Introduction
Obesity, especially excess visceral lipid
deposition, increases the risks of numerous diseases, including
type 2 diabetes, cardiovascular disease, and some cancers (1–4).
Generally, obesity involves hypertrophy and hyperplasia of excess
adipocytes (5,6). Adipocyte hyperplasia is dependent on
preadipocyte proliferation and differentiation. Research on
adipocyte hyperplasia has been focused largely on deciphering the
molecular mechanisms underlying obesity and developing novel
therapeutics for obesity.
Circular RNAs (circRNAs) are non-coding RNAs that
form a closed circular loop by back-splicing circularization
(7), and they exhibit higher
stability and resistance against RNA exonucleases compared with
linear RNAs (8). Recent research
has revealed that circRNAs regulate gene expression via multiple
mechanisms, such as regulating gene transcription and splicing
(9,10), acting as microRNA (miRNA) sponges
(11), and forming RNA-protein
complexes (12). Moreover, some
circRNAs can be transcribed into proteins (13). In mammals, circRNA expression is
tissue- and developmental stage-specific (14). Numerous studies have reported that
circRNAs participate in the regulation of various physiological and
pathological processes, such as regulating myogenesis and
tumorigenesis (13,15,16).
However, the role of circRNAs in visceral adipogenesis has not been
investigated, and no circRNA examined to date has been associated
with visceral adipogenesis.
Examination of the genes differentially expressed in
preadipocytes and adipocytes should identify novel factors
promoting or inhibiting lipid deposition. To identify the circRNAs
associated with visceral adipocyte hyperplasia, the expression
profiles of circRNAs in human preadipocytes derived from visceral
fat tissue (HPA-v) and adipocytes were analyzed using circRNA
microarrays. The results revealed that HPA-v and visceral
adipocytes had different circRNA expression patterns, and the
parental genes of the differentially expressed circRNAs were
related to lipid metabolism; moreover, the candidate circRNAs were
revealed to target many potential miRNA sites.
Materials and methods
Preadipocyte differentiation
HPA-v (cat. no. 7210; ScienCell Research
Laboratories, Inc.) were isolated from human visceral fat tissue
and cultured in preadipocyte medium (cat.no 7211; ScienCell
Research Laboratories, Inc.) containing 5% fetal bovine serum, 100
IU/ml penicillin-streptomycin, and 1% preadipocyte growth
supplement (cat.no. 7252; ScienCell Research Laboratories, Inc.).
After reaching confluence, the HPA-v were induced to differentiate
for 3 days in DMEM containing 0.1 mM 3-isobutyl-1-methylxanthine, 1
µM dexamethasone, and 5 µg/ml insulin. The differentiated HPA-v
were then maintained in DMEM containing 5 µg/ml insulin for 6
days.
Oil Red O staining
Cellular lipids were detected using Oil Red O
staining. Briefly, upon reaching 100% confluence or
differentiation, the preadipocytes were washed three times with
phosphate-buffered saline and fixed in 10% formalin for 15 min at
room temperature. After fixation, the cells were stained with Oil
Red O for 20 min at room temperature. Stained cells were visualized
using a Leica DMI 4000 B fluorescent microscope on the white light
setting (magnification, ×100).
Total RNA isolation
Total RNA was isolated from 5×106 HPA-v
cells and adipocytes, which were differentiated from HPA-v cells,
using TRIzol® reagent (cat. no. 15596018; Invitrogen;
Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA
using the HiScript III 1st Strand cDNA Synthesis kit (cat. no.
R312-01; Vazyme Biotech Co., Ltd.), according to the manufacturer's
protocol. RNA integrity was evaluated by electrophoresis on 2%
(w/v) denaturing agarose gels. The concentration and purity of RNA
were determined according to the OD260/OD280
values using the NanoDrop1000 Spectrophotometer (Thermo Fisher
Scientific, Inc.).
CircRNA microarray analysis
A human circRNA microarray (Agilent Technologies,
Inc.) containing 170,340 human circRNA probes was used. Six samples
(three HPA-v and three adipocyte samples) were detected by
CapitalBio Corporation using circRNA microarrays. CircRNAs were
purified, amplified, labeled with Cy3-dCTP, and hybridized onto the
circRNA array according to the manufacturer's protocol. The circRNA
expression data were normalized using the GeneSpring GX software
version 13.0 (https://www.agilent.com).
Differentially expressed circRNAs between HPA-v and adipocytes were
selected according to the following thresholds: |fold change| ≥5
and P-value <0.01. Volcano plots were generated to
visualize the circRNAs differentially expressed between HPA-v and
adipocytes. Hierarchical cluster analysis was used to evaluate
differential circRNA expression patterns across the six samples.
The parental genes of the differentially expressed circRNAs were
analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG)
database within the Database for Annotation, Visualization and
Integrated Discovery (https://david.ncifcrf.gov). The homology of circRNAs
between human and mice was analyzed using CIRCpedia version 2
software (http://circatlas.biols.ac.cn). The miRNA response
elements (MERs) within circRNAs were predicted using MiRanda
version 3.3 software (http://www.microrna.org).
Quantitative PCR (qPCR)
The expression levels of peroxisome
proliferator-activated receptor gamma 2 (PPARG2), CCAAT
enhancer binding protein alpha (CEBPA), fatty acid binding
protein 4 (FABP4), hsa_circ_0136134, hsa_circ_0017650, and
hsa-circRNA9227-1 were detected by qPCR. Ribosomal protein lateral
stalk subunit P0 (RPLP0) was used as an invariant control.
qPCR was performed using the ChamQ SYBR® qPCR Master mix
(cat. no. Q311-02; Vazyme Biotech Co., Ltd.), according to the
manufacturer's instructions, on an ABI 7300 instrument (ABI; Thermo
Fisher Scientific, Inc.). The primers used for qPCR were as
follows: PPARG2 forward, 5′-CGGATTGATCTTTTGCTA-3′ and
reverse, 5′-CTTTCTGGGTCAATAGGAG-3′; CEBPA forward,
5′-CGTGGAGACGCAGCAGAA-3′ and reverse, 5′-GGCCTTGACCAAGGAGCT-3′;
FABP4 forward, 5′-CAGCACCCTCCTGAAAAC-3′ and reverse,
5′-GCAAAGCCCACTCCTACT-3′; RPLP0 forward,
5′-CTCTGCATTCTCGCTTCC-3′ and reverse, 5′-GACTCGTTTGTACCCGTTG-3′;
hsa_circ_0136134 forward, 5′-AAGGCACCTGCGGTATTT-3′ and reverse,
5′-AGCCACGGACTCTGCTACT-3′; hsa_circ_0017650 forward,
5′-AAGACCTTCCTCCTTTACCC-3′ and reverse, 5′-GCAACAGTCTGACTTGCCTC-3′;
and hsa-circRNA9227-1 forward, 5′-CCGACGCACCATCAGTTT-3′ and
reverse, 5′-GAGCGAGGCACAGAAAGG-3′. The thermocycling conditions for
the qPCR were as follows: Initial denaturation at 95°C for 30 sec;
45 cycles of denaturation at 95°C for 5 sec; and annealing and
extension at 60°C for 30 sec. The relative expression levels of RNA
were analyzed with the 2−ΔΔCq method (17) and normalized to the loading control
RPLP0.
Statistical analysis
The data are presented as the means ± standard
deviation. The significance of the differences was analyzed using
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
HPA-v differentiation
To obtain mature visceral adipocytes, HPA-v were
induced to differentiate in medium containing 0.1 mM
3-isobutyl-1-methylxanthine, 1 µM dexamethasone, and 5 µg/ml
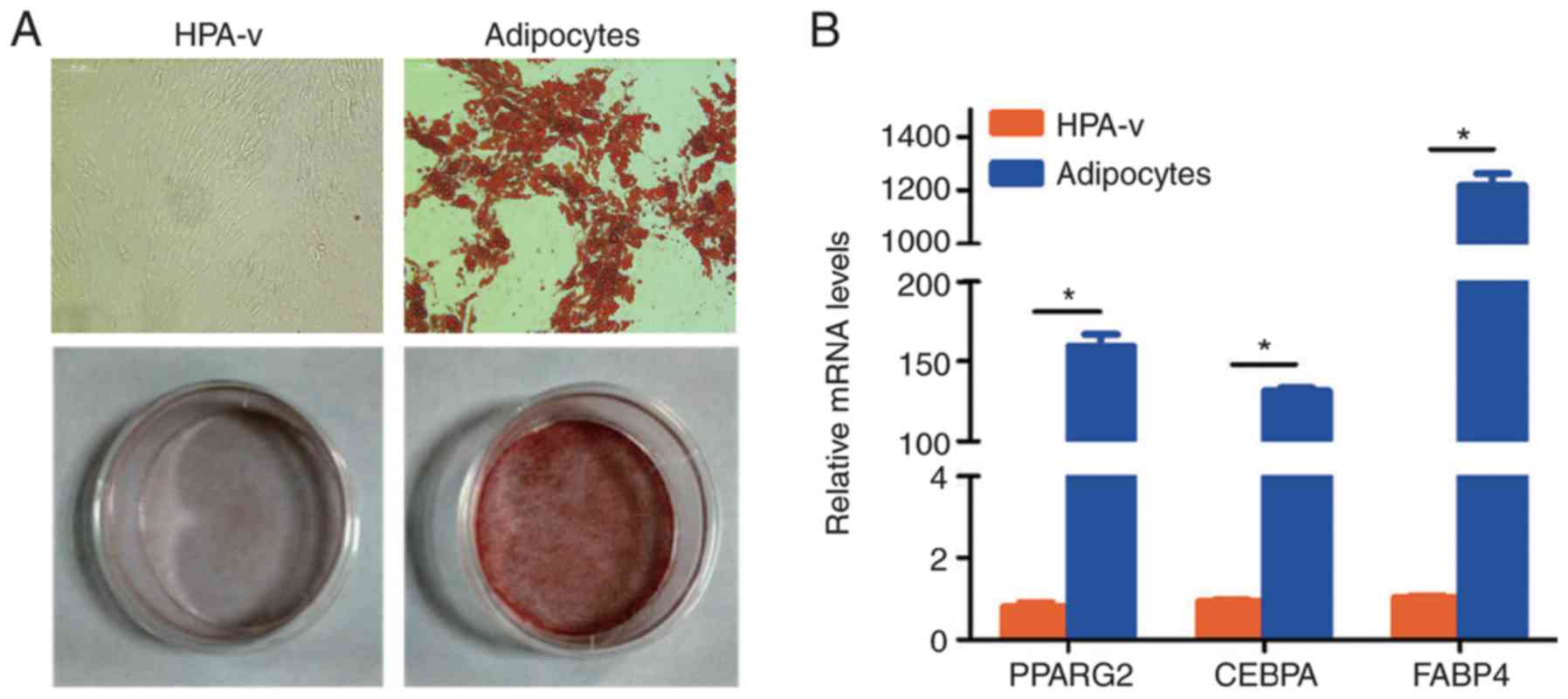
insulin. The characteristics of HPA-v differentiation were
confirmed by Oil Red O staining and evaluation of adipogenic marker
gene expression (Fig. 1). Oil Red
O staining revealed substantial lipid deposition in the cytoplasm
after differentiation (Fig. 1A).
In addition, PPARG2, CEBPA and FABP4 mRNA expression
levels were significantly increased in mature visceral adipocytes
(Fig. 1B).
Expression profiles of circRNAs in
HPA-v and adipocytes
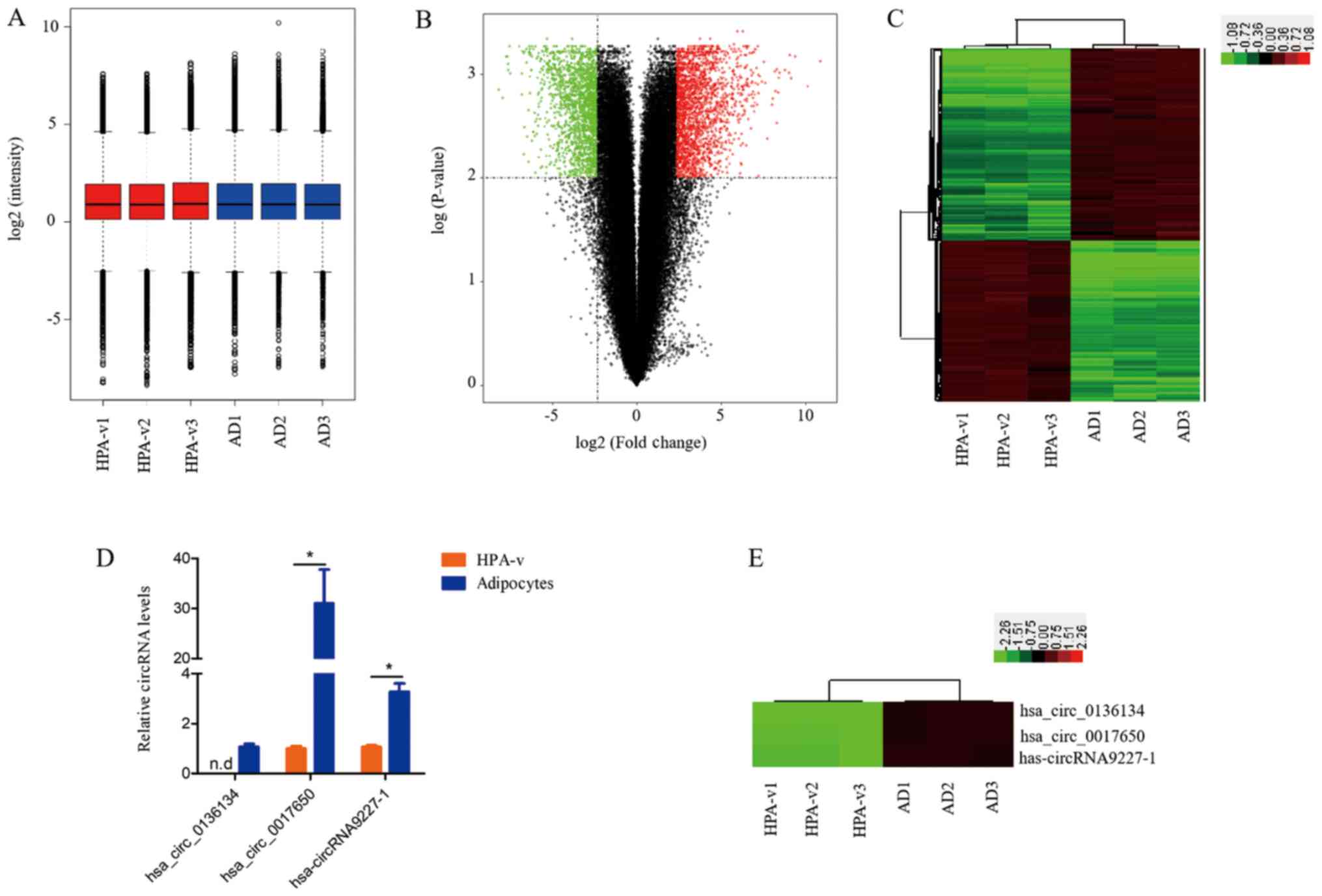
To investigate whether circRNAs are associated with
lipid deposition, a human circRNA microarray (version 2.0; Agilent
Technologies, Inc.) was used to assess circRNA expression profiles
in HPA-v and adipocytes. The distribution of circRNA expression was
illustrated in a box plot after normalization using the GeneSpring
GX software (Fig. 2A), which
revealed that the distribution of log2 ratios was
similar among all samples. Volcano plots were generated to compare
the circRNA expression profiles between HPA-v and adipocytes
(Fig. 2B). The circRNAs
differentially expressed between HPA-v and adipocytes were
identified as those with a fold change ≥5.0 and a P-value ≤0.01. In
total, 2,215 up- and 1,865 downregulated circRNAs were identified
in adipocytes compared with HPA-v (Table S1). Table I lists the top 10 up- and
downregulated circRNAs, and the homologous circRNAs of
hsa_circ_0094183, hsa_circ_0116913 in mice were
MMU_CIRCpedia_216382, MMU_CIRCpedia_14213, respectively, while the
other 18 circRNAs did not find their homologous circRNAs in mice
(Table I). The expression patterns
of the differentially expressed circRNAs were visualized by
hierarchical cluster analysis (Fig.
2C), which indicated different expression circRNA patterns
between visceral adipocytes and HPA-v.
 | Table I.Top 10 up- and downregulated circRNAs
in adipocytes. |
Table I.
Top 10 up- and downregulated circRNAs
in adipocytes.
| circRNA ID | Fold change | Regulation | Conservation |
|---|
|
hsa_circ_0136134 | 1925.3240 | Up |
Species-specific |
|
hsa_circ_0136132 | 1141.0890 | Up |
Species-specific |
|
hsa_circ_0136131 | 680.3070 | Up |
Species-specific |
|
hsa_circ_0067409 | 581.1932 | Up |
Species-specific |
|
hsa_circ_0094183 | 546.6634 | Up |
MMU_CIRCpedia_216382 |
|
hsa_circ_0060972 | 526.6423 | Up |
Species-specific |
|
hsa_circ_0017650 | 447.0942 | Up |
Species-specific |
|
hsa_circ_0128428 | 355.4433 | Up |
Species-specific |
|
hsa-circRNA9227-1 | 343.7039 | Up |
Species-specific |
|
hsa_circ_0060971 | 336.8149 | Up |
Species-specific |
|
hsa-circRNA9333-2 | 292.9276 | Down |
Species-specific |
|
hsa-circRNA1786-2 | 248.5333 | Down |
Species-specific |
|
hsa_circ_0116913 | 214.6204 | Down |
MMU_CIRCpedia_14213 |
|
hsa_circ_0023242 | 207.2891 | Down |
Species-specific |
|
hsa_circ_0032023 | 197.7922 | Down |
Species-specific |
|
hsa-circRNA9333-9 | 197.0217 | Down |
Species-specific |
|
hsa-circRNA2910-9 | 187.1047 | Down |
Species-specific |
|
hsa_circ_0032024 | 184.9652 | Down |
Species-specific |
|
hsa_circ_0052586 | 125.1419 | Down |
Species-specific |
|
hsa_circ_0003543 | 113.2247 | Down |
Species-specific |
To examine the reliability of the circRNA microarray
data, three circRNAs were selected for validation by qPCR.
According to the qPCR results, hsa_circ_0136134 was detected in
adipocytes exclusively, hsa_circ_0017650 and hsa-circRNA9227-1 were
upregulated 30.0- and 2.3-fold in adipocytes compared with HPA-v,
respectively (Fig. 2D). The qPCR
results were consistent with those of the circRNA microarrays
(Fig. 2D and E).
General characteristics of the
differentially expressed circRNAs
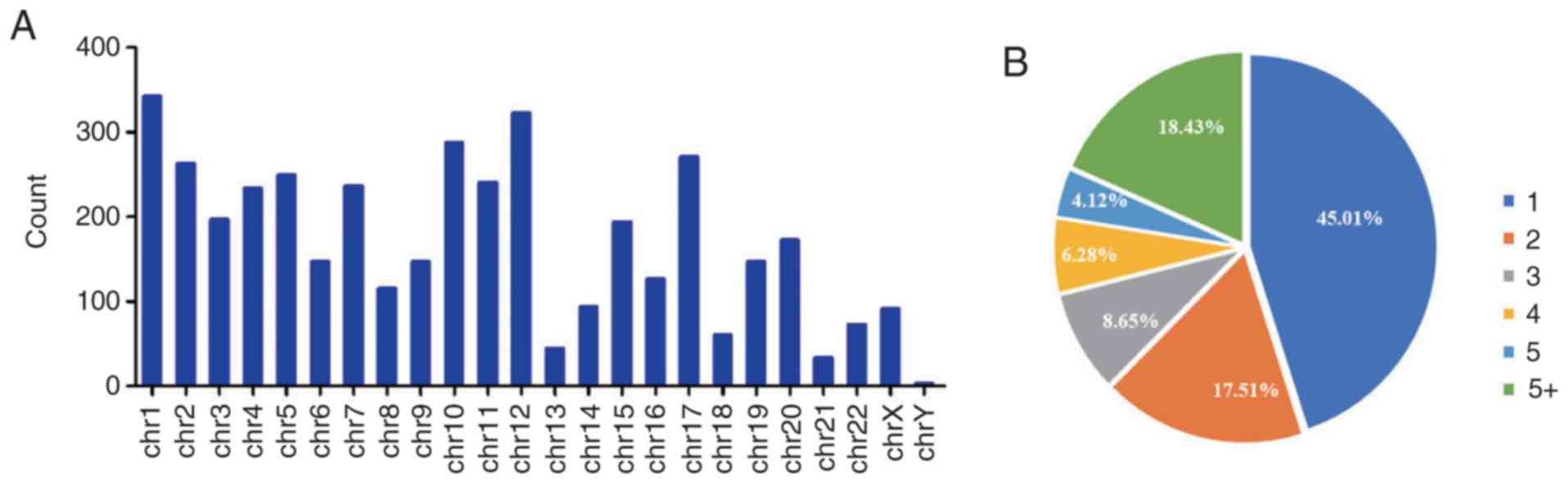
The distribution of the differentially expressed
circRNAs on human chromosomes was analyzed, and the that 4,080
differentially expressed circRNAs were derived from genes located
on all chromosomes, although rarely on chromosomes 13, 18, 21, and
Y (Fig. 3A). When the distribution
of these circRNAs was analyzed among the parental genes, it was
revealed that 3,968 (97.25%) circRNAs were mapped to 971 parental
genes, with 437 (45.01%) parental genes generating one circRNA and
179 (18.43%) parental genes generating more than five circRNAs
(Fig. 3B).
KEGG analysis of the circRNA parental
genes
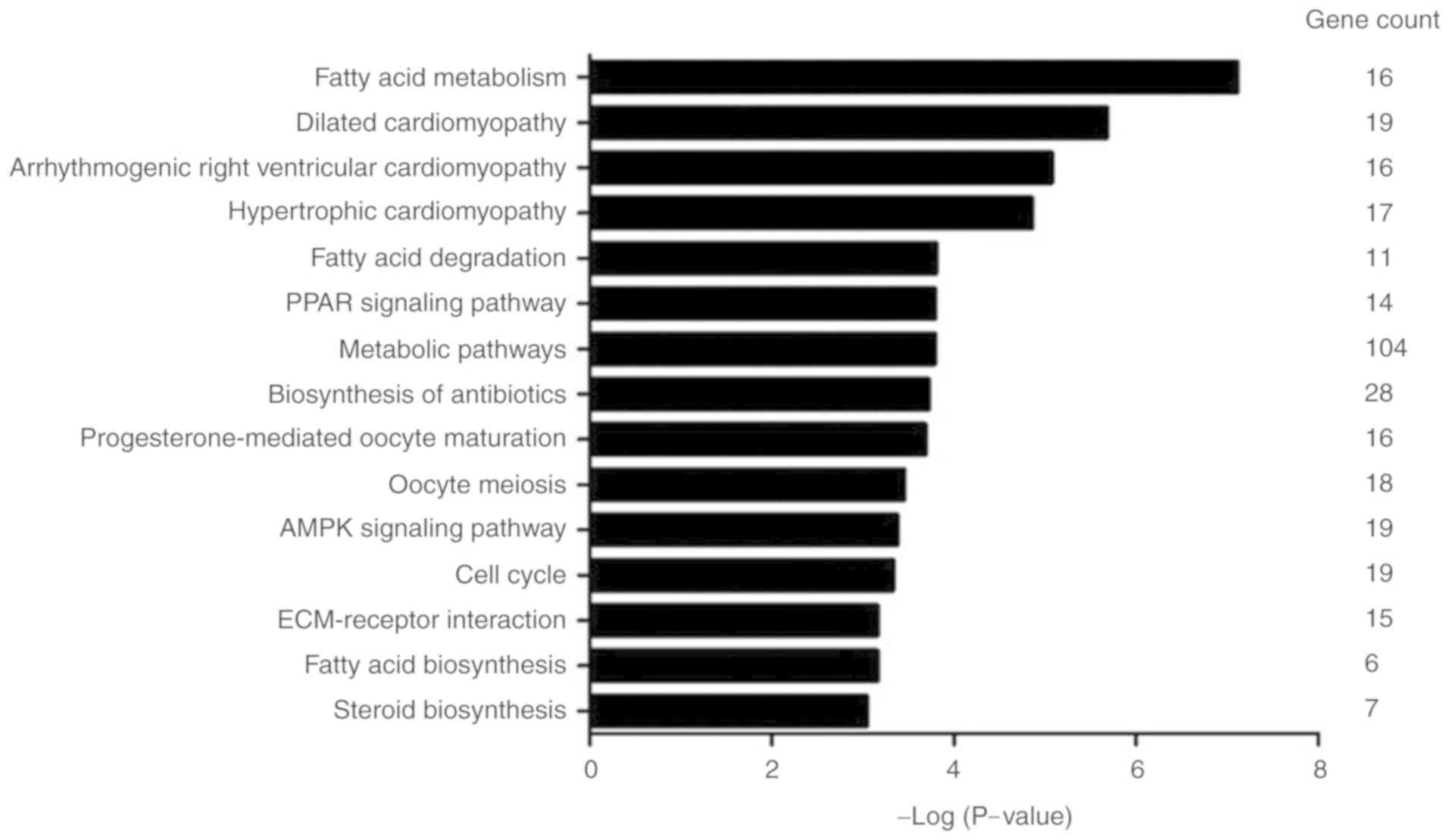
To investigate the potential functions of the
differently expressed circRNAs, 971 parental genes were analyzed
using the KEGG database. The top 15 most significantly enriched
pathways, which included fatty acid metabolism, fatty acid
degradation, fatty acid biosynthesis, and PPAR signaling pathways
are presented in Fig. 4. The most
significantly enriched pathway was fatty acid metabolism
(P=7.83E-08), and most genes were involved in metabolic pathways
(Gene count=104).
CircRNA-miRNA interactions
To dissect the potential functions of differentially
expressed circRNAs, the MERs of top 10 up- and downregulated
circRNAs were predicted. Table II
lists the miRNAs with more than two MERs targeting the top 10 up-
and downregulated circRNAs. hsa_circ_0136134, hsa_circ_0067409,
hsa-circRNA9227-1, hsa_circ_0060971, hsa-circRNA9333-2,
hsa_circ_0052586, and hsa-circRNA2910-9 potentially interact with
1, 1, 1, 1, 2, 3, and 5 MERs, respectively. The interaction between
circRNAs and miRNAs requires further study.
 | Table II.miRNAs with >2 miRNA response
elements targeting the top 10 upregulated and downregulated
circRNAs. |
Table II.
miRNAs with >2 miRNA response
elements targeting the top 10 upregulated and downregulated
circRNAs.
| miRNA ID | circRNA ID |
|---|
| hsa-miR-3138 |
hsa_circ_0136134 |
|
hsa-miR-4717-5p |
hsa_circ_0067409 |
| hsa-miR-665 |
hsa-circRNA9227-1 |
|
hsa-miR-6791-5p |
hsa_circ_0060971 |
|
hsa-miR-4725-3p |
hsa-circRNA9333-2 |
|
hsa-miR-6824-5p |
hsa-circRNA9333-2 |
|
hsa-miR-6808-5p |
hsa-circRNA2910-9 |
| hsa-miR-4514 |
hsa-circRNA2910-9 |
|
hsa-miR-6752-5p |
hsa-circRNA2910-9 |
|
hsa-miR-6757-5p |
hsa-circRNA2910-9 |
|
hsa-miR-7112-5p |
hsa-circRNA2910-9 |
| hsa-let-7e-5p |
hsa_circ_0052586 |
|
hsa-miR-6840-3p |
hsa_circ_0052586 |
|
hsa-miR-7851-3p |
hsa_circ_0052586 |
Discussion
Adipocytes are traditionally classified into white,
brown, and beige adipocytes (18,19).
White adipocytes are involved mainly in energy storage and can
trans-differentiate into beige adipocytes and de-differentiate into
preadipocyte-like precursors (20–22),
while brown and beige adipocytes are involved in adaptive
thermogenesis (23). Removing
visceral fat (white adipose tissue) (24,25)
or increasing the activity or number of beige adipocytes can
reverse or reduce metabolic dysfunction, including insulin
resistance and obesity (20,26).
In the present study, the expression profiles of circRNAs between
HPA-v and visceral adipocytes were compared, which were produced by
HPA-v differentiation, to reveal the potential molecular mechanisms
of visceral fat accumulation and provide clues for the treatment of
visceral obesity.
CircRNAs participate in numerous physiological and
pathological processes (13,15,16).
However, it is not clear whether circRNAs are associated with
adipogenesis and lipid metabolism. Li et al reported that
circRNAs have different expression profiles in the subcutaneous
adipose tissues of the Laiwu pig and Large White pig, which implies
that circRNAs participate in subcutaneous adipose deposition
(27). In this study, the circRNA
expression profiles in HPA-v and adipocytes were first analyzed by
microarray analysis, which identified 4,080 circRNAs differently
expressed circRNAs in HPA-v and adipocytes, suggesting that HPA-v
and adipocytes have different circRNA expression patterns, and that
these circRNAs may be associated with visceral adipocyte
hyperplasia.
Some circRNAs regulate the expression of their
parental gene (e.g., ci-ankrds regulate the ankyrin repeat domain)
(28), sometimes by affecting the
alternative splicing of the parental gene (9,29,30).
The parental genes of hsa_circ_0136134 and hsa_circ_0017650 are
lipoprotein lipase (LPL) and inter-alpha-trypsin inhibitor
heavy chain 5 (ITIH5), respectively. LPL, a key enzyme in
adipose tissue triglyceride metabolism, is an adipocyte
differentiation marker and upregulated during preadipocytes
differentiation (31,32). ITIH5 is a secreted protein, and the
ITIH5 expression in adipose tissue is increased in obesity
and reduced after diet-induced weight loss, but the role of
ITIH5 in preadipocyte differentiation has not been reported
(33). Thus, hsa_circ_0136134 and
hsa_circ_0017650 may influence HPA-v differentiation by regulating
the expression of their parental genes. Further studies are
required to confirm the regulatory relationship between circRNAs
and their parental genes.
Visceral adipocyte hyperplasia is a complex process
that involves multiple intracellular signaling pathways. In the
present study, the signaling pathways related to fatty acids, which
are the substrates of triglyceride synthesis (34), such as ‘fatty acid metabolism’,
‘fatty acid degradation’, and ‘fatty acid biosynthesis’ were
enriched. This indicated that circRNAs influence the expression of
genes associated with fatty acid metabolism to regulate the
accumulation of triglycerides. The ‘PPAR signaling pathway’, ‘AMPK
signaling pathway’, ‘Metabolic pathways’, and ‘Cell cycle’ serve
important roles in preadipocyte differentiation, fatty acid
oxidation, fatty acid transport, fatty acid synthesis, lipolysis,
gluconeogenesis, glycolysis, and cell growth, and may induce the
expression of genes related to preadipocyte differentiation or
alter the activities of enzymes related to lipid metabolism,
contributing to lipid storage. The circRNAs related to these
pathways may play critical roles in visceral adipocyte
hyperplasia.
CircRNAs can recruit miRNAs to regulate target gene
expression (11), and most
circRNAs have more than one miRNA binding site; for example, ciRS-7
contains over 60 target sites for miR-7 and can function as a miR-7
sponge and influence miR-7 target gene expression (11). In the present study, it was
revealed that the top 10 up- and downregulated circRNAs had many
potential miRNA binding sites; for example, hsa-circRNA9227-1,
which was upregulated in visceral adipocytes (Table I), contained at least two target
sites for hsa-miR-665 (Table II).
Previous studies revealed that hsa-miR-665 is downregulated during
adipocyte differentiation of human mesenchymal stem cells, and
Seipin, which promotes adipocyte differentiation (35), is a potential target gene of
miR-665 (36). These results
indicated that hsa-circRNA9227-1 was involved in regulating
adipogenesis by recruiting hsa-mir-665. However, more research is
required to elucidate the function of circRNAs as miRNA sponges in
visceral lipid deposition.
The present study assessed the circRNA expression
profiles in human visceral preadipocytes and adipocytes. The
markedly different circRNA expression profiles between the two cell
types reflect the close association between circRNAs and
adipogenesis. Further research is required to clarify the function
of circRNAs in visceral preadipocyte differentiation and lipid
deposition to develop novel therapeutics for obesity.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
National Natural Science Foundation of China (grant nos. 81502803
and 31801196), The Natural Science Research Program of Jiangsu
Province (grant no. 15KJB330005), and The Shenzhen Science and
Technology Innovation Committee (grant no. JCYJ2018030712).
Availability of data and materials
The datasets used/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FD and GX conceived and designed the experiments; WS
performed the experiments and drafted the manuscript; and WS, XS,
SY and WC performed the data analysis.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cefalu WT, Wang ZQ, Werbel S, Bell-Farrow
A, Crouse JR III, Hinson WH, Terry JG and Anderson R: Contribution
of visceral fat mass to the insulin resistance of aging.
Metabolism. 44:954–959. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fujimoto WY, Bergstrom RW, Boyko EJ, Chen
KW, Leonetti DL, Newell-Morris L, Shofer JB and Wahl PW: Visceral
adiposity and incident coronary heart disease in Japanese-American
men. The 10-year follow-up results of the Seattle Japanese-American
Community Diabetes Study. Diabetes Care. 22:1808–1812. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakaguchi M, Fujisaka S, Cai W, Winnay JN,
Konishi M, O'Neill BT, Li M, García-Martín R, Takahashi H, Hu J, et
al: Adipocyte dynamics and reversible metabolic syndrome in mice
with an inducible adipocyte-specific deletion of the insulin
receptor. Cell Metab. 25:448–462. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Silva HM, Bafica A, Rodrigues-Luiz GF, Chi
J, Santos PDA, Reis BS, Hoytema van Konijnenburg DP, Crane A, Arifa
RDN, Martin P, et al: Vasculature-associated fat macrophages
readily adapt to inflammatory and metabolic challenges. J Exp Med.
216:786–806. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Avram MM, Avram AS and James WD:
Subcutaneous fat in normal and diseased states 3. Adipogenesis:
From stem cell to fat cell. J Am Acad Dermatol. 56:472–492. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fajas L: Adipogenesis: A cross-talk
between cell proliferation and cell differentiation. Ann Med.
35:79–85. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Munschauer M, Nguyen CT, Sirokman K,
Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP,
Subramanian V, Chen J, et al: The NORAD lncRNA assembles a
topoisomerase complex critical for genome stability. Nature.
561:132–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao J, Li L, Wang Q, Han H, Zhan Q and Xu
M: CircRNA expression profile in early-stage lung adenocarcinoma
patients. Cell Physiol Biochem. 44:2138–2146. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37 e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel Role of FBXW7
Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer
Inst. 110:2018. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chaurasia B, Kaddai VA, Lancaster GI,
Henstridge DC, Sriram S, Galam DL, Gopalan V, Prakash KN, Velan SS,
Bulchand S, et al: Adipocyte ceramides regulate subcutaneous
adipose browning, inflammation, and metabolism. Cell Metab.
24:820–834. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roh HC, Tsai LTY, Shao M, Tenen D, Shen Y,
Kumari M, Lyubetskaya A, Jacobs C, Dawes B, Gupta RK and Rosen ED:
Warming induces significant reprogramming of Beige, but not brown,
adipocyte cellular identity. Cell Metab. 27:1121–1137.e5. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shao M, Ishibashi J, Kusminski CM, Wang
QA, Hepler C, Vishvanath L, MacPherson KA, Spurgin SB, Sun K,
Holland WL, et al: Zfp423 maintains white adipocyte identity
through suppression of the beige cell thermogenic gene program.
Cell Metab. 23:1167–1184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang QA, Song A, Chen W, Schwalie PC,
Zhang F, Vishvanath L, Jiang L, Ye R, Shao M, Tao C, et al:
Reversible de-differentiation of mature white adipocytes into
preadipocyte-like precursors during lactation. Cell Metab.
28:282–288.e3. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bi P, Yue F, Karki A, Castro B, Wirbisky
SE, Wang C, Durkes A, Elzey BD, Andrisani OM, Bidwell CA, et al:
Notch activation drives adipocyte dedifferentiation and tumorigenic
transformation in mice. J Exp Med. 213:2019–2037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang W, Ishibashi J, Trefely S, Shao M,
Cowan AJ, Sakers A, Lim HW, O'Connor S, Doan MT, Cohen P, et al: A
PRDM16- driven metabolic signal from adipocytes regulates precursor
cell fate. Cell Metab. 30:174–189.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gabriely I, Ma XH, Yang XM, Atzmon G,
Rajala MW, Berg AH, Scherer P, Rossetti L and Barzilai N: Removal
of visceral fat prevents insulin resistance and glucose intolerance
of aging: An adipokine-mediated process? Diabetes. 51:2951–2958.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barzilai N, She L, Liu BQ, Vuguin P, Cohen
P, Wang J and Rossetti L: Surgical removal of visceral fat reverses
hepatic insulin resistance. Diabetes. 48:94–98. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seki T, Hosaka K, Fischer C, Lim S,
Andersson P, Abe M, Iwamoto H, Gao Y, Wang X, Fong GH and Cao Y:
Ablation of endothelial VEGFR1 improves metabolic dysfunction by
inducing adipose tissue browning. J Exp Med. 215:611–626. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li A, Huang W, Zhang X, Xie L and Miao X:
Identification and characterization of CircRNAs of two pig breeds
as a new biomarker in metabolism-related diseases. Cell Physiol
Biochem. 47:2458–2470. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kelly S, Greenman C, Cook PR and
Papantonis A: Exon skipping is correlated with exon
circularization. J Mol Biol. 427:2414–2417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gong H, Ni Y, Guo X, Fei L, Pan X, Guo M
and Chen R: Resistin promotes 3T3-L1 preadipocyte differentiation.
Eur J Endocrinol. 150:885–892. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding ST, McNeel RL and Mersmann HJ:
Expression of porcine adipocyte transcripts: Tissue distribution
and differentiation in vitro and in vivo. Comp Biochem Physiol B
Biochem Mol Biol. 123:307–318. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anveden A, Sjoholm K, Jacobson P,
Palsdottir V, Walley AJ, Froguel P, Al-Daghri N, McTernan PG,
Mejhert N, Arner P, et al: ITIH-5 expression in human adipose
tissue is increased in obesity. Obesity (Silver Spring).
20:708–714. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Beale EG, Harvey BJ and Forest C: PCK1 and
PCK2 as candidate diabetes and obesity genes. Cell Biochem Biophys.
48:89–95. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bi J, Wang W, Liu Z and Huang X, Jiang Q,
Liu G, Wang Y and Huang X: Seipin promotes adipose tissue fat
storage through the ER Ca2+-ATPase SERCA. Cell Metab.
19:861–871. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yi X, Liu J, Wu P, Gong Y, Xu X and Li W:
The key microRNA on lipid droplet formation during adipogenesis
from human mesenchymal stem cells. J Cell Physiol. 235:328–338.
2020. View Article : Google Scholar : PubMed/NCBI
|