEK® 2 Compact according to the
manufacturer's protocol. The assay was repeated in triplicate.
Determination of the expression of
blaKPC-2 via reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNAs were extracted from the NM1049 E.
coli strains harboring the pHS092839 or
ΔKlcAHSpHS092839 plasmids using the
E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek, Inc.) and
treated with RNase-free DNase (Omega Bio-Tek, Inc.). A total of 0.5
µg of each total RNA was reverse transcribed into complementary DNA
with the PrimeScript™ RT reagent kit with gDNA Eraser (Takara
Biotechnology Co., Ltd.) according to the manufacturer's
instructions. RT-qPCR analysis was then performed on an ABI 7500
Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with SYBR Green RT-PCR kit
(Shanghai Yeasen Biotech Co., Ltd.) in a total volume of 20 µl,
containing 10 ng RNA and 10 pmol each primer [P15/P16 for
blaKPC-2, P17/P18 for 16S ribosomal RNA (rRNA);
Table I]. The following cycling
conditions were used for all amplifications: 2 min at 50°C, 10 min
at 95°C, and 40 cycles of 95°C for 15 sec and 60°C for 60 sec,
followed by a dissociation step at 95°C for 15 sec, 60°C for 15 sec
and 95°C for 15 sec. The baselines were adjusted manually to the
same level for the two groups with SDS software (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The messenger RNA
(mRNA) of the housekeeping 16S rRNA gene was selected as an
endogenous reference RNA to adjust the variation of RNA content and
amplification efficiency for relative quantification. Relative
quantities of blaKPC-2 mRNA were determined with
the comparative quantification cycle (Cq) method. The Cq value
indicates the number of PCR cycles at which the fluorescence
intensity begins to increase exponentially. The equation
2−ΔΔCq (20) was used
to calculate the differences in blaKPC-2 mRNA
expression levels between E. coli NM1049 strains harboring
pHS092839 and ΔKlcAHSpHS092839, where
ΔΔCq=ΔCq-blaKPC-2-ΔCq−16S rRNA. All
experiments were performed in triplicate from three RNA
preparations. The expression levels are presented as the mean of
three independent experiments. Negative controls were performed
using the purified RNA without reverse transcription as the
templates. The assay was repeated in triplicate. Data are presented
as the mean ± standard deviation.
Statistical analysis
Data were analyzed using SPSS 17.0 (SPSS, Inc.,
Chicago, IL, USA). The assay was repeated in triplicate. The
variables are described as the mean ± standard deviation. The
comparison tests were performed using non-parametric methods with
the Mann-Whitney U test. P<0.05 was considered to indicate a
statistically significant difference.
Results
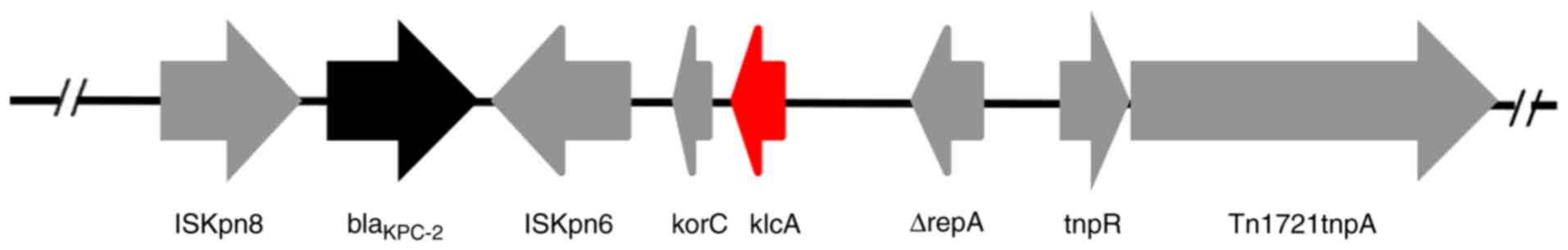
Analysis of the genetic structure of
blaKPC-2-harboring plasmids
The backbone region of the extracted pHS10842 and
pHS092839 plasmids extracted was confirmed, in which the
KlcAHS and blaKPC-2 genes
coexisted (Fig. 3).
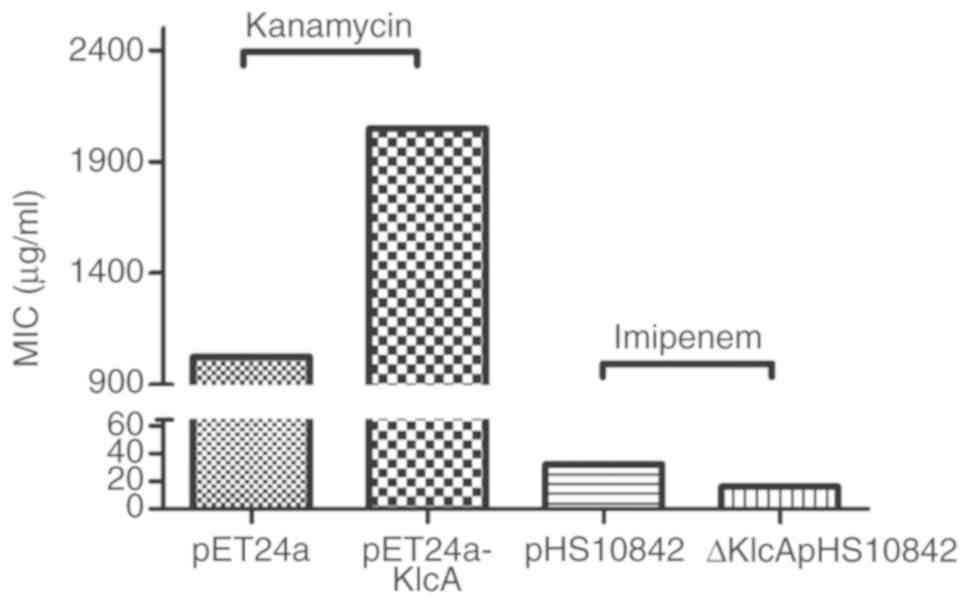
Examination of the association between
KlcAHS and antibiotic MICs
The imipenem MIC of the transformants harboring
ΔKlcAHSpHS10842 was lower (16 µg/ml) than that of
the wild-type pHS10842 (32 µg/ml), and the Kan MIC of transformants
harboring pET24a was lower (1,024 µg/ml) than that of
pET24a-KlcAHS (2,048 µg/ml) (Fig. 4). In addition, the imipenem MICs of
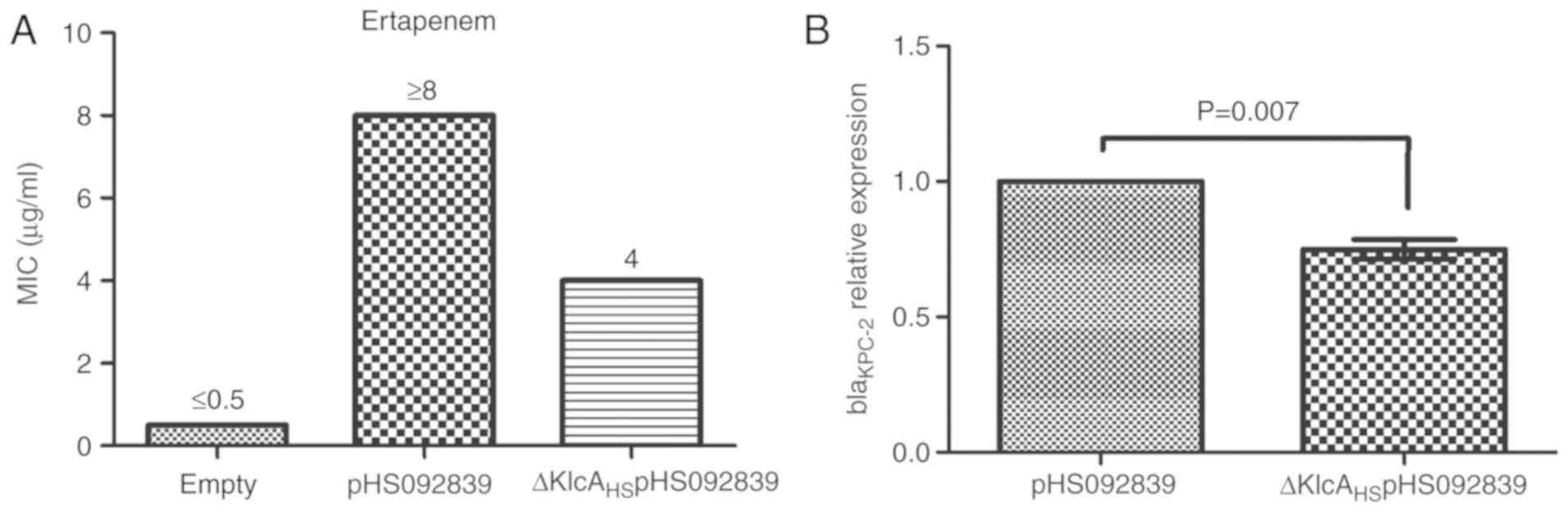
the two NM1049 E. coli strains carrying plasmids pHS092839
or ΔKlcAHSpHS092839 exceeded 16 µg/ml, and the
ertapenem MIC of host strains harboring
ΔKlcAHSpHS092839 was 4 µg/ml, compared with ≥8
µg/ml for the host strains carrying pHS092839 (Fig. 5A). Taken together, it was confirmed
that KlcAHS contributed to the decreased
susceptibility to the corresponding antibiotics. However, the MIC
alteration was not large, with only a 2-fold decrease in
susceptibility to carbapenems in the presence of the
KlcAHS gene.
Expression of blaKPC-2 is
affected by the KlcAHS gene
The results of RT-qPCR analysis revealed that the
mRNA expression levels of blaKPC-2 in the
transformants carrying ΔKlcAHSpHS092839 were
significantly downregulated (P=0.007) compared with those observed
in the transformants carrying pHS092839 (Fig. 5B).
Discussion
It was reported that 27 KPC-producing K.
pneumoniae isolates exhibit a range of carbapenem MICs, with 11
exhibiting low-level carbapenem resistance to imipenem or meropenem
(MIC <4 µg/ml), two exhibiting an intermediate level (MIC=8
µg/ml) and 14 a high level (MIC >16 µg/ml) (7). Other studies have reported that the
susceptibility rates to ertapenem, imipenem and meropenem for
KPC-producing K. pneumoniae were 0–6, 26–29 and 16–52%,
respectively (4,5,21).
The growth of KPC-producing isolates may be inhibited at carbapenem
MIC values lower than the recommended breakpoint concentrations. In
our previous study, there was a fraction of isolates showing a
lower MIC than the breakpoint concentration in KPC-producing K.
pneumoniae, namely 4.5% for imipenem in contrast to 3.6% for
meropenem (unpublished data), which indicated that plasmid encoding
carbapenemases were not the unique factor influencing MICs.
Therefore, it is more difficult to detect KPC-producing K.
pneumoniae clinical isolates with low carbapenem MICs in
clinical laboratories. Proper identification of KPC-producing
isolates is critical to the therapeutic schedule, as carbapenems
are prohibited from being used to control the infections due to
KPC-producing pathogens. Although the carbapenem-resistant
phenotype is caused by combined mechanisms associated with extended
spectrum β-lactamase (ESBL) and outer-membrane permeability
defects, the clinical isolates are not KPC-producing pathogens, the
use of carbapenems remains controversial (3). Therefore, the CLSI carbapenem
breakpoint concentrations were revised in 2010. At the same time,
EUCAST (http://www.eucast.org/) recommended a
lower cut-off point to identify potential carbapenemase-producing
Enterobacteriaceae (CPE). Numerous factors are able to alter
carbapenems MIC values. Carbapenems MICs were increased when
carbapenems are degraded in clinical strains producing additional
ESBLs or AmpC β-lactamases (22).
In addition, alterations or losses of porins in K.
pneumoniae (22,23) or E. coli (24) and the enhanced activity of efflux
pumps (25) have been shown to
reduce susceptibility to carbapenems. In the present study,
KlcAHS was shown to elevate the MIC of antibiotics not
only for imipenem, but also for any other antibiotics. The results
of this study showed that KlcAHS upregulated the
expression of mRNA of blaKPC-2. The variation in
carbapenems MIC values in CPE may involve diverse combinations of
resistance mechanisms in each clinical isolate (7,12).
An understanding of the underlying mechanisms associated with the
alteration of carbapenem MICs will facilitate anti-infective
therapy, particularly for patients infected with KPC-producing
pathogens.
In the present study, it was demonstrated that the
KlcAHS gene, in addition to the
blaKPC-2 gene, was located at the backbone region
of several plasmids in carbapenem-resistant K. pneumonia
clinical isolates. The KlcAHS gene upregulated
the expression of blaKPC-2 and reduced the
susceptibility to antibiotics such as carbapenems. The present
study also had limitations; for example, the E. coli BL21
(DE3) strain was selected as a host strain for
ΔKlcAHSpHS10842 and pHS10842, whereas the host E.
coli strain NM1049 was selected as ΔKlcAHSpHS092839
and pHS092839; although host background is identical, the MIC of
each is comparable and it is better to transform the two plasmids
and their derivants into the same strains. In addition, when
studying the function of a gene, knockout and complementation
experiments are classical experimental methods, when the mutation
point is located on the bacterial chromosome. The complementation
experiment can be completed by introducing an expression plasmid
carrying the target gene into the host cell, but the gene in the
present study was located on the plasmid carried by the host cell,
which limited the use of the complementation experiment.
Additionally, there remains limited understanding of the exact
function of the anti-restriction KlcAHS; in the present
study, it was found that KlcAHS upregulated the
transcription of blaKPC-2 gene and improving this
knowledge is aimed in future studies.
The present findings showed that
KlcAHS conferred increased resistance to
carbapenems in the host strains. The survival probability of
clinical pathogens may be enhanced by the presence of the
KlcAHS gene in antibiotics used on a large
scale.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. NSFC 81571365 and
81372141), the Natural Science Foundation of Jiangsu Province
(grant no. BK20191210), the Jiangsu Commission of Health (grant no.
H2018073), the Lianyungang Science and Technology Bureau Project
(grant no. SH1526) and the Bengbu Medical College Research Project
(grant no. BYKY17182).
Availability of data and materials
The datasets used in the present study are available
from the corresponding author on reasonable request.
Authors' contributions
HS and GHH conceived and designed the experiments.
WL, CZ, YW, WJZ, WCZ, YPS and YZ performed the experiments. WL, CZ,
LY, JH, YPS, LY and XL analyzed the data. WL and CZ interpreted the
data and WL wrote the first draft of the manuscript. CZ, YW and WJZ
developed the structure and arguments for the paper. WCZ and YPS
made critical revisions. All authors have reviewed and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tzouvelekis LS, Markogiannakis A,
Psichogiou M, Tassios PT and Daikos GL: Carbapenemases in
Klebsiella pneumoniae and other Enterobacteriaceae: An evolving
crisis of global dimensions. Clin Microbiol Rev. 25:682–707. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karaiskos I and Giamarellou H:
Multidrug-resistant and extensively drug-resistant Gram-negative
pathogens: Current and emerging therapeutic approaches. Expert Opin
Pharmacother. 15:1351–1370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nordmann P, Cuzon G and Naas T: The real
threat of Klebsiella pneumoniae carbapenemase-producing bacteria.
Lancet Infect Dis. 9:228–236. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tenover FC, Kalsi RK, Williams PP, Carey
RB, Stocker S, Lonsway D, Rasheed JK, Biddle JW, McGowan JE Jr and
Hanna B: Carbapenem resistance in Klebsiella pneumoniae not
detected by automated susceptibility testing. Emerg Infect Dis.
12:1209–1213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anderson KF, Lonsway DR, Rasheed JK,
Biddle J, Jensen B, McDougal LK, Carey RB, Thompson A, Stocker S,
Limbago B and Patel JB: Evaluation of methods to identify the
Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin
Microbiol. 45:2723–2725. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Naas T, Cuzon G, Truong HV and Nordmann P:
Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob
Agents Chemother. 56:4753–4759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kitchel B, Rasheed JK, Endimiani A, Hujer
AM, Anderson KF, Bonomo RA and Patel JB: Genetic factors associated
with elevated carbapenem resistance in KPC-producing Klebsiella
pneumoniae. Antimicrob Agents Chemother. 54:4201–4207. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pages JM, Peslier S, Keating TA, Lavigne
JP and Nichols WW: Role of the outer membrane and porins in
susceptibility of β-lactamase-producing enterobacteriaceae to
ceftazidime--avibactam. Antimicrob Agents Chemother. 60:1349–1359.
2016. View Article : Google Scholar :
|
|
9
|
Landman D, Bratu S and Quale J:
Contribution of OmpK36 to carbapenem susceptibility in
KPC-producing Klebsiella pneumoniae. J Med Microbiol. 58:1303–1308.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin XM, Yang JN, Peng XX and Li H: A novel
negative regulation mechanism of bacterial outer membrane proteins
in response to antibiotic resistance. J Proteome Res. 9:5952–5959.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Santajit S and Indrawattana N: Mechanisms
of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int.
2016:24750672016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saw HT, Webber MA, Mushtaq S, Woodford N
and Piddock LJ: Inactivation or inhibition of AcrAB-TolC increases
resistance of carbapenemase-producing Enterobacteriaceae to
carbapenems. J Antimicrob Chemother. 71:1510–1519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roth AL, Kurpiel PM, Lister PD and Hanson
ND: bla(KPC) RNA expression correlates with two transcriptional
start sites but not always with gene copy number in four genera of
Gram-negative pathogens. Antimicrob Agents Chemother. 55:3936–3938.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang W, Xie Y, Xiong W, Tang Y, Li G,
Jiang X and Lu Y: Anti-restriction protein, KlcAHS, promotes
dissemination of carbapenem resistance. Front Cell Infect
Microbiol. 7:1502017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Serfiotis-Mitsa D, Herbert AP, Roberts GA,
Soares DC, White JH, Blakely GW, Uhrín D and Dryden DT: The
structure of the KlcA and ArdB proteins reveals a novel fold and
antirestriction activity against Type I DNA restriction systems in
vivo but not in vitro. Nucleic Acids Res. 38:1723–1737. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gust B, Challis GL, Fowler K, Kieser T and
Chater KF: PCR-targeted Streptomyces gene replacement identifies a
protein domain needed for biosynthesis of the sesquiterpene soil
odor geosmin. Proc Natl Acad Sci USA. 100:1541–1546. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Werling U and Edelmann W:
Seamless Ligation Cloning Extract (SLiCE) cloning method. Methods
Mol Biol. 1116:235–244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Derbise A, Lesic B, Dacheux D, Ghigo JM
and Carniel E: A rapid and simple method for inactivating
chromosomal genes in Yersinia. FEMS Immunol Med Microbiol.
38:113–116. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Methods for antimicrobial susceptibility
testing of anaerobic bacteria. 9th. CLSI standard M11. Clinical and
Laboratory Standards Institute; Wayne, PA: 2018
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marchaim D, Navon-Venezia S, Schwaber MJ
and Carmeli Y: Isolation of imipenem-resistant Enterobacter
species: Emergence of KPC-2 carbapenemase, molecular
characterization, epidemiology, and outcomes. Antimicrob Agents
Chemother. 52:1413–1418. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Elliott E, Brink AJ, van Greune J, Els Z,
Woodford N, Turton J, Warner M and Livermore DM: In vivo
development of ertapenem resistance in a patient with pneumonia
caused by Klebsiella pneumoniae with an extended-spectrum
beta-lactamase. Clin Infect Dis. 42:e95–e98. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jacoby GA, Mills DM and Chow N: Role of
beta-lactamases and porins in resistance to ertapenem and other
beta-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother.
48:3203–3206. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lartigue MF, Poirel L, Poyart C,
Reglier-Poupet H and Nordmann P: Ertapenem resistance of
Escherichia coli. Emerg Infect Dis. 13:315–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szabo D, Silveira F, Hujer AM, Bonomo RA,
Hujer KM, Marsh JW, Bethel CR, Doi Y, Deeley K and Paterson DL:
Outer membrane protein changes and efflux pump expression together
may confer resistance to ertapenem in Enterobacter cloacae.
Antimicrob Agents Chemother. 50:2833–2835. 2006. View Article : Google Scholar : PubMed/NCBI
|